Abstract
In todays' environment, it is becoming increasingly difficult to ignore the role of cancer in social health. Although a huge budget is allocated on cancer research every year, cancer remains the second global cause of death. And, exclusively, less than 50% of patients afflicted with advanced cancer live one year subsequent to standard cancer treatments. RNA interference (RNAi) is a mechanism for gene silencing. Such mechanism possesses uncanny ability in targeting cancer-related genes. A majority of gene products involved in tumorigenesis have recently been utilized as targets in RNAi based therapy. The evidence from these studies indicates that RNAi application for targeting functional carcinogenic molecules, tumor resistance to chemotherapy and radiotherapy is required in today's cancer treatment. Knock downing of gene products by RNAi technology exerts antiproliferative and proapoptotic effects upon cell culture systems, animal models and in clinical trials in the most studies. The recognition of RNAi mechanism and the progress in this field leaded several new RNAi-based drugs to Clinical Trial phases. This has also developed genome based personalized cancer therapeutics. Hopefully, this type of treatment will work as one of the efficient one for cancer patients.
Keywords: RNAi, Cancer, Treatment
Introduction
RNAi, which is commonly understood as RNA interference, refers to a member of non-coding RNA (ncRNA). The term non-coding RNA (ncRNA) is used for RNA that are not translated into protein; however, this does not mean that non-coding RNA delivers no performance.1 New evidence suggests that a majority of the mammalian genome is transcribed into ncRNA and exclusively 2% of it is transcribed into mRNA and translated into protein.2-10 RNA sequencing studies showed that the origin of ncRNAs is in the transcript antisense protein-coding genes, bidirectional promoter transcripts, enhancer and repeated sequences areas transcription, Intronic transcripts.11 Viruses and other double-stranded RNA microorganisms insert their genome into their host cells or artificially synthesize double-stranded RNA.12-14 Non-coding RNA is divided into two groups: 1- Small regulatory RNA and 2- Long non-coding RNA (Table 1). RNA interference (RNAi) is part of a small regulatory RNA, including siRNA and miRNA.15 The discovery of RNA interference molecules indebted to Mr. Fire's and Mello's research into the C.elegans in 1998.12 The advances in RNAi were made in following years leading eventually to the Physiology and Medicine Nobel Prize for Fire and Mello in 2006. Specific gene expression silencing by RNAi is a mechanism of transcriptional regulation in the eukaryotic cell. This is mediated by small RNA with 21-23 nucleotides length called siRNA and is conserved in terms of evolution among eukaryotes. RNAi seems to protect against not only exogenous genes such as microbial organisms genes including viral, bacterial genes, but also endogenous genes such as transposons. The other roles of these molecules in cells involving gene expression regulation and cell growth control have been demonstrated.16-19 On this regard, 3 mechanisms have been identified embodying: 1) heterochromatin formation changes 2) Inhibition of translation of target mRNA 3) Degradation of the target mRNA. The second and third mechanisms are more divulged. Cancer is one of the main targets for RNAi-based therapy. Several studies conducted invivo and invitro showed that RNAi- based therapy can be used for treating single-gene disorders and those with overexpression of proteins.20 There are different types of small synthetic RNA used in cancer therapy, that is, siRNA, shRNA and bishRNA. Such cancer therapy outweighs the others due to the silencing mechanism, specificity and lack of side effects.21
Table 1. Various classes of ncRNA in mammalian.16-19,27-42 .
| Established ncRNA classes | Common abbreviation | Size in nucleotides | Characteristics |
| Long non-coding RNAs | lncRNAs | more than 200 nucleotides | The widest class, action as sequence-specific tethers for protein complexes, epigenetic regulation performance and determination of the cell compartments and localization. |
| Small interfering RNAs | siRNAs | 21–22 nucleotides | produced by Dicer cleavage of 200-500 nucleotides length dsRNA duplexes, siRNAs form complexes with RISC involved in gene silencing, viral defense and transposon control. |
| microRNAs | miRNAs | 22 nucleotides | produced by Dicer and drosha cleavage of imperfect RNA hairpins encoded in long primary transcripts. Associated with RISC and primarily involved in post-transcriptional gene regulation. |
| PIWI-interacting RNAs | piRNAs | 26–30 nucleotides | Dicer-independent small RNAs, restricted to the somatic cells and germline bordering the germline, associated with PIWI and Argonaute proteins to regulation of transposon activity and chromatin state. |
| Promoter-associated RNAs | PARs | - | Inclusion of a set of long and short RNAs, promoter-associated RNAs (PASRs) and transcription initiation RNAs (tiRNAs) that overlap promoters and TSSs, which may regulate gene expression. |
| Small nucleolar RNAs | snoRNAs | - | This guide rRNA methylation and pseudouridylation, and gene-regulatory roles in some studies. |
| Small nuclear RNAs | snRNAs | approximately 150 nucleotides | Known as a U-RNA, the primary function of the processing of pre-mRNA in the nucleus, regulation of transcription factors, RNA polymerase II and maintaining the telomeres. |
| X-inactivation RNAs | xiRNAs | - | Dicer-dependent small RNAs processed from Xist and Tsix the two lncRNAs, responsible for X-chromosome inactivation in placental mammals. |
| Sno-derived RNAs | sdRNAs | - | Small RNAs, being Dicer-dependent of some of them, and processed from small nucleolar RNAs (snoRNAs), action as miRNA-like and translation regulation by some of them. |
| microRNA-offset RNAs | moRNAs | 20 nucleotides | Small RNAs, derived from the regions related to miRNA , unknown function. |
| tRNA-derived RNAs | - | - | Small RNAs processed by a conserved RNase (angiogenin) from tRNAs, induction of translational repression. |
| MSY2-associated RNAs | MSY-RNAs | 26-30 nucleotides | associated with the germ cell-specific DNA/RNA binding protein MSY2. restricted to the germline, unknown function. |
| Telomere small RNAs | tel-sRNAs | 22 nucleotides | Dicer-independent RNAs, derived from the G-rich sequence of telomeric repeats. a role in telomere maintenance. |
| Centrosome-associated RNAs | crasiRNAs | 34-42 nucleotides | Small RNAs, derived from centrosomes, a role in local chromatin modifications. |
Mechanisms of gene silencing by siRNA
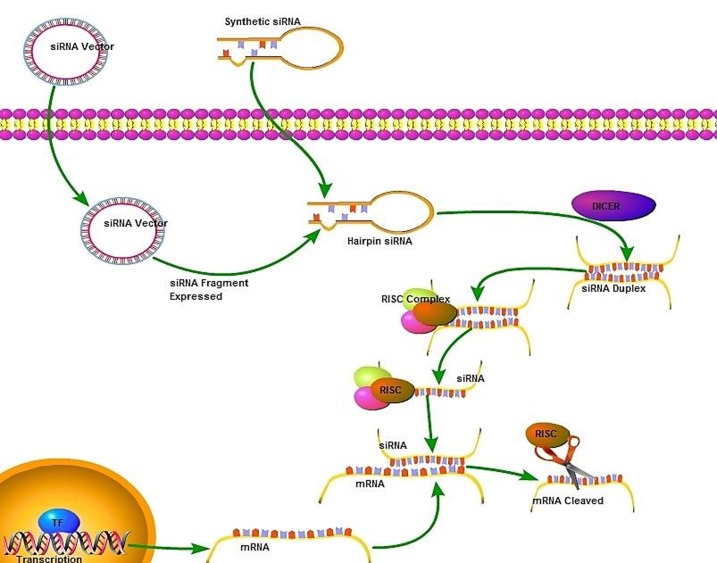
SiRNA is produced in two stages, that is, starting and effecting stages. In the starting stage, a long double-stranded RNA (500-200 bp) is cleaved into fragments with a length of 23-21 nucleotides by Dicer and siRNA is produced. SiRNA is an functional molecule. The double-stranded RNA can originate from the viral genome, bacterial DNA or synthetic RNA produced by using bioinformatic data (Figure 1).22,23 In the effecting stage, double-stranded siRNA are separated by helicase and then sense strand is demarcated by endogenous endonucleases and antisense strand is directed to the RNA induced silencing complex (RISC). This complex is then directed to the target mRNA. Argonate, as a member of RISC, by its ribonuclease activity from piwi region degrades the target mRNA. Gene expression terminates as long as target mRNA is broken by RISC. There are two pathways through which mRNA is broken. First, they might be broken by ribonuclease. Second, they can be connected to the homologous strand and RNA polymerase forms double stranded RNA leading to the continued interference pathway (Figure 1). By degradation of mRNA, expression of the target gene is suppressed, which is known as post-transcriptional gene silencing (PTGS).22,23 After the discovery of the fact that siRNA targets gene expression unstably and transiently and finding the evidence that these molecules stimulate the innate immune response such as interferon, another post-transcriptional silencing technique managed by vector called shRNA was developed. This technique functions through frequently gene silencing after transfection into genome by vectors.24-26
Figure 1 .
Long double-stranded RNA is cleaved into small interfering RNAs (siRNAs) by Dicer. This reaction needs ATP. SiRNA is then directed to RISC. The separation of the two strands needs ATP. Antisense strand is then directed to RISC
Mechanisms of gene silencing by shRNA
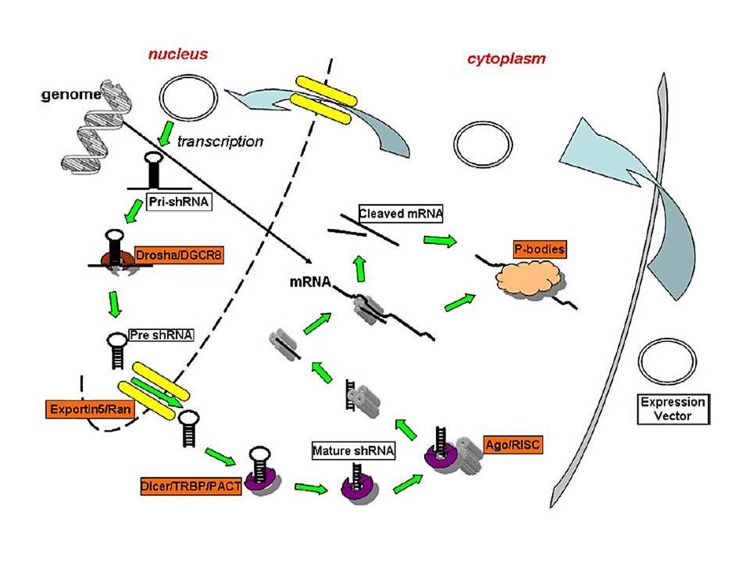
Unlike siRNA, shRNA is synthesized in the nucleus and then is transported into the cytoplasm for final processing; thereafter, it binds to RISC and performs its activities.43 ShRNA activity within the cell is shown schematically in Figure 2. Processing and maturation of shRNA is similar to those of miRNA.44 ShRNA is transcribed by RNA polymerase II or III and by the means of RNA polymerase II or III promoters on the expression cassette. Initial transcription of RNA polymerase II promoter is produced by stem-loop-like structure which is processed by complex containing RNaseIII family, Drosha and double-stranded RNA binding protein domain (DGCR8).45 ShRNA hairpin complex is processed by the abovementioned enzyme and makes individual shRNAs by 2 over hang nucleotide at the 3' terminal.46 This stage processes a molecule called pre-shRNA that is transported into cytoplasm by exportin5/RanGTPase.47,48 In the cytoplasm, pre-shRNA loaded onto RNaseIII complexes containing Dicer, TRBP and PACT and the loop is cleaved and double-stranded siRNA with two nucleotide over hang in the 3' is produced.48,49 The complex containing Dicer helps loading of siRNA onto the RISCs Argo2 protein. Pre-shRNA is a component of RLC; therefore, pre-shRNA is potentially associated with the RLC. After loaded onto RLC, the passenger strand becomes separated. This also occurs in siRNA. Argonaute family constitutes the major portions of RISC complex.50,51 from argonaute family, only Ago2 have an endonuclease activity, which is necessary for cleavage and double stranded stem passenger release.14,52,53 Other members of argonaute are Ago1, Ago3, Ago4, which do not have endonuclease activity, but enters RISC and act independent of the cleavage. RISC complex have two functions, that is to say, function independent of the cleavage and function dependent of the cleavage52 (Figure 2). Argonaute proteins of RISC complex not only are involved in loading onto siRNA and miRNA, but also in transcriptional and post-transcriptional silencing. Ago protein loading onto siRNA, passenger strand and miRNA are targeted on mRNA and results in of endonuclease activity of Ago2 and cleavage of target mRNA54,55 (Figure 2).Ago1, Ago3 and Ago4, which do not possess endonuclease activity; are located on 3' UTR of mRNA and inhibit translation by the processing target mRNA in P bodies.56,57 Details the suppression mechanisms of mRNA translation in the P bodies mRNA release from these bodies are still unknown. Adenylation of target mRNA causing mRNA instability occurs in P bodies. The Coimmunoprecipitation experiments have shown that the RISC complex is strongly associated with polyribosomes and small ribosomal subunits.58 Cleavage and conformation changes may be made in p bodies.31,59
Figure 2 .
Schematic diagram of the shRNA gene silencing pathway. ShRNA is entered the cytoplasm by using expressing vector. Vector is transferred to the nucleus for transcription. Initial transcription is processed by Drosha-DGCR8 complex. Pre-shRNA is generated from pri-shRNA. Pre-shRNA is transported into cytoplasm by Ran exportin 5. Pre-shRNA is loaded onto Dicer, TRBP, PACT complex. Mature shRNA is generated from Pre-shRNA. The mature shRNA is loaded onto Dicer, TRBP, PACT complex. The complex is joined to argounate protein in RISC, and provides RNA interference (41).
Similarities and differences between the siRNA and shRNA
The similarities between these two pathways exist in the function, which is involvement in post-transcriptional silencing. Both siRNA and shRNA use common molecules in their pathways including Dicer, RISC complex and their aim is degradation of target mRNA. However, there are significant differences shown in Table 2.21
Table 2. Differences between siRNA and shRNA21 .
| scale | siRNA | shRNA |
| Source | Synthetic | Nuclear expression |
| Delivery | Delivery to the cytoplasm using natural and synthetic polymers or lipids | Delivery to the nuclear using viral or plasmid vectors |
| Durability | Degradation of 99% of them in 46 hours | Expression up to 3 years |
| Usage | Limited local or systemic injection | local or systemic injection |
| Dose | High(nm) | Low(5 copies) |
| Possibility of Specific Off-target effects | More than shRNA | Lower than siRNA |
| Possibility of nonspecific Off-target effects | High stimulation of immune system, inflammation, and cytotoxicity | Low stimulation of immune system, inflammation, and cytotoxicity |
| Application | Acute diseases | Chronic disease |
Bifunctional shRNA (bishRNA)
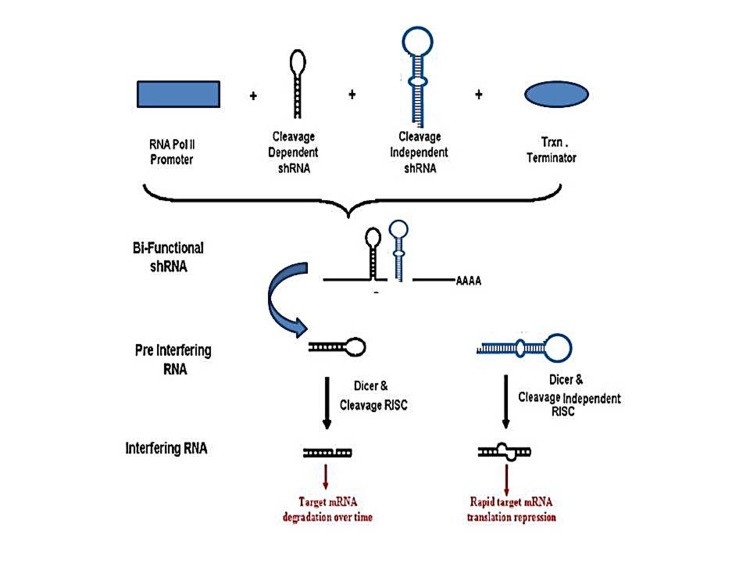
BishRNA, the newly designed RNA interference, is being increasingly used in the post-transcriptional silencing. BishRNA increases efficacy and durability of RNA interference and hastens gene expression silencing. Compared to shRNA, bishRNA is able to loaded onto various RISC complexes. Such property leads to target mRNA degradation and inhibits translation of the mRNA individually.21 BishRNA shows high efficiency and effectiveness because the leading (antisense) strand in bishRNA is loaded onto at least two RISC complexes, which increases the activity of gene silencing. Moreover, RISC complex loading activates both cleavage dependent and independent pathways, and causes target mRNA degradation and inhibits target mRNA translation21 (Figure 3).
Figure 3.
Schematic diagram regarding the bifunctional shRNA. It is designed from two shRNA for an target mRNA, including : 1) one with perfect match and 2) one with mismatches at the central location (bases 9–12). The aim is to activate both RISC dependent cleavage and RISC independent cleavage pathways (41).
The roles of RNA interference in cancer therapy
Effectiveness of RNA interference in cancer therapy Has been characterized by high efficiency and potential, induction of silencing in the advanced stages of growth, transmission of silenced gene to the next generation, low cost compared to the other methods of gene therapy60,61 and high specificity compared to the other methods of cancer therapy such as chemotherapy. As mentioned above, cancer is one of the main targets for RNAi-based therapy. Oncogenes, mutated tumor suppressor genes and several other genes involved in tumor progression are good targets for gene silencing by RNAi-based therapy due to the precise functional mechanism, high potential, and high specificity of gene silencing by RNAi, and lack side effects compared to chemotherapies. The major advantage of RNAi in cancer therapy is targeting multiple genes of various cellular pathways involved in tumor progression.62 Simultaneous inhibition of multiple genes is an effective approach to treat cancer as well as reduction of the possibility of multiple drugs’ resistance caused by overdose of chemical drugs. Another advantage of this type of treatment is developing suitable personalized drugs for a specific patient. Personalized drugs are likely to be more effective than others in controlling tumor growth.62 Many studies have been done in the field of RNAi-based drugs leading to introduction of several RNAi based drugs, which are now studied in the clinical trials. The studies conducted on animal models suggest that targeting essential proteins in the cell cycle, such as kinesin spindle protein (KSP) and polo-like kinase 1 (PLK1) by using specific siRNA have exhibited potent antitumor activity observed potent antitumor activity in both subcutaneous and hepatic tumor models.58 KSP inhibition causes cell cycle termination and apoptosis induction63 that is similar to PLK1 inhibition inducing apoptosis.62 Protein kinase N3 (PKN3) has been introduced as a valid therapeutic target in cancer. The inhibition of PKN3 reduces lymph node metastases in orthotopic prostate cancer models.64 Atu027 (siRNA-lipoplex targeted against PKN3) has been administrated to mice, rats and non-human primates. And, PKN3 expression silencing resulted in significant tumor growth and lymph node metastasis inhibition.63 Atu027 were used in animal models of metastatic lung cancer and the results showed the inhibition of metastasis after administration.63 In addition, Atu027 was evaluated in the animal model of breast cancer metastasis to the lung and the results showed inhibition of lung metastasis.65 A phase I clinical trial for treatment of solid cancers ended in late 2012 and the next phase of clinical trials has recently been started. Table 3 provides detailed information about the current status of RNAi-based drugs.66-68 CALAA-01 is another RNAi-based drug that is being evaluated in phase I clinical trial. It is a specific siRNA against transferrin encapsulated by non-chemical nanoparticles and targets the M2 subunit of ribonucleotide reductase (RRM2). The gene is involved in DNA replication. CALAA-01 binds to the transferrin receptor and siRNA releases RRM2-specific after endocytosis, leading eventually to inhibition of RRM2 expression and inhibition of proliferation of tumor cells expressing the transferrin receptor. In a phase I clinical trial of the drug, biopsies from patients with melanoma treated with the drug were collected and the results showed the nanoparticles inside biopsies and a reduction in RRM2 mRNA and RRM2 protein levels. At the end of phase I clinical trial, it was observed that administration of siRNA can systemically silence carcinogenic genes with specific targeting of tumor cells.69 ATN-RNA, a 160-bp double stranded RNA for targeting Tenasion-c, which is administered locally, is currently in phase I clinical trial.68 It is directly administrated into the neoplastic tissues during glioma operation. It was found that the survival of patients promoted from 48.2 weeks to 106.8 weeks after treatment. The authors also observed no neurological toxicity for this drug. FANGTM vaccine, designed by bishRNA technology, can be used for ex vivo silencing of Furin. Furin is a non-functional and calcium-dependent proprotein and is essential for proteolytic processing maturation of TGF-β isoform (β1, β2). FANGTM vaccine silences Furin and boosts GM-CSF. The administration of FANGTM vaccine into body causes 3 actions, that is, extensive presentation of antigens, Immune stimulation by GM-CSF, and inhibition of Immunosuppressive protein (TGF-β1, β2). The drug is still in clinical trial studies. In clinical trials, cancer cells are collected from body and GM-CSF/bishRNA furin is transfected into expression plasmid by Electroporation method.68 bishRNA-STMN1 is a RNAi-based drug, whose phase I clinical trial has recently been started, and targets Stathmin1. Stathmin1 is a 149-amino acid protein that is increased in tumor tissues and is involved in tubulin-microtubule compartmentalization. It plays a role in M-phase entrance and exit, and cell motility. SiRNA and ribozyme can target stathmin1 and it was shown that inhibition of stathmin1 by siRNA and ribozyme increases population of cells arrested in G2/M phase as well as inhibition of cell cloning and apoptosis induction.68
Table 3. Current Status of cancer siRNA based drug66-68 .
| Company | Drug | Target | Vehicle | Disease | Phase |
| Calando Pharmaceuticals | CALAA-01 | RRM2 | Cyclodextrin nanoparticle, TF, and PEG | Solid tumors | I |
| Silence Therapeutics AG | Atu027 | PKN3 | siRNA-lipoplex | Advanced solid cancer | I |
| Alnylam Pharmaceuticals | ALN-VSP02 | VEGF, KSP | SNALP | Solid tumors | I |
| Silenseed Ltd. | siG12D LODER | KRAS | LODER polymer | PDAC | I |
| Sataris Pharma and Enzon Pharmaceuticals | EZN-2968 | HIF-1, survivin | Nacked | Advanced solid tumor or lymphoma | I |
| Tekmira | SNALP-PLK1 | PLK1 | SNALP | Solid tumors | I |
| Duisburg University | Bcr-Abl siRNA | Bcr-Abl | Anionic liposome | CML | |
| Gradalis Inc. | FANG vaccine | Furin and GM-CSF | Electroporation | Solid tumors | I |
| Duke University | iPsiRNA | LMP2, LMP7, MECL1 | Transfection | Metastatic melanoma | I |
| Polish academy of sciences | ATN-RNA | Tenascin-c | Nacked | Astrocytic tumor | I |
KSP, kinesin spindle protein; PKN3, protein kinase N3; RRM2, M2 subunit of ribonucleotide reductase; VEGF, vascular endothelial growth factor; HIF-1, hypoxia-induced factor; KRAS, V-ki-ras2 Kirsten rat sarcoma viral oncogene homolog; PLK1, polo-like kinase 1; IND, investigational new drug. Bcr-Abl, breakpoint cluster region- Abelson; GM-CSF, Granulocyte-macrophage colony-stimulating factor; LMP, latent membrane protein; MECL1, Multicatalytic Endopeptidase Complex-Like 1
The future of cancer therapy by RNA interference
Information theory is a new strategic and helpful approach to cancer treatment.70 Clinical responses by siRNA-based drug were transient and often cause tumor recurrence or progression.71 Heterogeneity of the tumor, multiple signaling pathways, cross talks and vertical and horizontal feedback loops challenge the gene targeting in most solid tumors. Drug resistance caused by self-treatment is also another problem.72 In order to minimize drug resistance, at least three different treatments are suggested. With this strategy, an effective therapy by targeting triple factors reduces the risk of cancer.68 Now researchers are developing an integrated database in a participatory process for the identification of dominant signaling pathways and involved genes based on formulation of targeting three different factors. The next generation of RNAi-based therapies for cancer are based on individual treatments (Figure 4). The approach to cancer will be RNAi based on disruption of signaling pathways and developing of RNAi-based triple vaccine. This approach acts through damages to tumor growth by modifying the ratio of tumor/ T-cell activation, induction of apoptosis to increase antigen presenting, and reducing local and systemic immunosuppression associated with immunotherapy. The effect of these drugs are not limited by time and can provide long time memory T-cell responses.68
Figure 4 .
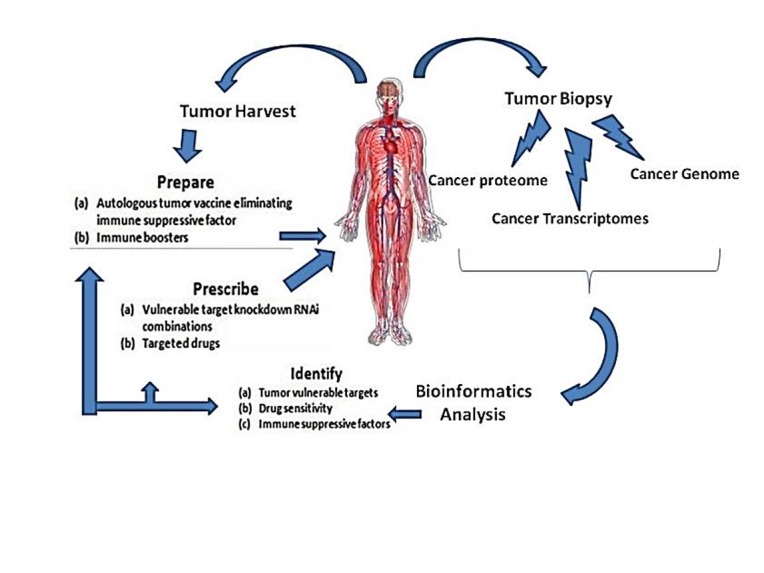
Schematic diagram of the mechanism implementing personalized drugs for cancer. The process is started by collecting a biopsy from the tumor, evaluating DNA, RNA and protein profiles. Tumor and normal tissue profiles are compared. By Using bioinformatic data, genetic abnormalities, impaired tumor-specific pathways are identified. Two-pronged approach (target therapy and vaccine) administered to each patient for having effective treatment (74).
Conclusion
Efficacy and clinical safety of several main components of RNAi technologies such as siRNA, shRNA and bishRNA have been demonstrated. In addition, specify to target and anticancer activity regarding RNAi technologies have been proven in animal models. To date, systemic delivery to tumor targets other than the liver has resulted in many problems; thus, researchers are now examining safe and effective delivery approaches of these molecules to their target tissues. The results of phase I clinical trial of several RNAi-based drugs are available and shows the safety of treatment. All in all, It is postulated that RNAi-based therapy will be used as a new method of cancer therapy in the near future.
Conflict of Interest
The authors report no conflicts of interest.
References
- 1.Gesteland RF, Cech TR, Atkins JF. The RNA World. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 2.Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature . 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N. et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J. et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet . 2006;38(6):626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S. et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science . 2005;308(5725):1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 6.Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK. et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods . 2008;5(7):613–9. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 7.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science . 2008;322(5909):1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet . 2005;21(2):93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science . 2007;316(5830):1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 10.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA. et al. Divergent transcription from active promoters. Science . 2008;322(5909):1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods . 2010;7(9):709–15. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature . 1998;391(6669):806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 13.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol . 2005;23(2):222–6. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 14.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell . 2005;123(4):607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 15.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell . 2003;115(2):209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 16.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet . 2006;22(5):268–80. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell . 2009;136(4):642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet . 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell . 2009;136(4):656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dykxhoorn DM, Lieberman J. Knocking down disease with siRNAs. Cell . 2006;126(2):231–5. doi: 10.1016/j.cell.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vsshRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61(9):746–59. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev . 2003;67(4):657–85. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet . 2005;6(1):24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 24.Cullen BR. Induction of stable RNA interference in mammalian cells. Gene Ther . 2006;13(6):503–8. doi: 10.1038/sj.gt.3302656. [DOI] [PubMed] [Google Scholar]
- 25.Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C. et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet . 2003;33(3):396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 26.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J. et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet . 2003;33(3):401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 27.Belostotsky D. Exosome complex and pervasive transcription in eukaryotic genomes. Curr Opin Cell Biol . 2009;21(3):352–8. doi: 10.1016/j.ceb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Bortoluzzi S, Biasiolo M, Bisognin A. MicroRNA-offset RNAs (moRNAs): by-product spectators or functional players? Trends Mol Med 2011;17(9):473-4. Trends Mol Med. 2011;17(9):473–4. doi: 10.1016/j.molmed.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W. et al. A human snoRNA with microRNA-like functions. Mol Cell . 2008;32(4):519–28. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Gerbi SA. Small nucleolar RNA. Biochem Cell Biol . 1995;73(11-12):845–58. doi: 10.1139/o95-092. [DOI] [PubMed] [Google Scholar]
- 31.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J . 2001;20(14):3617–22. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langenberger D, Bermudez-Santana C, Hertel J, Hoffmann S, Khaitovich P, Stadler PF. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics . 2009;25(18):2298–301. doi: 10.1093/bioinformatics/btp419. [DOI] [PubMed] [Google Scholar]
- 33.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet . 1999;21(4):400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 34.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol . 2007;8(3):209–20. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 35.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet . 2009;10(3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 36.Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science . 2008;320(5881):1336–41. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi W, Hendrix D, Levine M, Haley B. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat Struct Mol Biol . 2009;16(2):183–9. doi: 10.1038/nsmb.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA . 2009;15(7):1233–40. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle . 2009;8(15):2332–8. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 40.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell . 2009;138(2):215–9. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev . 2009;23(13):1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol . 2009;11(3):228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 43.Cullen BR. RNAi the natural way. Nat Genet . 2005;37(11):1163–5. doi: 10.1038/ng1105-1163. [DOI] [PubMed] [Google Scholar]
- 44.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J . 2005;24(1):138–48. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature . 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J . 2002;21(21):5875–85. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J . 2002;21(17):4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ. et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell . 2004;117(1):69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 49.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev . 2003;17(24):3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science . 2001;293(5532):1146–50. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 51.Sontheimer EJ, Carthew RW. Molecular biology. Argonaute journeys into the heart of RISC. Science. 2004;305(5689):1409–10. doi: 10.1126/science.1103076. [DOI] [PubMed] [Google Scholar]
- 52.Preall JB, Sontheimer EJ. RNAi: RISC gets loaded. Cell . 2005;123(4):543–5. doi: 10.1016/j.cell.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 53.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell . 2005;123(4):621–9. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 54.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science . 2002;297(5589):2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 55.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science . 2004;304(5670):594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 56.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A . 2005;102(47):16961–6. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature . 2007;447(7146):875–8. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 58.Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A. et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest . 2009;119(3):661–73. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell . 2007;25(5):635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 60.Yazdi Samadi B, Valizadeh M. Genetics A molecular approach. Iran: Tehran Univercity Publishers; 1388. [Google Scholar]
- 61.Fire AZ. Gene silencing by double-stranded RNA. Cell Death Differ . 2007;14(12):1998–2012. doi: 10.1038/sj.cdd.4402253. [DOI] [PubMed] [Google Scholar]
- 62.Bora RS, Gupta D, Mukkur TK, Saini KS. RNA interference therapeutics for cancer: challenges and opportunities (review) Mol Med Rep . 2012;6(1):9–15. doi: 10.3892/mmr.2012.871. [DOI] [PubMed] [Google Scholar]
- 63.Tao W, South VJ, Zhang Y, Davide JP, Farrell L, Kohl NE. et al. Induction of apoptosis by an inhibitor of the mitotic kinesin KSP requires both activation of the spindle assembly checkpoint and mitotic slippage. Cancer cell . 2005;8(1):49–59. doi: 10.1016/j.ccr.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 64.Leenders F, Mopert K, Schmiedeknecht A, Santel A, Czauderna F, Aleku M. et al. PKN3 is required for malignant prostate cell growth downstream of activated PI 3-kinase. EMBO J . 2004;23(16):3303–13. doi: 10.1038/sj.emboj.7600345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santel A, Aleku M, Roder N, Mopert K, Durieux B, Janke O. et al. Atu027 prevents pulmonary metastasis in experimental and spontaneous mouse metastasis models. Clin Cancer Res . 2010;16(22):5469–80. doi: 10.1158/1078-0432.CCR-10-1994. [DOI] [PubMed] [Google Scholar]
- 66.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol . 2012;19(1):60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J . 2011;6(9):1130–46. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao DD, Wang Z, Senzer N, Nemunaitis J. RNA interference and personalized cancer therapy. Discov Med . 2013;15(81):101–10. [PubMed] [Google Scholar]
- 69.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA. et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature . 2010;464(7291):1067–70. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paninski L. Estimation of entropy and mutual information. Neural Comput . 2003;15(6):1191–253. [Google Scholar]
- 71.Duxbury MS, Whang EE. RNA interference: a practical approach. J Surg Res . 2004;117(2):339–44. doi: 10.1016/j.jss.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 72.Kerbel RS. A cancer therapy resistant to resistance. Nature . 1997;390(6658):335–6. doi: 10.1038/36978. [DOI] [PubMed] [Google Scholar]