Abstract
Detection of protein–protein interactions (PPIs) is important for understanding numerous processes in mammalian cells; however, existing PPI detection methods often give significant background signals. Here, we propose a novel PPI-detection method based on kinase-mediated growth induction of mammalian cells. In this method, target proteins are fused to the intracellular domain of c-kit (c-kit ICD) and expressed in interleukin-3-dependent mammalian cells. The PPI induces dimerization and activation of c-kit ICDs, which leads to cell growth in the absence of interleukin-3. Using this system, we successfully detected the ligand-dependent homo-interaction of FKBPF36V and hetero-interaction of FKBP and FRBT2098L, as well as the constitutive interaction between MDM2 and a known peptide inhibitor. Intriguingly, cells expressing high-affinity peptide chimeras are selected from the mixture of the cell populations dominantly expressing low-affinity peptide chimeras. These results indicate that this method can detect PPIs with low background levels and is suitable for peptide inhibitor screening.
Since protein–protein interactions (PPIs) regulate various cellular processes, understanding PPIs in living cells is important for biomedical studies. Many PPIs depend on specific modifications and structural alterations of proteins, as well as the existence of adaptor proteins. Therefore, a tool for analysing PPIs in living mammalian cells is necessary1.
Various methods have been developed to investigate PPIs in living mammalian cells. In the method based on fluorescence resonance energy transfer (FRET), proteins of interest (bait and prey) are fused to a pair of fluorescent proteins (donor and acceptor) and expressed in cells. The interaction between bait and prey leads to the donor and acceptor moieties being in close proximity and this causes FRET2,3,4. In protein-fragment complementation assay (PCA), bait and prey are fused to two fragments of a reporter protein that has been rationally dissected into two fragments using protein-engineering strategies. Through the association of bait and prey, the reporter protein fragments are brought into proximity and reconstitute the activity5,6,7,8. Using these methods, numerous PPIs have been successfully detected with sufficient sensitivities, which have substantially contributed to the understanding of molecular behavior of target proteins in living cells. However, the reporter activity depends on conformation and intermolecular distance of reporter proteins, which potentially leads to false negatives9,10. Another problem of these methods lies in high background signals. In FRET, background fluorescence is due to the direct excitation of the acceptor and the leakage of fluorescence from the donor11. In PCA, a weak affinity between two fragments intrinsically contributes to the background signal12,13. The background levels of these methods depend on the expression levels of the fusion proteins10. Therefore it is difficult to identify promising candidates that interact with bait from a prey library composed of various proteins with different stabilities.
c-kit belongs to the type III receptor tyrosine kinase family and controls the fate of a number of cell types, including hematopoietic stem cells, mast cells, melanocytes and germ cells14. The binding of the stem cell factor to the extracellular domain of c-kit results in dimerization and subsequent activation of the tyrosine kinase domain in the intracellular domain. Once activated, the kinase domain autophosphorylates intracellular tyrosine residues. Signal-transducing molecules including Src family kinase (SFK), growth factor receptor-bound protein 2 (Grb2) and phosphoinositide 3-kinase (PI3K) are recruited to the phospho-tyrosine residues, which induces cell survival, proliferation and differentiation15,16,17,18. Intriguingly, the intracellular juxtamembrane domain of c-kit inhibits the kinase activity in an unliganded monomeric state19,20.
Here, we aimed to develop a novel PPI-detection method based on mammalian cell growth. In this method, bait and prey are fused to the intracellular domain of c-kit (c-kit ICD) and expressed in interleukin-3 (IL-3)-dependent mammalian cells. Bait–prey interactions induce the dimerization and activation of c-kit ICDs and lead to cell growth in the absence of IL-3. Since the activity of c-kit ICD is repressed in the monomeric form, low background cell growth, i.e., low false-positives, would be expected.
To demonstrate the feasibility of this method, we first focused on cytoplasmic proteins FK506-binding protein 12 (FKBP) and FKBP12-rapamycin binding domain (FRB). The F36V mutant of FKBP (FKBPF36V) is homodimerized by a small-molecule ligand AP2018721,22,23, whereas FKBP and the T2098L mutant of FRB (FRBT2098L) are hererodimerized by another small-molecule ligand AP2196724,25. Therefore, we investigated whether AP20187-dependent homodimer formation of FKBPF36V and AP21967-dependent heterodimer formation of FKBP and FRBT2098L could be detected by our method. Second, the intracellular domain of c-kit was engineered so that the hetero-interaction between bait and prey can be evaluated without detecting the homo-oligomeric prey. Third, to propose the application of this method for evaluation of peptide inhibitors, we focused on an E3 ubiquitin ligase MDM2, which inhibits the functions of a tumor suppressor protein p5326. We investigated whether the constitutive interaction between MDM2 and a known peptide inhibitor could be detected. Finally, we demonstrated that the cells expressing high-affinity peptide chimeras were selected from the mixture of the cell populations dominantly expressing low-affinity peptide chimeras.
Results
Detection of AP20187-dependent homodimer formation of FKBPF36V
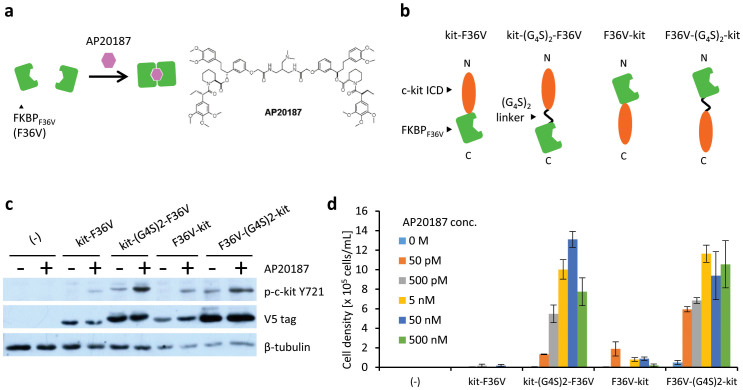
To demonstrate the feasibility of our method, we focused on AP20187-dependent homodimer formation of FKBPF36V (Fig. 1a). We constructed chimeras by fusing c-kit ICD to the N- and C-terminus of FKBPF36V (kit-F36V and F36V-kit, respectively). In addition, two variants with a flexible (G4S)2 linker between c-kit ICD and FKBPF36V were also designed (kit-(G4S)2-F36V and F36V-(G4S)2-kit) (Fig. 1b). Murine IL-3-dependent Ba/F3 cells were transduced with the retroviral vector encoding each chimera, followed by drug-resistance selection to establish stably transduced cells. Western blot analysis revealed that these cells expressed the corresponding chimeras (Fig. 1c, blots with V5 tag).
Figure 1. Detection of AP20187-dependent homodimer formation of FKBPF36V.
(a) Illustration for AP20187-dependent interaction of FKBPF36V. (b) The constructs of chimeras with FKBPF36V and c-kit ICD. (c) Tyrosine phosphorylation of c-kit ICD in chimeras. Cells were stimulated for 15 min with or without 50 nM AP20187 and lysed. Western blot analysis was performed with anti-phospho Y721 of c-kit to detect the phosphorylated form of the chimeras, and with an anti-V5 tag to detect the expression level of the chimeras. The blots for β-tubulin are indicated as a loading control. (d) AP20187 dependency of cell growth. Cells expressing chimeras were cultured with AP20187 at indicated concentrations. The initial cell density was 1 × 105 cells/mL. The viable cell densities on day 6 are indicated as mean ± SD (n = 3, biological replicates).
In the intrinsic mechanism, c-kit ICDs auto-phosphorylate their own tyrosine residues and recruit signalling molecules after trans-activation. Phosphorylation of Y721 is important for recruitment and activation of PI3K, which is involved in subsequent downstream signalling for cell survival and proliferation14,27. Therefore, we investigated whether AP20187, which induces homodimerization of FKBPF36V, could induce the phosphorylation of this tyrosine residue of the chimeras. As a result, AP20187-dependent phosphorylation of Y721 of the c-kit ICDs was clearly detected in all chimeras, particularly in kit-(G4S)2-F36V and F36V-(G4S)2-kit (Fig. 1c). This suggests that the flexible linker enabled the c-kit ICDs of the chimeras to trans-activate themselves efficiently.
These cells were then washed to remove IL-3 and subsequently cultured in the presence of AP20187. As expected, cell proliferation was observed in all of the transduced cells (Fig. 1d, and Supplementary Figs. 1–5 for the detailed time-dependent measurements). Consistent with the result of phospho-tyrosine detection (Fig. 1c), kit-(G4S)2-F36V and F36V-(G4S)2-kit induced faster cell growth than the chimeras without the flexible linker. These results indicate that the AP20187-dependent homodimeric interaction of FKBPF36V was successfully detected based on cell growth.
Detection of AP21967-dependent heterodimer formation of FKBP and FRBT2098L
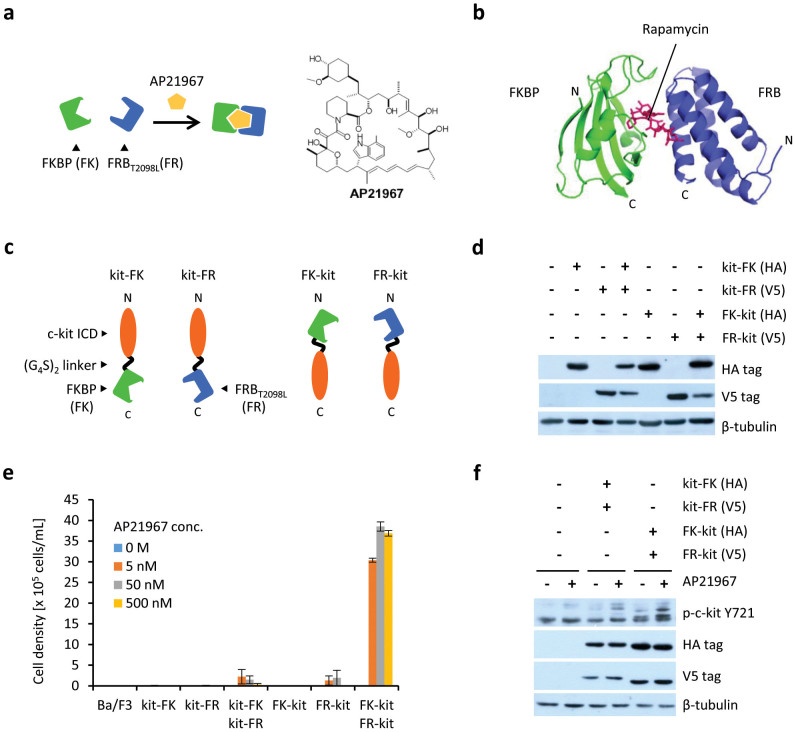
To investigate whether hetero-interactions can be detected by our method, we next focused on the AP21967-inducible FKBP-FRBT2098L interaction (Fig. 2a). Because AP21967 is a derivative of rapamycin, the structure of the FKBP–AP21967–FRBT2098L complex may be similar to that of FKBP–rapamycin–FRB28 (Fig. 2b). c-kit ICDs were fused to the N- or C-termini of FKBP (FK) and FRBT2098L (FR) with a flexible (G4S)2 linker inserted between these domains (kit-FK, kit-FR, FK-kit and FR-kit, respectively) (Fig. 2c). These chimeras were expressed in Ba/F3 cells, and their expression was verified by western blotting (Fig. 2d). Growth assays revealed that the cells co-expressing FK-kit and FR-kit, as well as those co-expressing kit-FK and kit-FR showed AP21967-dependent proliferation (Fig. 2e). In these cells, AP21967 stimulation led to tyrosine phosphorylation of c-kit ICD, suggesting that the chimeras were activated (Fig. 2f). The co-expression of FK-kit and FR-kit induced much more vigorous cell growth than that of kit-FK and kit-FR. This result suggests that c-kit ICDs fused to the C-termini of FK and FR are proximal and efficiently activate each other because of the proximity of the two C-termini in the FKBP–AP21967–FRBT2098L complex. Another possibility is that c-kit ICDs are readily activated when dimerization occurs in a fashion similar to the native receptor, in which the N-terminal extracellular domain is responsible for ligand binding and dimerization.
Figure 2. Detection of AP21967-dependent heterodimer formation of FKBP and FRBT2098L.
(a) Illustration for the AP21967-dependent interaction between FKBP and FRBT2098L. (b) Crystal structure of the FKBP–Rapamycin–FRB complex (PDB ID: 1FAP)28. (c) The constructs of FKBP-fused chimeras and FRBT2098L-fused chimeras. (d) The expression of FKBP-fused chimeras (HA tag) and FRBT2098L-fused chimeras (V5 tag). The blots for β-tubulin are indicated as a loading control. (e) AP21967 dependency of cell growth. Cells expressing chimeras were cultured with AP21967 at indicated concentrations. The initial cell density was 1 × 105 cells/mL. The viable cell densities on day 6 are indicated as mean ± SD (n = 3, biological replicates). (f) Tyrosine phosphorylation of c-kit ICD of chimeras. Cells were stimulated for 15 min with or without 500 nM AP21967 and lysed. Western blot analysis was performed with anti-phospho Y721 of c-kit to detect the phosphorylated form of the chimeras, and with an anti-HA or an anti-V5 tag to detect the expression level of the chimeras. The blots for β-tubulin are indicated as a loading control.
Engineering c-kit ICD for the detection of hetero-interactions
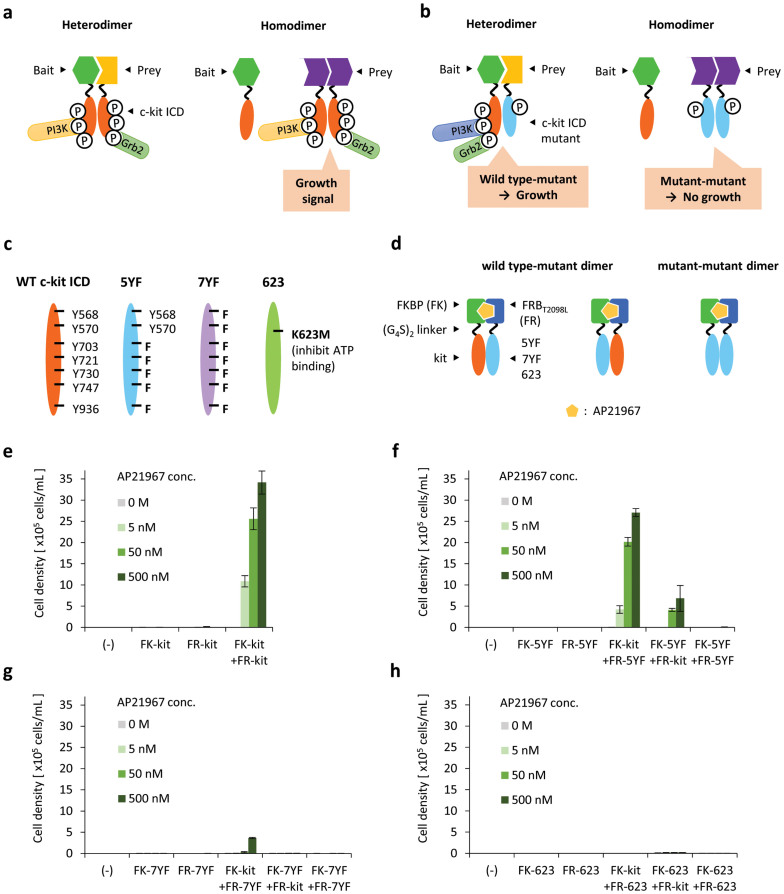
We aimed to apply our method for screening prey that interacts with bait from a protein/peptide library. There may be proteins or peptides that form homodimers in such libraries. In our method using wild-type c-kit ICDs, not only bait–prey interactions, but also prey–prey interactions induce the activation of chimeras (Fig. 3a). Therefore, bait–prey interactions cannot be correctly screened.
Figure 3. Engineering c-kit ICD for exclusive detection of hetero-interactions.
(a) Prey–prey interactions can also be detected if c-kit ICD is fused to prey. (b) Wild-type (WT) and mutant c-kit ICDs are fused to bait and prey, respectively. WT–mutant heterodimerization induces cell growth, whereas mutant–mutant homodimerization does not. (c) Illustration for WT and three mutant c-kit ICDs. (d) Using AP21967-dependent interaction between FKBP and FRBT2098L, WT–mutant and mutant–mutant dimers are induced. (e, f, g, h) AP21967 dependency of cell growth. Cells expressing chimeras were cultured with AP21967 at indicated concentrations. The initial cell density was 1 × 105 cells/mL. The viable cell densities on day 6 are indicated as mean ± SD (n = 3, biological replicates).
To overcome this problem, we designed c-kit ICD mutants in which tyrosine residues are replaced with phenylalanine residues. In this strategy, the c-kit ICD mutant is fused to prey, while the bait is fused to the wild-type (WT) c-kit ICD. The mutant–mutant dimer formed by the homo-interaction of prey cannot transduce a growth signal owing to the mutations in the tyrosine residues, which are critical for recruitment of key signalling molecules. On the other hand, the mutant–WT dimer formed by the bait–prey interaction can recruit signalling molecules to WT c-kit ICD and lead to cell growth (Fig. 3b).
c-kit ICD has nine key tyrosine residues (Y568, Y570, Y703, Y721, Y730, Y747, Y823, Y900, Y936), which are mainly involved in recruiting signalling molecules14,29. We designed two c-kit ICD mutants. One mutant has five Y-to-F mutations and was named 5YF (Y703F, Y721F, Y730F, Y747F, Y936F), whereas the other mutant has seven Y-to-F mutations and was named 7YF (Y568F, Y570F, Y703F, Y721F, Y730F, Y747F, Y936F) (Fig. 3c). Y823 and Y900 remained unchanged in both 5YF and 7YF mutants, because Y823 and Y900 are located within the kinase domain of c-kit ICD. In a previous report, the homodimer formation of the 5YF and 7YF mutants of murine c-kit did not lead to cell growth, although the 5YF mutant induced cell survival30. It has also been reported that Y568 and Y570 located in the juxtamembrane domain of c-kit ICD contributes to autoinhibition of the kinase activity, and that this inhibitory state is released by the phosphorylation of Y568 and Y57019,20. In addition, Y568 and Y570 are important binding sites for a number of signal transduction molecules involved in both positive signaling (Src family kinases, SHP2, Shc) and negative signaling (SHP1, Lnk, APS, Cbl, Chk)14,29,31. Therefore, we investigated the effect of these tyrosine residues by comparing 5YF with 7YF. As a negative control, a kinase-dead mutant K623M32,33 was also constructed (Fig. 3c).
Based on the FK-kit and FR-kit constructs, the mutated chimeras with 5YF, 7YF or K623M were constructed and introduced into Ba/F3 cells (Fig. 3d). The expression of the chimeras was then confirmed by western blotting (Supplementary Figs. 6–8). Next, the cells were washed for depriving IL-3 and cultured in the presence of AP21967 to form mutant–WT heterodimers and mutant–mutant homodimers. The 5YF–WT heterodimer was found to induce cell growth, whereas the 7YF–WT and K623M–WT heterodimers induced negligible and no growth, respectively (Fig. 3e, f, g, h). In all mutants, homodimerization caused no cell growth. These results indicate that 5YF is activated only when heterodimerized with WT and this feature is suitable for exclusively capturing hetero-interactions.
Evaluation of the peptide PPI inhibitor
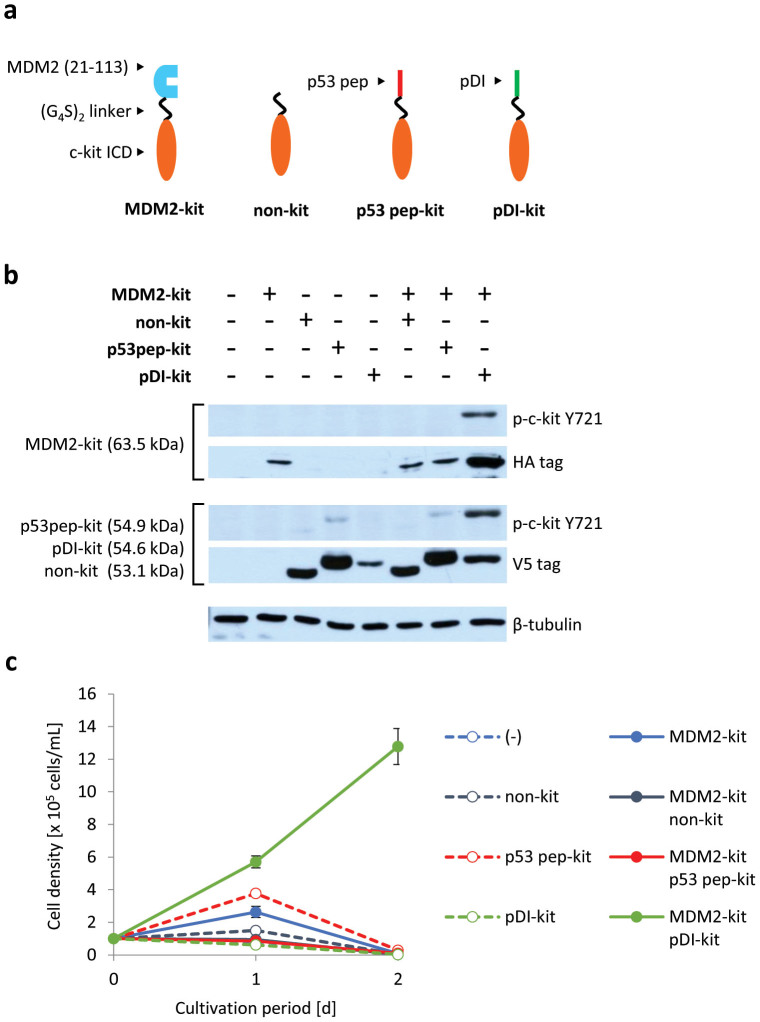
We applied this method to evaluate peptides that inhibit specific PPIs. MDM2 binds to a tumour suppressor protein p53 and induces ubiquitin-dependent degradation of p53, which inhibits p53 functions26. Hu et al. found a peptide (pDI) that binds to the p53-binding domain of MDM2 and inhibits the p53–MDM2 interaction34. Pazgier et al. determined the affinity of pDI towards MDM2 (Kd = 19.6 nM) and a p53-derived peptide (p53 pep) towards MDM2 (Kd = 140 nM)35.
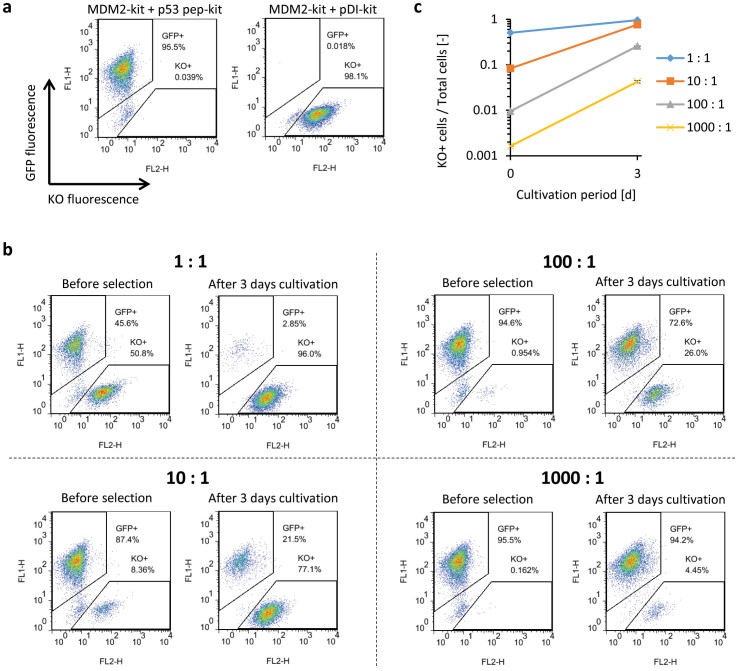
We constructed chimeras in which WT c-kit ICD was fused to the C-termini of MDM2 and these peptides (Fig. 4a), and introduced these constructs into Ba/F3 cells. The western blotting result showed strong tyrosine-phosphorylation of c-kit ICDs in the cells co-expressing MDM2-kit and pDI-kit, but not in the cells co-expressing MDM2-kit and p53 pep-kit (Fig. 4b). These cells were then cultured in the absence of IL-3. Only the cells co-expressing MDM2-kit and pDI-kit proliferated (Fig. 4c). These results indicate that the MDM2–pDI interaction is sufficiently strong to activate c-kit ICDs of the chimeras, whereas the MDM2–p53 pep interaction is not.
Figure 4. Detection of the interaction between MDM2 and its peptide inhibitor.
(a) Illustration for the constructed chimeras. The N-terminal domain of MDM2 and its peptide inhibitors (p53 pep and pDI) were fused to c-kit ICD. (b) Tyrosine phosphorylation of c-kit ICD of chimeras. Western blot analysis was performed with anti-phospho Y721 of c-kit to detect the phosphorylated form of the chimeras, and with an anti-HA or an anti-V5 tag to detect the expression level of the MDM2-fused chimeras (HA tag) or the others (V5 tag). The blots for β-tubulin are indicated as a loading control. (c) Growth of the cells expressing chimeras. Initial cell density was 1 × 105 cells/mL. The viable cell densities are indicated as mean ± SD (n = 3, biological replicates).
Enrichment of the high-affinity peptide
To examine whether this system can positively select cells expressing high-affinity peptide chimeras from a mixture of the cell populations dominantly expressing low-affinity peptide chimeras, EGFP-positive cells co-expressing MDM2-kit and p53 pep-kit and Kusabira Orange (KO)-positive cells co-expressing MDM2-kit and pDI-kit (Fig. 5a) were mixed at ratios of 1:1, 10:1, 100:1 and 1000:1, and cultured in the absence of IL-3. After 3 d cultivation, the enrichment of KO-positive cells was clearly observed (Fig. 5b, c). The KO-positive cells were enriched by (1.89 ± 0.00), (9.17 ± 0.06), (27.0 ± 0.3) and (26.2 ± 1.6)-fold for the mixed ratios of 1:1, 10:1, 100:1 and 1000:1, respectively (Fig. 5c). This result suggests that this method is suitable for high-affinity peptide screening.
Figure 5. Enrichment of cells expressing the high-affinity peptide against MDM2.
(a) EGFP and Kusabira orange (KO) fluorescence of the cells co-expressing MDM2-kit, p53 pep-kit and EGFP, and those co-expressing MDM2-kit, pDI-kit and KO. (b, c) The EGFP-positive cells and the KO-positive cells were mixed in ratios of 1:1, 10:1, 100:1 and 1000:1. Mixed cells were cultured for 3 days, and the ratios were measured with flow cytometry. In (c), the logarithm of the KO-positive cell ratios was plotted as mean ± SD (n = 3, biological replicates).
Discussion
A method for analysing PPIs in living mammalian cells is necessary, because many PPIs are dependent on the cellular environment. To date, a number of PPI detection methods have been developed, and have significantly contributed to our knowledge on cellular events. Pull-down assay with following mass-spectrometric analysis is useful for identifying binding partners in molecular complexes within cells36,37, although multiple washing steps may exclude weak binders10,38. This weakness could be overcome by coupling with promiscuous biotin ligase assay, where the biotin ligase fused with a target protein covalently attaches biotin to the interacting partners, thereby enabling high-affinity capture of them through streptavidin-conjugated beads39. Proximity ligation assay is a tool to sensitively detect PPIs using oligonucleotide-conjugated antibodies and subsequent signal amplification by PCR40,41. While these assays can entrap PPIs among endogenous proteins, cells need to be lysed or fixed before analysis. To detect PPIs in living cells throughout analysis, bait- and prey-fused genetic probes need to be introduced into cells. Such genetic methods include FRET2,3,4, bioluminescence resonance energy transfer (BRET)42,43, PCA5,6,7,8, transcription factor-based two-hybrid44,45, and JAK-STAT-based two-hybrid called mammalian protein–protein interaction trap (MAPPIT)46,47. While the principles of these methods differ among one another, reporter activity is finally detected as fluorescence or bioluminescence, which is a readily detectable readout. The reporter activity, however, is often affected by conformational effects derived from fusing the reporter with bait or prey, which limits generalization of these methods9,10. Therefore, the development of PPI detection methods with distinct principles is desired for analysing various types of PPIs. To develop a novel PPI detection method in living mammalian cells, we engineered receptor tyrosine kinase c-kit. In this method, the proteins of interest (bait and prey) are fused to the c-kit ICD and expressed in IL-3-dependent mammalian cells. Bait–prey interactions induce dimerization and activation of the c-kit ICD and this dimerization leads to cell growth. Since c-kit is activated only by dimerization, background growth is negligible.
To investigate whether our method can detect PPIs, we constructed chimeras fusing FKBPF36V, FKBP or FRBT2098L as a model protein and introduced these proteins into IL-3-dependent Ba/F3 cells. Ligand-dependent homodimeric FKBPF36V and heterodimeric FKBP–FRBT2098L interactions were successfully detected based on cell growth (Figs. 1 and 2). Remarkably, almost no cell growth was observed in the absence of the ligands, suggesting that background growth is significantly suppressed in our method.
We engineered c-kit ICD for screening prey that interacts with bait from a protein- or peptide-library. In these libraries, there may be proteins and peptides that form homodimers or homo-oligomers. If WT c-kit ICDs were fused to both bait and prey, not only bait–prey interactions but also prey–prey interactions would induce cell growth and be detected. To overcome this problem, we designed c-kit ICD mutants called 5YF and 7YF, in which, 5 and 7 tyrosine residues, respectively, were replaced with phenylalanine to prevent recruitment of signalling molecules. As a result, 5YF induces cell growth when heterodimerized with WT c-kit ICD but not when homodimerized (Fig. 3). Based on this knowledge, fusing 5YF and WT c-kit ICD to prey and bait, respectively, can specifically evaluate bait–prey interactions. In this design, however, homodimeric bait protein cannot be used, which is a limitation of our current system.
Since PPI inhibitors are promising for therapy, PPI-detection methods including FRET have been used for evaluating PPI inhibitors. To investigate whether our method can evaluate peptide inhibitors, p53 pep and pDI, two binding peptides against MDM2, were used as model interactions. Although low-affinity interaction between MDM2 and p53 pep was not detected, high-affinity interaction between MDM2 and pDI was clearly detected (Fig. 4). This result suggests that our method can enrich peptides with high affinity towards a target protein.
Peptide inhibitor screening has largely relied on phage display and yeast two-hybrid48,49. While these are useful as robust screening methods, it is uncertain whether peptides selected in these methods function in living mammalian cells. Therefore, the selected peptides need to be assessed in additional experimental settings using living mammalian cells. Through the model peptide-enrichment experiment, we demonstrated that our method can select cells expressing the chimera with a high-affinity peptide from a mixture of the cells that are dominantly expressing the chimera with a low-affinity peptide (Fig. 5). This suggests that our method is potentially suitable for screening a peptide library directly in living mammalian cells. In a peptide library screening, the gene encoding a target protein is fused with the signaling domain of c-kit. Ba/F3 cells are transduced with the retroviral vector encoding the chimeric gene to establish a stable transductant. The transductant is further transduced with retroviral vectors encoding a genetic library for diverse peptides fused with the signaling domain of c-kit. Because the transductants with peptide binders to the target protein are growth-selected in the absence of IL-3, genomic PCR analysis of the selected cells identifies the sequence of the binders. Functional analyses of the binders may be subsequently performed by another appropriate system to determine whether the binders exert loss- or gain-of-function on the target protein.
As demonstrated in Fig. 4, our system may miss weak and transient interactions in its current format. One possible solution is to introduce helper interactions in the constructs. Previous reports demonstrated that introduction of leucine zipper interaction (derived from Jun and Fos) or domain–peptide interaction (based on the WW or SH3 domain and its interacting peptide) facilitated the interaction between FRET probes, leading to enhanced FRET efficiency9,50. Based on this strategy, increased sensitivity of antigen detection was attained in antibody-fused FRET probes50, and transient weak interaction between Raf1 and B-Raf was successfully detected9. Therefore, such engineering may enhance chimeric c-kit-mediated growth signaling, which may enable the detection of weak and transient interactions in our method. While this study demonstrated applicability of our method to cytosolic proteins, another issue to be addressed is applicability to membrane proteins, which are attractive targets in drug development. Because native c-kit is a membrane protein originally, signal transduction through c-kit may rather be enhanced by membrane localization. This context would be worth testing in future studies for demonstrating versatility of our method.
In summary, we successfully developed a novel method for PPI detection based on kinase-mediated growth induction of mammalian cells. This method detects PPI through dimer formation and subsequent activation of c-kit kinase domains, which allows low-background PPI detection with a simple readout of “growth or death” using a factor-dependent cell line. This simplicity could facilitate application of our method to a robust screening of chemical and peptide libraries for finding drug candidates in the future.
Methods
Plasmid construction
Primer sequences are summarized in Supplementary Table 1. The correspondence between the constructed plasmids and names of the chimeras is summarized in Supplementary Table 2.
pMK-D2TM-stuffer-IP, which encodes the extracellular D2 and transmembrane domain of the erythropoietin receptor (EpoR) and an internal ribosomal entry site (IRES)-puromycin cassette, was linearized by polymerase chain reaction (PCR) using two primers (F36V-ID-Myc_f, mpl-GST-F36V_r). As an insert, FKBPF36V was amplified with PCR using two primers (F36V_f, F36V_r) and pC4M-Fv2E (Ariad Pharmaceuticals, Cambridge, MA) as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme (Clontech, Mountain View, CA) to make pMK-D2TM-stuffer-F36V-IP. pMK-D2TM-stuffer-F36V-IP was linearized by PCR using two primers (kit-GST-F36V_f, EpoRTM-RT-kit_r). As an insert, the intracellular domain of human c-kit was amplified with PCR using two primers (kitIC_f, kitIC_r) and pMK-SKK-Flag-IG, which is a variant of pMK-SEmK-Flag-IG51, as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to give pMK-D2TM-kit-del-F36V-IP. To make pMK-Met-D2TM-kit-del-F36V-IP, a signal sequence was removed from pMK-D2TM-kit-del-F36V-IP by PCR using two primers (QC-Met-V5_f, QC-Met-V5_r). EpoR D2 domain and transmembrane domain were removed from pMK-Met-D2TM-kit-del-F36V-IP by PCR using two primers (QC-V5-GSG-kit_f, QC-V5-GSG-kit_r) to obtain pMK-kit-del-F36V-IP.
The c-kit intracellular domain was removed from pMK-kit-del-F36V-IP by PCR using two primers (QC-V5-GSG-36V_f, QC-V5-GSG-36V_r) to make pMK-F36V-IP. pMK-F36V-IP was linearized by PCR using two primers (IF-kit-ID-myc_f, IF-36-GST-kit-r). As an insert, the c-kit intracellular domain was amplified with PCR using two primers (kitIC_f, kitIC_r) and pMK-SKK-Flag-IG as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pMK-F36V-del-kit-IP.
pMK-kit-del-F36V-IP was linearized by PCR using two primers (G4S-F36V_f, kit-G4S_r). The linearized plasmid was phosphorylated by T4 polynucleotide kinase (PNK) and circularized by T4 DNA ligase to obtain pMK-kit-(G4S)2-F36V-IP.
pMK-F36V-del-kit-IP was linearized by PCR using two primers (G4S-kit_f, F36V-G4S_r). The linearized plasmid was phosphorylated by T4 PNK and circularized by T4 DNA ligase to obtain pMK-F36V-(G4S)2-kit-IP.
pFB-HA-FKBP-IN, which encodes a hemagglutinin (HA) tag and human FKBP12, was linearized by PCR using two primers (IF-kit-GST-FK_f, IF-HA-GSG-kit_r). As an insert, the intracellular domain of c-kit was amplified with PCR using two primers (kitIC_f, kitIC_r) and pMK-SKK-Flag-IG as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pFB-kit-del-FKBP-IN. pFB-kit-del-FKBP-IN was linearized by PCR using two primers (G4S-F36V_f kit-G4S_r). The linearized plasmid was phosphorylated by T4 PNK and circularized by T4 DNA ligase to obtain pFB-kit-(G4S)2-FKBP-IN.
pFB-HA-FKBP-IN was linearized by PCR using two primers (IF-kit-I-flag_f, IF-FK-GST-kit_r). As an insert, the c-kit intracellular domain was amplified with PCR using two primers (kitIC_f, kitIC_r) and pMK-SKK-Flag-IG as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pFB-FKBP-del-kit-IN. pFB-FKBP-del-kit-IN was linearized by PCR using two primers (G4S-kit_f, F36V-G4S_r). The linearized plasmid was phosphorylated by T4 PNK and circularized by T4 DNA ligase to obtain pFB-FKBP-(G4S)2-kit-IN.
pMK-kit-del-F36V-IP was linearized by PCR using two primers (IF-FR-ID-myc_f, IF-kit-GST-FR_r). As an insert, FRBT2098L was amplified with PCR using two primers (FRB_f, FRB_r) and pC4-RHE (Ariad Pharmaceuticals) as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pMK-kit-del-FRBT2098L-IP. pMK-kit-del-FRBT2098L-IP was linearized by PCR using two primers (G4ST-FR_f, kit-G4S_r). The linearized plasmid was phosphorylated by T4 PNK and circularized by T4 DNA ligase to obtain pMK-kit-(G4S)2-FRBT2098L-IP.
pMK-F36V-del-kit-IP was linearized by PCR using two primers (IF-FR-GST-kit_f, IF-V5-GST-FR_r). As an insert, FRBT2098L was amplified with PCR using two primers (FRB_f, FRB_r) and pC4-RHE as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pMK-FRBT2098L-del-kit-IP. pMK-FRBT2098L-del-kit-IP was linearized by PCR using two primers (G4S-kit_f, FR-GSSG4S_r). The linearized plasmid was phosphorylated by T4 PNK and circularized by T4 DNA ligase to obtain pMK-FRBT2098L-(G4S)2-kit-IP.
5YF, which is a mutant of the intracellular domain of c-kit (Y703F, Y721F, Y730F, Y747F, Y936F), was synthesized by GenScript (Piscataway, NJ, USA) and cloned into pUC55, resulting in pUC55-kit5YF. As an insert, 5YF was amplified with PCR using two primers (kitIC_f, kitIC_r) and pUC55-kit5YF as a template. pMK-FRBT2098L-(G4S)2-stuffer-IP and pFB-FKBP-(G4S)2-stuffer-IN were linearized by PCR using two-primer sets (IF-kit-ID-myc_f, IF-G4S2-kit_r) and (IF-kit-I-flag_f, IF-G4S2-kit_r), respectively. Each linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pMK-FRBT2098L-(G4S)2-5YF-IP and pFB-FKBP-(G4S)2-5YF-IN, respectively.
7YF is a mutant of the intracellular domain of c-kit (Y568F, Y570F, Y703F, Y721F, Y730F, Y747F, Y936F). Two additional mutations were introduced by PCR using two primers (QC-Y568,570F_f, QC-Y568,570F_r) with pMK-FRBT2098L-(G4S)2-5YF-IP and pMK-FRBT2098L-(G4S)2-5YF-IP as templates to obtain pMK-FRBT2098L-(G4S)2-7YF-IP and pFB-FKBP-(G4S)2-7YF-IN, respectively.
K623M is a kinase-dead mutant of c-kit. The mutation was introduced by PCR using two primers (QC-K623M_f, QC-K623M_r) with pMK-FRBT2098L-(G4S)2-kit-IP as a template to obtain pMK-FRBT2098L-(G4S)2-K623M-IP.
pFB-FKBP-(G4S)2-stuffer-IN was linearized by PCR using two primers (IF-kit-I-flag_f, IF-G4S2-kit_r). As an insert, K623M was amplified with PCR using two primers (kitIC_f, kitIC_r) and pMK-FRBT2098L-(G4S)2-K623M-IP as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pFB-FKBP-(G4S)2-K623M-IN.
pFB-FKBP-(G4S)2-kit-IN was linearized by PCR using two primers (GSS-G4S2_f, HA-GSG_r). As an insert, human MDM2 (amino acid residues ranged from 21 to 113) was amplified with PCR using two primers (HA-GSG-MDM2_f, MDM2-GSS-G4S2_r) and HEK293 cell-derived cDNA as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pFB-MDM2(21-113)-kit-IN.
FRBT2098L was removed from pMK-FRBT2098L-(G4S)2-kit-IP by PCR using two primers (V5-GSG-G4S2_f, V5-GSG-G4S2_r) to obtain pMK-non-kit-IP.
To replace FRBT2098L of pMK-FRBT2098L-(G4S)2-kit-IP with human p53 (amino acid residues ranged from 15 to 29, SQETFSDLWKLLPEN), pMK-FRBT2098L-(G4S)2-kit-IP was linearized by PCR using two primers (p53pep-G4S2_f, V5-p53pep_r). The linearized plasmid was phosphorylated by T4 PNK and circularized by T4 DNA ligase to obtain pMK-p53pep-kit-IP.
To replace FRBT2098L of pMK-FRBT2098L-(G4S)2-kit-IP with the pDI peptide (LTFEHYWAQLTS), pMK-FRBT2098L-(G4S)2-kit-IP was linearized by PCR using two primers (p53like-G4S2_f, V5-p53like_r). The linearized plasmid was phosphorylated by T4 PNK and circularize by T4 DNA ligase to obtain pMK-pDI-kit-IP.
The DNA fragment of the chimera composed of a V5 tag, p53 (15–29), a (G4S)2 linker, the intracellular domain of c-kit and a myc tag was prepared by digesting pMK-p53pep-kit-IP with EcoRI and BamHI. This fragment was ligated with pMK-stuffer-IG digested with EcoRI and BamHI to obtain pMK-p53pep-kit-IG, which encodes an IRES-EGFP cassette.
To replace the puromycin resistance gene with the Kusabira Orange (KO) gene, pMK-pDI-kit-IP was linearized by PCR using two primers (pMK-IK-host_f3, pMK-IK-inser_r2). As an insert, the KO gene was amplified by PCR using two primers (pMK-IK-insert_f, pMK-IK-host_r) and pGCDNsam-stuffer-IK as a template. The linearized plasmid and the insert were fused by the In-Fusion HD enzyme to obtain pMK-pDI-kit-IK, which encodes an IRES-KO cassette.
Cell lines
A murine IL-3-dependent pro-B cell line, Ba/F3 (RCB0805, RIKEN Cell Bank, Tsukuba, Japan), was cultured in RPMI 1640 medium (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 10% foetal bovine serum (FBS; Biowest, Paris, France) and 1 ng/ml murine IL-3 (R&D Systems, Cambridge, MA, USA). A retroviral packaging cell line, Plat-E52, was cultured in Dulbecco's modified Eagle's medium (Nissui Pharmaceutical) supplemented with 10% FBS, 1 μg/ml puromycin (Sigma, St Louis, MO, USA) and 10 μg/ml blasticidin (Kaken Pharmaceutical, Tokyo, Japan).
Vector transduction
Plat-E cells were transfected with the constructed plasmids using Lipofectamine LTX (Invitrogen, Groningen, The Netherlands) according to the manufacturer's protocol. The culture supernatant on day 2 was used for retroviral transduction of Ba/F3 cells in the presence of 1 ng/ml IL-3 in a 24-well plate, using RetroNectin (Takara Bio, Otsu, Japan) according to the manufacturer's instructions. Ba/F3 cells transduced with vectors were cultured in the presence of 800 μg/mL G418 (Calbiochem, La Jolla, CA, USA) and/or 2 μg/mL puromycin.
Starvation and AP20187- and AP21967-stimulation of cells
The transductants cultured in 1 ng/ml IL-3 were washed twice with phosphate-buffered saline (PBS), cultured in a depletion medium without IL-3 for 6 h, and then stimulated with AP20187 or AP21967 (Ariad Pharmaceuticals) at 37°C for 15 min. Equal volume of ice-cold 2 mM Na3VO4 in PBS was added to the cells to stop the reaction and inhibit dephosphorylation. The cells were collected by centrifugation at 800 g, followed by preparation of lysates and western blotting.
Western blotting
The transductants (1 × 106) were washed with PBS, lysed with 100 μl of lysis buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin) and incubated on ice for 10 min. After centrifugation at 21,500 g at 4°C for 10 min, the supernatant was mixed with Laemmli's sample buffer and boiled. The lysate was resolved by sodium dodecyl sulphate polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (GE Healthcare, Buckinghamshire, UK). After the membrane was blocked either with 5% skim milk (Wako Pure Chemical Industries, Osaka, Japan) for the detection of the HA tag, V5 tag and β-tubulin or with 3% bovine serum albumin (Sigma) for the detection of p-c-kit Y721. The blot was probed with rabbit primary antibodies, followed by horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Biosource, Camarillo, CA, USA). The primary antibodies used are: rabbit p-c-kit Y721 (which corresponds to Y719 of mouse origin) (Cell Signaling Technology, Danvers, MA, USA), rabbit anti-HA (BETHYL, Montgomery, TX, USA), rabbit anti-V5 (Millipore, Billerica, MA, USA) and rabbit anti-β-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Detection was performed using Luminata Western HRP Substrates (Millipore).
Cell proliferation assay
The transductants were washed twice with PBS and seeded into 24-well plates at 1 × 105 cells/well with serial concentrations of AP20187 or AP21967. The viable cell numbers were counted with a FACSCalibur flow cytometer (Becton-Dickinson, Lexington, KY, USA) using Flow-Count (Beckman Coulter, Fullerton, CA, USA).
Enrichment experiment
The cells co-expressing MDM2-kit, p53 pep-kit and EGFP and those co-expressing MDM2-kit, pDI-kit and Kusabira orange (KO) were washed twice with PBS and mixed in the ratios of 1:1, 10:1, 100:1 and 1000:1. These cells were seeded into 60 mm dishes at 1 × 106 cells/dish. After 3 days cultivation, the EGFP and KO fluorescence intensities of these cells were measured with a FACSCalibur flow cytometer with excitation at 488 nm and fluorescence detection at 530 ± 15 nm and 585 ± 21 nm for detecting EGFP and KO, respectively.
Author Contributions
M.K. conceived the project. S.M. and M.K. designed and performed the experiments, and analysed the data. S.M., T.N. and M.K. wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to T. Kitamura (The University of Tokyo) for the retroviral expression system, and Ariad Pharmaceuticals for the ARGENT dimerization kit and ligands. This work was supported by Grant-in-Aid for Challenging Exploratory Research 23656516 (M.K.) from the Japan Society for the Promotion of Science, by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN) (M.K.) and by the Global COE Program for Chemistry Innovation.
References
- Lievens S., Lemmens I. & Tavernier J. Mammalian two-hybrids come of age. Trends Biochem. Sci. 34, 579–588 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. H. & Stagljar I. Strategies for membrane interaction proteomics: no mass spectrometry required. Proteomics 12, 1519–1526 (2012). [DOI] [PubMed] [Google Scholar]
- Harter K., Meixner A. J. & Schleifenbaum F. Spectro-microscopy of living plant cells. Mol. Plant 5, 14–26 (2012). [DOI] [PubMed] [Google Scholar]
- Padilla-Parra S. & Tramier M. FRET microscopy in the living cell: different approaches, strengths and weaknesses. Bioessays 34, 369–376 (2012). [DOI] [PubMed] [Google Scholar]
- Michnick S. W., Ear P. H., Manderson E. N., Remy I. & Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat. Rev. Drug Discov. 6, 569–582 (2007). [DOI] [PubMed] [Google Scholar]
- Ventura S. Bimolecular fluorescence complementation: illuminating cellular protein interactions. Curr. Mol. Med. 11, 582–598 (2011). [DOI] [PubMed] [Google Scholar]
- Shekhawat S. S. & Ghosh I. Split-protein systems: beyond binary protein-protein interactions. Curr. Opin. Chem. Biol. 15, 789–797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A., Ozawa T. & Umezawa Y. Detection of protein-protein interactions in bacteria by GFP-fragment reconstitution. Methods Mol. Biol. 705, 251–258 (2011). [DOI] [PubMed] [Google Scholar]
- Grunberg R. et al. Engineering of weak helper interactions for high-efficiency FRET probes. Nat. Methods 10, 1021–1027 (2013). [DOI] [PubMed] [Google Scholar]
- Stynen B., Tournu H., Tavernier J. & Van Dijck P. Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol. Mol. Biol. Rev. 76, 331–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B. H. et al. Optical sensors for monitoring dynamic changes of intracellular metabolite levels in mammalian cells. Nat. Protoc. 6, 1818–1833 (2011). [DOI] [PubMed] [Google Scholar]
- Kerppola T. K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston D. W. & Kremers G. J. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem. Sci. 32, 407–414 (2007). [DOI] [PubMed] [Google Scholar]
- Ronnstrand L. Signal transduction via the stem cell factor receptor/c-Kit. Cell. Mol. Life Sci. 61, 2535–2548 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartsson J. & Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol. Rev. 92, 1619–1649 (2012). [DOI] [PubMed] [Google Scholar]
- Edling C. E. & Hallberg B. c-Kit--a hematopoietic cell essential receptor tyrosine kinase. Int. J. Biochem. Cell Biol. 39, 1995–1998 (2007). [DOI] [PubMed] [Google Scholar]
- Liang J. et al. The C-kit receptor-mediated signal transduction and tumor-related diseases. Int. J. Biol. Sci. 9, 435–443 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P., Krishnamoorthy N., Oriss T. B. & Ray A. Signaling of c-kit in dendritic cells influences adaptive immunity. Annals N.Y. Acad. Sci. 1183, 104–122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S. R. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell Biol. 5, 464–471 (2004). [DOI] [PubMed] [Google Scholar]
- Lemmon M. A. & Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. Investigating protein-ligand interactions with a mutant FKBP possessing a designed specificity pocket. J. Med. Chem. 43, 1135–1142 (2000). [DOI] [PubMed] [Google Scholar]
- Yang W. et al. Regulation of gene expression by synthetic dimerizers with novel specificity. Bioorg. Med. Chem. Lett. 13, 3181–3184 (2003). [DOI] [PubMed] [Google Scholar]
- Clackson T. et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. USA 95, 10437–10442 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle J. H. et al. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem. Biol. 13, 99–107 (2006). [DOI] [PubMed] [Google Scholar]
- Grunberg R., Ferrar T. S., van der Sloot A. M., Constante M. & Serrano L. Building blocks for protein interaction devices. Nucleic Acids Res. 38, 2645–2662 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Lain S., Verma C. S., Fersht A. R. & Lane D. P. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer 9, 862–873 (2009). [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P. et al. Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3′-kinase is essential for male fertility. Nat. Genet. 24, 157–162 (2000). [DOI] [PubMed] [Google Scholar]
- Choi J., Chen J., Schreiber S. L. & Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273, 239–242 (1996). [DOI] [PubMed] [Google Scholar]
- Gilfillan A. M. & Rivera J. The tyrosine kinase network regulating mast cell activation. Immunol. Rev. 228, 149–169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Munugalavadla V. & Kapur R. c-Kit-mediated overlapping and unique functional and biochemical outcomes via diverse signaling pathways. Mol. Cell. Biol. 24, 1401–1410 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R. Jr Signaling by Kit protein-tyrosine kinase--the stem cell factor receptor. Biochem. Biophys. Res. Commun. 337, 1–13 (2005). [DOI] [PubMed] [Google Scholar]
- Mancini A., Koch A., Stefan M., Niemann H. & Tamura T. The direct association of the multiple PDZ domain containing proteins (MUPP-1) with the human c-Kit C-terminus is regulated by tyrosine kinase activity. FEBS Lett. 482, 54–58 (2000). [DOI] [PubMed] [Google Scholar]
- Tetsu O. et al. Mutations in the c-Kit gene disrupt mitogen-activated protein kinase signaling during tumor development in adenoid cystic carcinoma of the salivary glands. Neoplasia 12, 708–717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Gilkes D. M. & Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 67, 8810–8817 (2007). [DOI] [PubMed] [Google Scholar]
- Pazgier M. et al. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc. Natl. Acad. Sci. USA 106, 4665–4670 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh G. C., Porras P., Aranda B., Hermjakob H. & Orchard S. E. Analyzing protein-protein interaction networks. J. Proteome Res. 11, 2014–2031 (2012). [DOI] [PubMed] [Google Scholar]
- Gavin A. C., Maeda K. & Kuhner S. Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22, 42–49 (2011). [DOI] [PubMed] [Google Scholar]
- Rajagopala S. V., Sikorski P., Caufield J. H., Tovchigrechko A. & Uetz P. Studying protein complexes by the yeast two-hybrid system. Methods 58, 392–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K. J., Kim D. I., Raida M. & Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibrecht I. et al. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev. Proteomics 7, 401–409 (2010). [DOI] [PubMed] [Google Scholar]
- Blokzijl A. et al. Protein biomarker validation via proximity ligation assays. Biochim. Biophys. Acta 1844, 933–939 (2014). [DOI] [PubMed] [Google Scholar]
- De A., Jasani A., Arora R. & Gambhir S. S. Evolution of BRET Biosensors from Live Cell to Tissue-Scale Imaging. Front. Endocrinol. 4, 131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger K. D. & Eidne K. A. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nat. Methods 3, 165–174 (2006). [DOI] [PubMed] [Google Scholar]
- Mendonca D. B., Mendonca G. & Cooper L. F. Mammalian two-hybrid assays for studies of interaction of p300 with transcription factors. Methods Mol. Biol. 977, 323–338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncivais K. & Zhang Z. J. Tetracycline repressor-based mammalian two-hybrid systems. Methods Mol. Biol. 812, 259–273 (2012). [DOI] [PubMed] [Google Scholar]
- Lievens S., Peelman F., De Bosscher K., Lemmens I. & Tavernier J. MAPPIT: a protein interaction toolbox built on insights in cytokine receptor signaling. Cytokine Growth Factor Rev. 22, 321–329 (2011). [DOI] [PubMed] [Google Scholar]
- Lievens S., Caligiuri M., Kley N. & Tavernier J. The use of mammalian two-hybrid technologies for high-throughput drug screening. Methods 58, 335–342 (2012). [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A., Tate S. A., Beswick R. W., Wagner S. D. & Ko Ferrigno P. A peptide aptamer to antagonize BCL-6 function. Oncogene 25, 2223–2233 (2006). [DOI] [PubMed] [Google Scholar]
- Schmidt S., Diriong S., Mery J., Fabbrizio E. & Debant A. Identification of the first Rho-GEF inhibitor, TRIPalpha, which targets the RhoA-specific GEF domain of Trio. FEBS Lett. 523, 35–42 (2002). [DOI] [PubMed] [Google Scholar]
- Ohiro Y., Arai R., Ueda H. & Nagamune T. A homogeneous and noncompetitive immunoassay based on the enhanced fluorescence resonance energy transfer by leucine zipper interaction. Anal. Chem. 74, 5786–5792 (2002). [DOI] [PubMed] [Google Scholar]
- Kaneko E., Kawahara M., Ueda H. & Nagamune T. Growth control of genetically modified cells using an antibody/c-Kit chimera. J. Biosci. Bioeng. 113, 641–646 (2012). [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima T. & Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information