Abstract
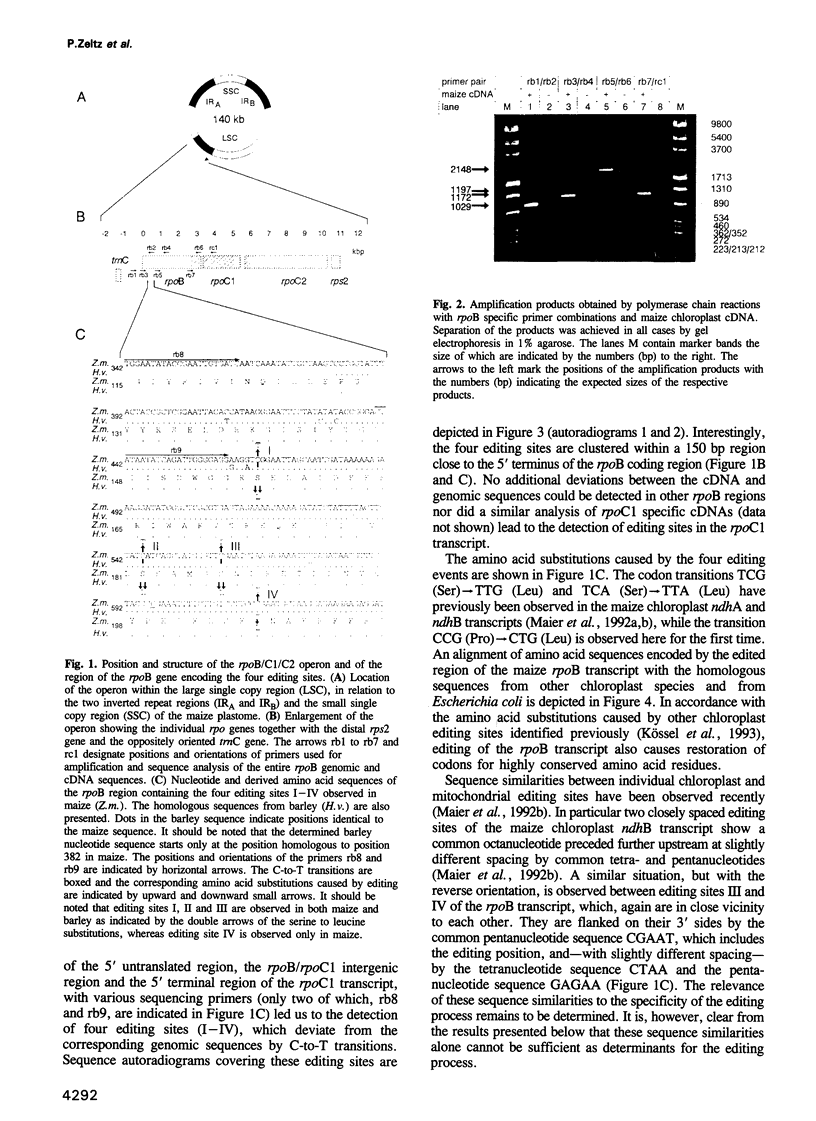
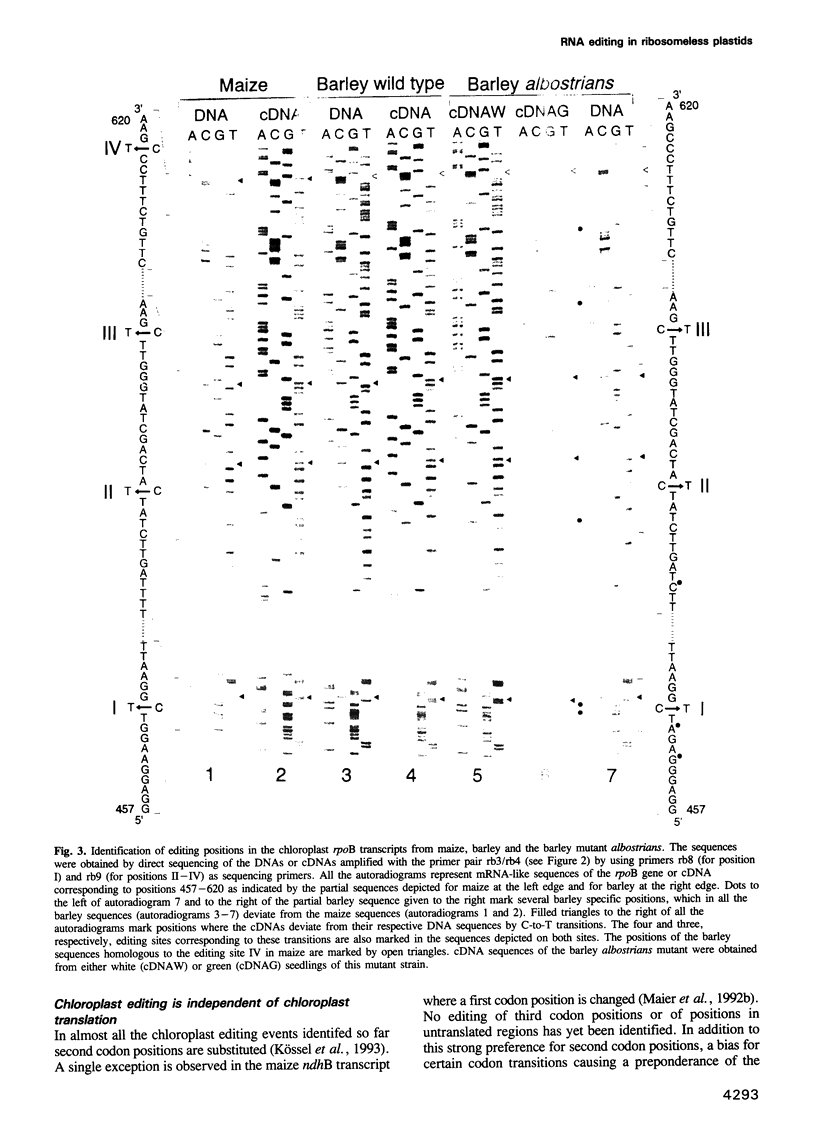
Sequence analysis of amplified cDNAs derived from the maize chloroplast rpoB transcript which encodes the beta subunit of a chloroplast specific, DNA dependent RNA polymerase reveals four C-to-U editing sites clustered within 150 nucleotides of the 5' terminal region of the rpoB message. These newly identified editing sites confirm the bias of chloroplast editing for certain codon transitions and for second codon positions which both appear suggestive for an involvement of the translational apparatus in the editing process. This supposition prompted us to investigate editing of the rpoB transcript from ribosome deficient, and hence protein synthesis deficient, plastids of the barley mutant albostrians. In this mutant editing is, however, not impaired at any of the editing sites functional in the barley wild type rpoB transcript. This demonstrates that chloroplast editing is neither linked to nor dependent on the chloroplast translational apparatus. As a further consequence any peptide components required for chloroplast editing must be encoded in the nuclear genome. In spite of strong sequence conservation only three of the four editing sites identified in the maize rpoB transcript are functional in barley. This indicates that sequences surrounding an editing site alone are not sufficient as determinants for the editing process in chloroplasts, but that trans-acting templates carrying the editing information for each individual site may also be required.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansorge W., Sproat B. S., Stegemann J., Schwager C. A non-radioactive automated method for DNA sequence determination. J Biochem Biophys Methods. 1986 Dec;13(6):315–323. doi: 10.1016/0165-022x(86)90038-2. [DOI] [PubMed] [Google Scholar]
- Bachmann B., Lüke W., Hunsmann G. Improvement of PCR amplified DNA sequencing with the aid of detergents. Nucleic Acids Res. 1990 Mar 11;18(5):1309–1309. doi: 10.1093/nar/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner B. J., Rapp J. C., Mullet J. E. Plastid Genes Encoding the Transcription/Translation Apparatus Are Differentially Transcribed Early in Barley (Hordeum vulgare) Chloroplast Development (Evidence for Selective Stabilization of psbA mRNA). Plant Physiol. 1993 Mar;101(3):781–791. doi: 10.1104/pp.101.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. Differences in editing at homologous sites in messenger RNAs from angiosperm mitochondria. Nucleic Acids Res. 1990 Sep 11;18(17):5189–5196. doi: 10.1093/nar/18.17.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S. E., Surzycki S. J. Chloroplast RNA polymerase genes of Chlamydomonas reinhardtii exhibit an unusual structure and arrangement. Curr Genet. 1992 May;21(6):485–497. doi: 10.1007/BF00351659. [DOI] [PubMed] [Google Scholar]
- Hess W. R., Prombona A., Fieder B., Subramanian A. R., Börner T. Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 1993 Feb;12(2):563–571. doi: 10.1002/j.1460-2075.1993.tb05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch B., Maier R. M., Appel K., Igloi G. L., Kössel H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991 Sep 12;353(6340):178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- Igloi G. L., Meinke A., Döry I., Kössel H. Nucleotide sequence of the maize chloroplast rpo B/C1/C2 operon: comparison between the derived protein primary structures from various organisms with respect to functional domains. Mol Gen Genet. 1990 May;221(3):379–394. doi: 10.1007/BF00259403. [DOI] [PubMed] [Google Scholar]
- Kudla J., Igloi G. L., Metzlaff M., Hagemann R., Kössel H. RNA editing in tobacco chloroplasts leads to the formation of a translatable psbL mRNA by a C to U substitution within the initiation codon. EMBO J. 1992 Mar;11(3):1099–1103. doi: 10.1002/j.1460-2075.1992.tb05149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R. M., Hoch B., Zeltz P., Kössel H. Internal editing of the maize chloroplast ndhA transcript restores codons for conserved amino acids. Plant Cell. 1992 May;4(5):609–616. doi: 10.1105/tpc.4.5.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R. M., Neckermann K., Hoch B., Akhmedov N. B., Kössel H. Identification of editing positions in the ndhB transcript from maize chloroplasts reveals sequence similarities between editing sites of chloroplasts and plant mitochondria. Nucleic Acids Res. 1992 Dec 11;20(23):6189–6194. doi: 10.1093/nar/20.23.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen H., Bogorad L. Diurnal and Circadian Rhythms in the Accumulation and Synthesis of mRNA for the Light-Harvesting Chlorophyll a/b-Binding Protein in Tobacco. Plant Physiol. 1988 Dec;88(4):1104–1109. doi: 10.1104/pp.88.4.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissinger B., Schuster W., Brennicke A. Species-specific RNA editing patterns in the mitochondrial rps13 transcripts of Oenothera and Daucus. Mol Gen Genet. 1990 Dec;224(3):389–395. doi: 10.1007/BF00262433. [DOI] [PubMed] [Google Scholar]
- Yepiz-Plascencia G. M., Radebaugh C. A., Hallick R. B. The Euglena gracilis chloroplast rpoB gene. Novel gene organization and transcription of the RNA polymerase subunit operon. Nucleic Acids Res. 1990 Apr 11;18(7):1869–1878. doi: 10.1093/nar/18.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]





