Summary
The tandem bromodomains (BD1 and BD2) of the BET family proteins BRD2, BRD3, BRD4 and BRDT, are structurally conserved but not functionally equivalent. In this issue of Chemistry & Biology, Zhou and colleagues report that a BD1-specific chemical inhibitor, Olinone, enhances oligodendrocyte differentiation, contrasting the reverse process triggered by broad BD1/BD2-targeting inhibitors, highlighting distinct roles of BD1 and BD2 in cell fate decision.
The amino acid sequences and three-dimensional structures of the tandem acetyl-lysine (Kac)-binding bromodomains (BD1 and BD2) present in the bromodomain and extraterminal (ET) domain-containing BET family proteins are evolutionarily conserved (Wu and Chiang, 2007; Filippakopoulos et al., 2012). Nevertheless, the finding of two additional conserved regions with one containing the N-terminal cluster of casein kinase II phosphorylation sites (NPS) and one harboring basic residues-enriched interaction domain (BID) situated downstream of the BD2 in all the BET family proteins (Figure 1A) predicts that BD1 and BD2 are differentially regulated by posttranslational modification that in turn recruits different partner proteins to regulate BET protein function in diverse cellular processes (Wu et al., 2013).
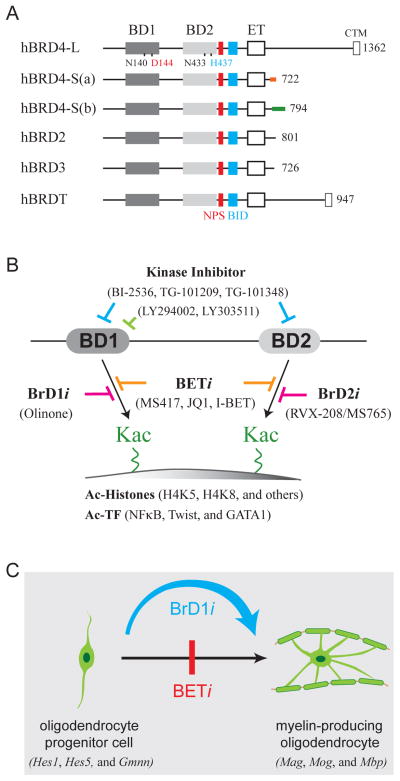
Figure 1. Differential Effects of BET Inhibitors on Oligodendrocyte Differentiation.
(A) Human BET family proteins. The evolutionarily conserved regions, including bromodomain I (BD1), bromodomain II (BD2), extraterminal domain (ET), C-terminal motif (CTM), N-terminal cluster of CK2 phosphorylation sites (NPS), and basic residues-enriched interaction domain (BID) are schematized with conserved N140 and N433 as well as diverged D144 and H437 residues between BD1 and BD2 also indicated. The full-length long (L) and two short (S) forms (a and b) of human BRD4 with common amino acids 1–719 but different 3′ regions generated by alternative splicing are also shown.
(B) Chemical inhibitors targeting BD1 and/or BD2 of BET proteins. BrD1i and BrD2i are respective BD1-specific and BD2-specific inhibitors, whereas BETi represents broad inhibitors binding to both BD1 and BD2. Acetylated (Ac) histones and transcription factors (TF) binding to BD1 and/or BD2 are indicated on the bottom.
(C) BrD1i promotes, but BETi inhibits, oligodendrocyte differentiation. Representative genes expressed in oligodendrocyte progenitor cells and myelin-producing cells are indicated in respective cells in parenthesis.
Using a structure-guided design to synthesize and screen for small compounds binding selectively to the BD1 of bromodomain-containing protein 4 (BRD4), Zhou and colleagues identified a high-affinity compound containing an acetyl moiety linked by four methylene groups to a tricyclic tetrahydro-pyrido indole scaffold that exhibits over 100-fold higher binding affinity to BD1 (Kd ~3.4 μM) than BD2 (Kd > 300 μM). This compound, named Olinone, shows comparable binding affinity to BRD2-BD1 and BRD3-BD1, and equal reluctance of association with the second bromodomains of the other BET proteins (Gacias et al., 2014). Olinone occupies the hydrophobic Kac-binding pocket in a configuration and surface contact, similar to that seen with a diacetyl histone H4 lysine 5 (K5) and lysine 8 (K8) peptide binding to BRD4-BD1, as revealed by X-ray crystallography at 0.94 Å. Although the asparagine 140 (N140) residue of BD1 crucial for contacting Kac and Olinone is likewise present in BD2 (N433, Figure 1A), a nonconserved histidine residue (H437) at BD2 poses a steric clash for Olinone contact complementarily mediated by aspartate 144 (D144) at the comparable position in BRD4-BD1. This structural insight elegantly explains the binding selectivity of Olinone for the BD1 of BET proteins. Conversely, H437 provides crucial contacts with K73/K76-diacetylated Twist transcription factor that recruits BRD4 via BD2-specific association to enhance epithelial-to-mesenchymal transition (EMT) in basal-like breast cancer cells and conversion of D144 to histidine allows BD1 interaction with acetylated Twist (Shi et al., 2014), further highlighting the importance of BD1-specific D144 and BD2-specific H437 for bromodomain selectivity in drug design and recruitment of transcription factors. Other than Twist, the structural similarity for BET bromodomain binding to acetylated histone tails and acetylated nonhistone proteins has also been illustrated with BRD3-BD1-specific contact with K312/K315-diacetylated hematopoietic transcription factor GATA1 (Gamsjaeger et al., 2011) and individual BD1 and BD2 of BRD4 association with the K310-acetylated RelA subunit of inflammatory transcription factor NFkB (Zou et al., 2014).
The availability of BD1-selective chemical inhibitor (BrD1i) Olinone, distinct from the previously characterized broad BET inhibitors (BETi) MS417, JQ1 and I-BET that target both BD1 and BD2, and a BD2-selective inhibitor (BrD2i) MS765/RVX-208 (Figure 1B) allows the use of these pharmacological agents to address the target selectivity and functional significance of BD1 and BD2. Using mouse oligodendrocyte progenitor cells (OPCs) that can differentiate into myelin-producing oligodendrocytes, Zhou and colleagues found that treating OPCs with Olinone promotes oligodendrocyte differentiation as reflected by enhanced myelin-specific Mag, Mog, and Mbp gene expression, accompanied by reduced progenitor Hes1, Hes5, and Gmnn marker expression; but surprisingly, treating oligodendrocytes with broad BET inhibitors such as MS417 that target both BD1 and BD2 actually hinders differentiation (Figure 1C). This observation was further confirmed via the use of additional bromodomain-selective BET inhibitors, including MS611 BrD1i and RVX-208/MS765 BrD2i (Figure 1B). Enhanced myelin formation by BrD1i, but not BrD2i and BETi, highlights the need to develop more selective bromodomain inhibitors to enrich our molecular understanding of BD1- and BD2-specific function in gene targeting and disease treatment. It would be interesting to determine whether oligodendrocyte lineage gene expression is indeed regulated by BRD2 that is predominantly expressed in these cells and whether BRD4 and BRD3 could independently or collaboratively regulate progenitor and differentiated oligodendrocyte gene expression with BRD2. The existence of other evolutionarily conserved regions (e.g., ET, NPS, and BID) that regulate chromatin binding and partner association of the BET family proteins also predicts new drug development targeting other functionally important regions of the BET proteins. The recent finding that many protein kinase inhibitors targeting PLK1 (e.g., BI-2536), JAK2 (e.g., TG-101209, and TG-101348), PI3K (e.g., LY294002, and LY303511), and other kinases also exhibit strong binding affinity to both BD1 and BD2 or specifically to BD1 (Ciceri et al., 2014; Dittmann et al., 2014; Ember et al., 2014; and see Figure 1B) raises not only interest in developing dual kinase/BET inhibitors for cancer therapeutics but also concerns of off-target effects that require further mechanistic studies of drug action in various biological systems.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ciceri P, Müller S, O’Mahony A, Fedorov O, Filippakopoulos P, Hunt JP, Lasater EA, Pallares G, Picaud S, Wells C, Martin S, Wodicka LM, Shah NP, Treiber DK, Knapp S. Nat Chem Biol. 2014;10:305–312. doi: 10.1038/nchembio.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann A, Werner T, Chung CW, Savitski MM, Savitski MF, Grandi P, Hopf C, Lindon M, Neubauer G, Prinjha RK, Bantscheff M, Drewes G. ACS Chem Biol. 2014;9:495–502. doi: 10.1021/cb400789e. [DOI] [PubMed] [Google Scholar]
- Ember SW, Zhu JY, Olesen SH, Martin MP, Becker A, Berndt N, Georg GI, Schönbrunn E. ACS Chem Biol. 2014;9:1160–1171. doi: 10.1021/cb500072z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacias M, Gerona-Navarro G, Plotnikov AN, Zhang G, Zeng L, Kaur J, Moy G, Rusinova E, Rodriguez Y, Matikainen B, Vincek A, Joshua J, Casaccia P, Zhou M-M. Chem Biol. 2014;21 doi: 10.1016/j.chembiol.2014.05.009. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, Mackay JP. Mol Cell Biol. 2011;31:2632–2640. doi: 10.1128/MCB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, Rusinova E, Zhang G, Wang C, Zhu H, Yao J, Zeng YX, Evers BM, Zhou MM, Zhou BP. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, Nair S, Chen LF. Oncogene. 2014;33:2395–2404. doi: 10.1038/onc.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]