Abstract
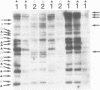
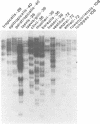
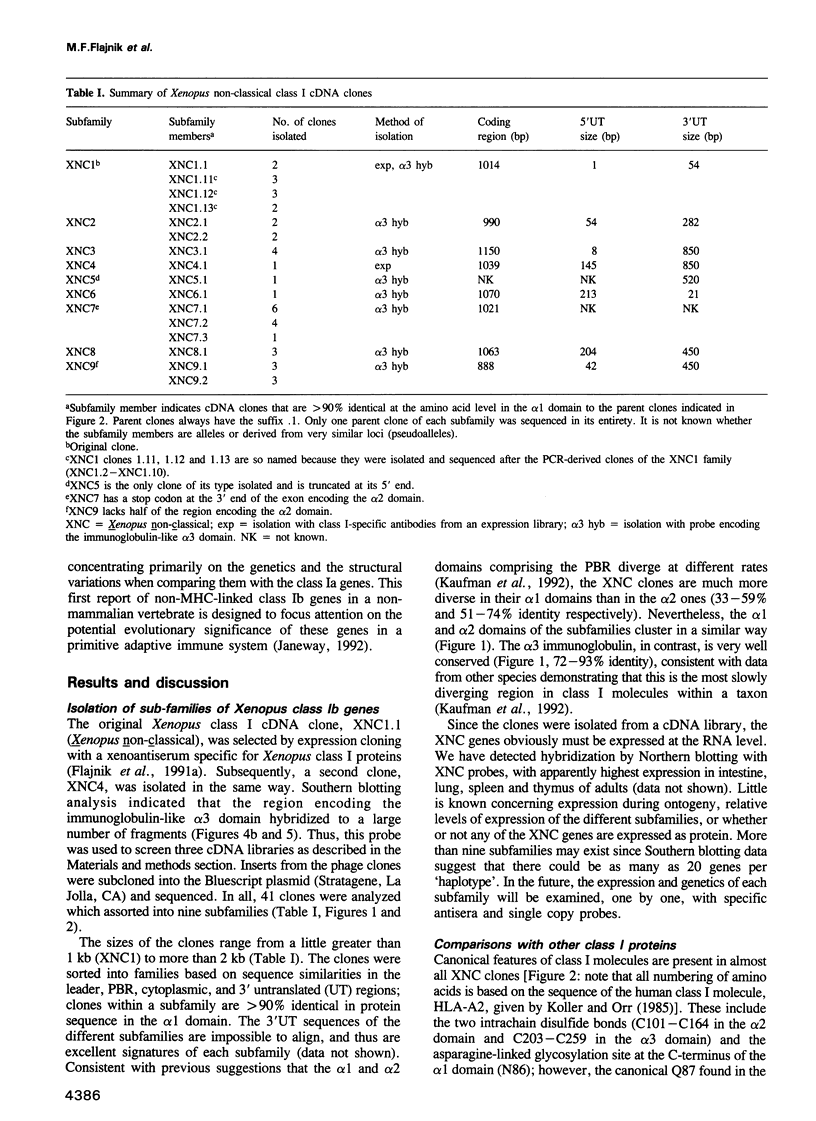
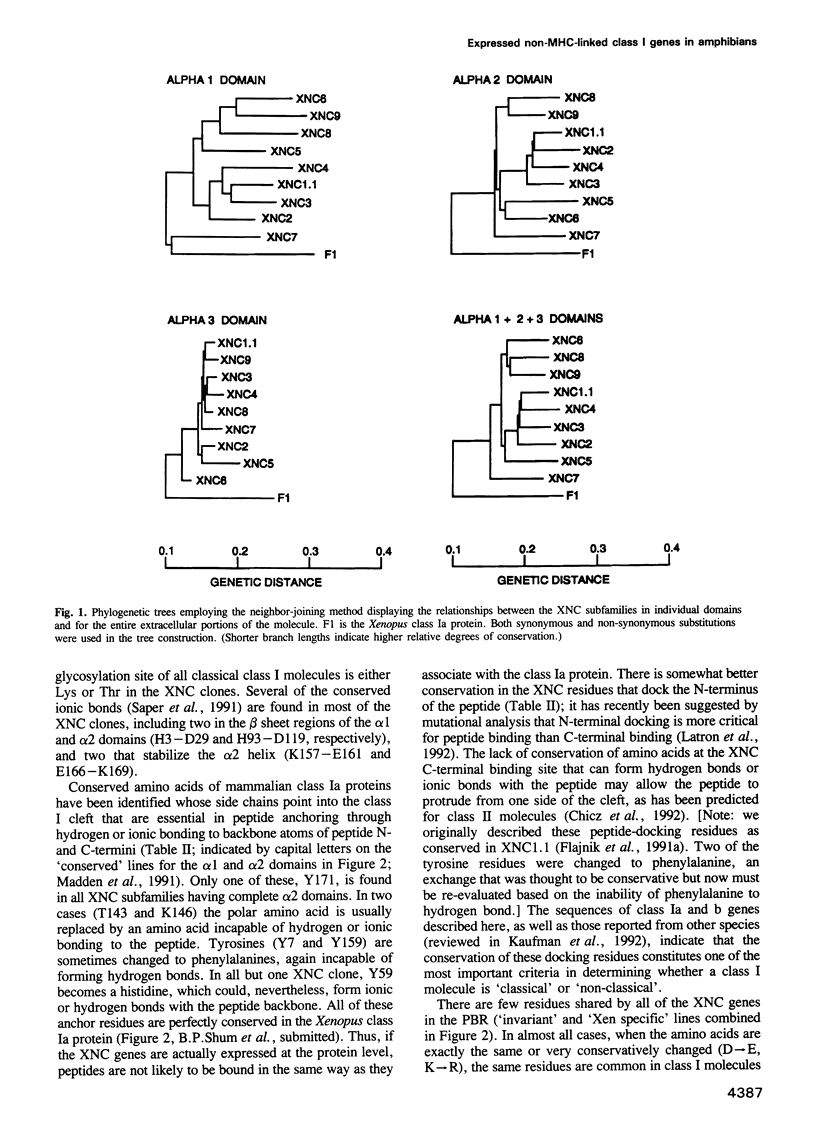
A Xenopus class I cDNA clone, isolated from a cDNA expression library using antisera, is a member of a large family of non-classical class I genes (class Ib) composed of at least nine subfamilies, all of which are expressed at the RNA level. The subfamilies are well conserved in their immunoglobulin-like alpha 3 domains, but their peptide-binding regions (PBRs) and cytoplasmic domains are very divergent. In contrast to the great allelic diversity found in the PBR of classical class I genes, the alleles of one of the Xenopus non-classical subfamilies are extremely well conserved in all regions. Several of the invariant amino acids essential for the anchoring of peptides in the classical class I groove are not conserved in some subfamilies, but the class Ib genes are nevertheless more closely related in the PBR to classical and non-classical genes linked to the MHC in mammals and birds than to any other described class I genes like CD1 and the neonatal rat intestinal Fc receptor. Comparison with the Xenopus MHC-linked class Ia protein indicate that amino acids presumed to interact with beta 2-microglobulin are identical or conservatively changed in the two major class I families. Genomic analyses of Xenopus species suggest that the classical and non-classical families diverged from a common ancestor before the emergence of the genus Xenopus over 100 million years ago; all of the non-classical genes appear to be linked on a chromosome distinct from the one harboring the MHC. We hypothesize that this class Ib gene family is under very different selection pressures from the classical MHC genes, and that each subfamily may have evolved for a particular function.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bjorkman P. J., Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987 Oct 8;329(6139):512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Bradbury A., Belt K. T., Neri T. M., Milstein C., Calabi F. Mouse CD1 is distinct from and co-exists with TL in the same thymus. EMBO J. 1988 Oct;7(10):3081–3086. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briles W. E., Goto R. M., Auffray C., Miller M. M. A polymorphic system related to but genetically independent of the chicken major histocompatibility complex. Immunogenetics. 1993;37(6):408–414. doi: 10.1007/BF00222464. [DOI] [PubMed] [Google Scholar]
- Brodsky F. M., Guagliardi L. E. The cell biology of antigen processing and presentation. Annu Rev Immunol. 1991;9:707–744. doi: 10.1146/annurev.iy.09.040191.003423. [DOI] [PubMed] [Google Scholar]
- Brown J. H., Jardetzky T., Saper M. A., Samraoui B., Bjorkman P. J., Wiley D. C. A hypothetical model of the foreign antigen binding site of class II histocompatibility molecules. Nature. 1988 Apr 28;332(6167):845–850. doi: 10.1038/332845a0. [DOI] [PubMed] [Google Scholar]
- Chicz R. M., Urban R. G., Lane W. S., Gorga J. C., Stern L. J., Vignali D. A., Strominger J. L. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992 Aug 27;358(6389):764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Flajnik M. F., Canel C., Kramer J., Kasahara M. Evolution of the major histocompatibility complex: molecular cloning of major histocompatibility complex class I from the amphibian Xenopus. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):537–541. doi: 10.1073/pnas.88.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik M. F., Canel C., Kramer J., Kasahara M. Which came first, MHC class I or class II? Immunogenetics. 1991;33(5-6):295–300. doi: 10.1007/BF00216688. [DOI] [PubMed] [Google Scholar]
- Flajnik M. F., Du Pasquier L. The major histocompatibility complex of frogs. Immunol Rev. 1990 Feb;113:47–63. doi: 10.1111/j.1600-065x.1990.tb00036.x. [DOI] [PubMed] [Google Scholar]
- Flajnik M. F., Kaufman J. F., Hsu E., Manes M., Parisot R., Du Pasquier L. Major histocompatibility complex-encoded class I molecules are absent in immunologically competent Xenopus before metamorphosis. J Immunol. 1986 Dec 15;137(12):3891–3899. [PubMed] [Google Scholar]
- Guild B. C., Erikson R. L., Strominger J. L. HLA-A2 and HLA-B7 antigens are phosphorylated in vitro by rous sarcoma virus kinase (pp60v-src) at a tyrosine residue encoded in a highly conserved exon of the intracellular domain. Proc Natl Acad Sci U S A. 1983 May;80(10):2894–2898. doi: 10.1073/pnas.80.10.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guild B. C., Strominger J. L. Human and murine class I MHC antigens share conserved serine 335, the site of HLA phosphorylation in vivo. J Biol Chem. 1984 Jul 25;259(14):9235–9240. [PubMed] [Google Scholar]
- Guillemot F., Billault A., Pourquié O., Béhar G., Chaussé A. M., Zoorob R., Kreibich G., Auffray C. A molecular map of the chicken major histocompatibility complex: the class II beta genes are closely linked to the class I genes and the nucleolar organizer. EMBO J. 1988 Sep;7(9):2775–2785. doi: 10.1002/j.1460-2075.1988.tb03132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U. B., Sundvall M., Erlich H. A. Allelic diversity is generated by intraexon sequence exchange at the DRB1 locus of primates. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3686–3690. doi: 10.1073/pnas.88.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., Nakanishi T., Kurosawa Y. Identification of a shark sequence resembling the major histocompatibility complex class I alpha 3 domain. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2209–2212. doi: 10.1073/pnas.89.6.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick S. M. Dawn of the hunt for nonclassical MHC function. Cell. 1992 Jul 24;70(2):177–180. doi: 10.1016/0092-8674(92)90092-q. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988 Sep 8;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Janeway C. A., Jr The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol Today. 1992 Jan;13(1):11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- Kaufman J., Andersen R., Avila D., Engberg J., Lambris J., Salomonsen J., Welinder K., Skjødt K. Different features of the MHC class I heterodimer have evolved at different rates. Chicken B-F and beta 2-microglobulin sequences reveal invariant surface residues. J Immunol. 1992 Mar 1;148(5):1532–1546. [PubMed] [Google Scholar]
- Klein J., Figueroa F. Evolution of the major histocompatibility complex. Crit Rev Immunol. 1986;6(4):295–386. [PubMed] [Google Scholar]
- Koller B. H., Orr H. T. Cloning and complete sequence of an HLA-A2 gene: analysis of two HLA-A alleles at the nucleotide level. J Immunol. 1985 Apr;134(4):2727–2733. [PubMed] [Google Scholar]
- Kurlander R. J., Shawar S. M., Brown M. L., Rich R. R. Specialized role for a murine class I-b MHC molecule in prokaryotic host defenses. Science. 1992 Jul 31;257(5070):678–679. doi: 10.1126/science.1496381. [DOI] [PubMed] [Google Scholar]
- Latron F., Pazmany L., Morrison J., Moots R., Saper M. A., McMichael A., Strominger J. L. A critical role for conserved residues in the cleft of HLA-A2 in presentation of a nonapeptide to T cells. Science. 1992 Aug 14;257(5072):964–967. doi: 10.1126/science.1380181. [DOI] [PubMed] [Google Scholar]
- Ljunggren H. G., Kärre K. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990 Jul;11(7):237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- Madden D. R., Gorga J. C., Strominger J. L., Wiley D. C. The structure of HLA-B27 reveals nonamer self-peptides bound in an extended conformation. Nature. 1991 Sep 26;353(6342):321–325. doi: 10.1038/353321a0. [DOI] [PubMed] [Google Scholar]
- Martin L. H., Calabi F., Milstein C. Isolation of CD1 genes: a family of major histocompatibility complex-related differentiation antigens. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9154–9158. doi: 10.1073/pnas.83.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986 Sep;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Pamer E. G., Wang C. R., Flaherty L., Lindahl K. F., Bevan M. J. H-2M3 presents a Listeria monocytogenes peptide to cytotoxic T lymphocytes. Cell. 1992 Jul 24;70(2):215–223. doi: 10.1016/0092-8674(92)90097-v. [DOI] [PubMed] [Google Scholar]
- Porcelli S., Morita C. T., Brenner M. B. CD1b restricts the response of human CD4-8- T lymphocytes to a microbial antigen. Nature. 1992 Dec 10;360(6404):593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper M. A., Bjorkman P. J., Wiley D. C. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991 May 20;219(2):277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- Sato K., Flajnik M. F., Du Pasquier L., Katagiri M., Kasahara M. Evolution of the MHC: isolation of class II beta-chain cDNA clones from the amphibian Xenopus laevis. J Immunol. 1993 Apr 1;150(7):2831–2843. [PubMed] [Google Scholar]
- Simister N. E., Mostov K. E. An Fc receptor structurally related to MHC class I antigens. Nature. 1989 Jan 12;337(6203):184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- Watkins D. I., Chen Z. W., Hughes A. L., Evans M. G., Tedder T. F., Letvin N. L. Evolution of the MHC class I genes of a New World primate from ancestral homologues of human non-classical genes. Nature. 1990 Jul 5;346(6279):60–63. doi: 10.1038/346060a0. [DOI] [PubMed] [Google Scholar]
- Wheeler C. J., Maloney D., Fogel S., Goodenow R. S. Microconversion between murine H-2 genes integrated into yeast. Nature. 1990 Sep 13;347(6289):192–194. doi: 10.1038/347192a0. [DOI] [PubMed] [Google Scholar]