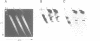Abstract
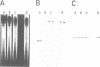
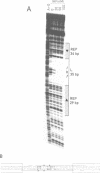
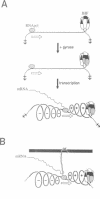
The prokaryotic REP (repetitive extragenic palindromes) or PU (palindromic units) sequences are often associated with other repetitive elements, forming arrangements which have been called 'BIMEs' (bacterial interspersed mosaic elements). It is estimated that the Escherichia coli chromosome carries approximately 300-500 BIMEs, whose biological role is at present unknown. We have identified a family of BIMEs consisting of two converging REP sequences flanking a 35 bp conserved segment which carries a static DNA bend and a binding site for IHF, the integration host factor of E.coli. We estimate that the E.coli genome harbors approximately 100 copies of this module, which we name 'RIB' (reiterative ihf BIME). We have analyzed by gel retardation and by footprinting the in vitro interaction of IHF with individual RIBs, and shown that the protein binds strongly and specifically to their center. We have also demonstrated binding of IHF to the chromosomal population of RIBs, using a new approach which combines two-dimensional bandshift and Southern blotting. RIB elements are at the end of transcription units, and thus define a new class of ihf sites. Possible implications for genome structure and DNA topology are discussed.
Full text
PDF
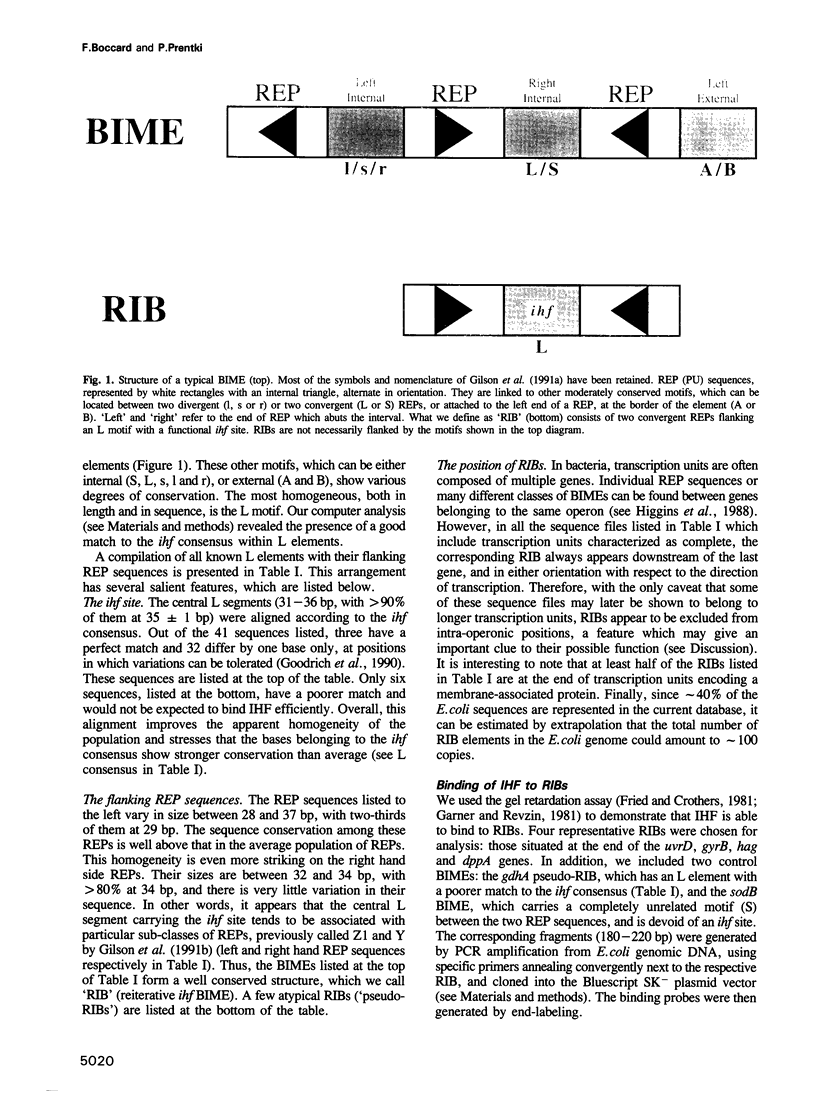




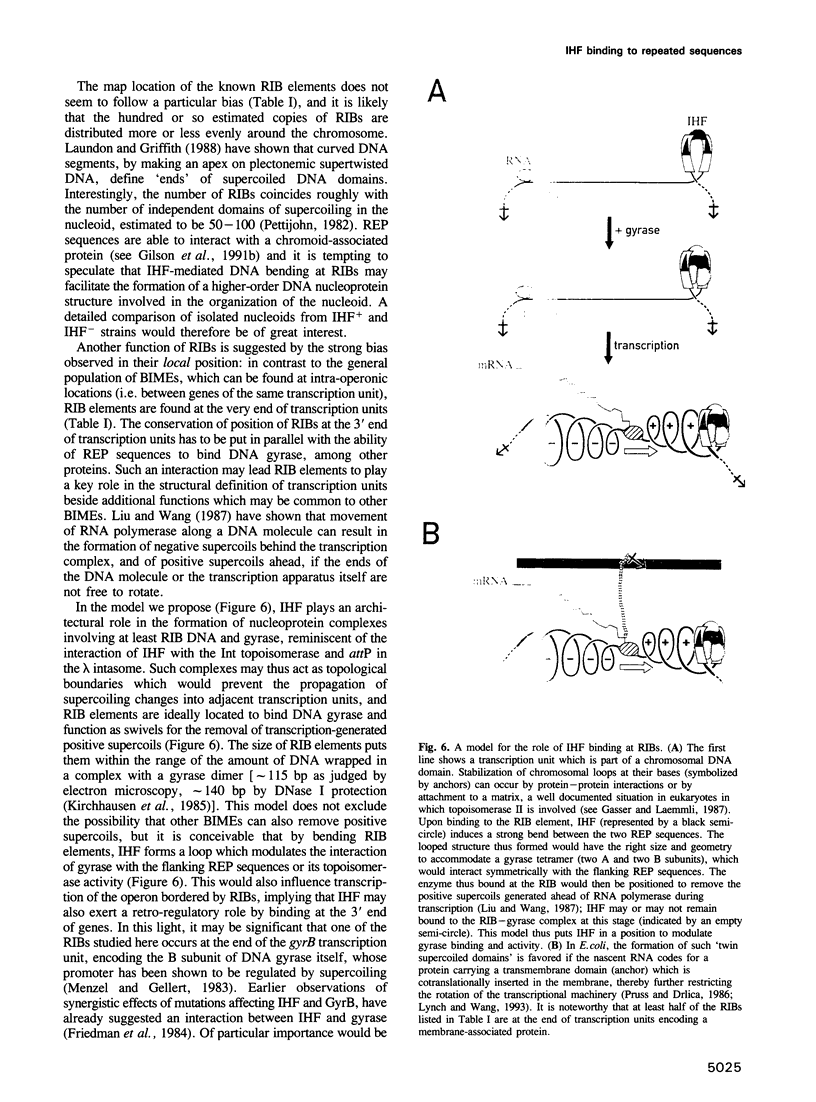


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong S. K., Pettis G. S., Forrester L. J., McIntosh M. A. The Escherichia coli enterobactin biosynthesis gene, entD: nucleotide sequence and membrane localization of its protein product. Mol Microbiol. 1989 Jun;3(6):757–766. doi: 10.1111/j.1365-2958.1989.tb00224.x. [DOI] [PubMed] [Google Scholar]
- Bachellier S., Perrin D., Hofnung M., Gilson E. Bacterial interspersed mosaic elements (BIMEs) are present in the genome of Klebsiella. Mol Microbiol. 1993 Feb;7(4):537–544. doi: 10.1111/j.1365-2958.1993.tb01144.x. [DOI] [PubMed] [Google Scholar]
- Boffini A., Prentki P. Identification of protein binding sites in genomic DNA by two-dimensional gel electrophoresis. Nucleic Acids Res. 1991 Apr 11;19(7):1369–1374. doi: 10.1093/nar/19.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987 Jan 12;15(1):247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G. P., Rudd K. E., Morgan M. K., Bayat H., Ames G. F. Physical mapping of repetitive extragenic palindromic sequences in Escherichia coli and phylogenetic distribution among Escherichia coli strains and other enteric bacteria. J Bacteriol. 1992 Jul;174(14):4583–4593. doi: 10.1128/jb.174.14.4583-4593.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Freundlich M., Ramani N., Mathew E., Sirko A., Tsui P. The role of integration host factor in gene expression in Escherichia coli. Mol Microbiol. 1992 Sep;6(18):2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. J., Carver D., Gellert M. Synergistic effect of himA and gyrB mutations: evidence that him functions control expression of ilv and xyl genes. J Bacteriol. 1984 Feb;157(2):484–489. doi: 10.1128/jb.157.2.484-489.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P., Chandler M. G., Prentki P., Galas D. J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987 May 20;195(2):261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Clément J. M., Brutlag D., Hofnung M. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 1984 Jun;3(6):1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Perrin D., Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990 Jul 11;18(13):3941–3952. doi: 10.1093/nar/18.13.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Saurin W., Perrin D., Bachellier S., Hofnung M. Palindromic units are part of a new bacterial interspersed mosaic element (BIME). Nucleic Acids Res. 1991 Apr 11;19(7):1375–1383. doi: 10.1093/nar/19.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Saurin W., Perrin D., Bachellier S., Hofnung M. The BIME family of bacterial highly repetitive sequences. Res Microbiol. 1991 Feb-Apr;142(2-3):217–222. doi: 10.1016/0923-2508(91)90033-7. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman N. D., Schneider T. D. High information conservation implies that at least three proteins bind independently to F plasmid incD repeats. J Bacteriol. 1992 Jun;174(11):3558–3560. doi: 10.1128/jb.174.11.3558-3560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Ames G. F., Barnes W. M., Clement J. M., Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982 Aug 19;298(5876):760–762. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., McLaren R. S., Newbury S. F. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene. 1988 Dec 10;72(1-2):3–14. doi: 10.1016/0378-1119(88)90122-9. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. The bacteriophage lambda int gene product. A filter assay for genetic recombination, purification of int, and specific binding to DNA. J Biol Chem. 1978 Oct 25;253(20):7149–7157. [PubMed] [Google Scholar]
- Kirchhausen T., Wang J. C., Harrison S. C. DNA gyrase and its complexes with DNA: direct observation by electron microscopy. Cell. 1985 Jul;41(3):933–943. doi: 10.1016/s0092-8674(85)80074-x. [DOI] [PubMed] [Google Scholar]
- Kosturko L. D., Daub E., Murialdo H. The interaction of E. coli integration host factor and lambda cos DNA: multiple complex formation and protein-induced bending. Nucleic Acids Res. 1989 Jan 11;17(1):317–334. doi: 10.1093/nar/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D., Prentki P., Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992 Dec;56(4):509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laundon C. H., Griffith J. D. Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell. 1988 Feb 26;52(4):545–549. doi: 10.1016/0092-8674(88)90467-9. [DOI] [PubMed] [Google Scholar]
- Lee E. C., MacWilliams M. P., Gumport R. I., Gardner J. F. Genetic analysis of Escherichia coli integration host factor interactions with its bacteriophage lambda H' recognition site. J Bacteriol. 1991 Jan;173(2):609–617. doi: 10.1128/jb.173.2.609-617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski J. R., Weinstock G. M. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992 Jul;174(14):4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch A. S., Wang J. C. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J Bacteriol. 1993 Mar;175(6):1645–1655. doi: 10.1128/jb.175.6.1645-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Mizuno T. Random cloning of bent DNA segments from Escherichia coli chromosome and primary characterization of their structures. Nucleic Acids Res. 1987 Sep 11;15(17):6827–6841. doi: 10.1093/nar/15.17.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G., Kavanagh T. Speeding-up the sequencing of double-stranded DNA. Nucleic Acids Res. 1988 Jun 10;16(11):5198–5198. doi: 10.1093/nar/16.11.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn D. E. Structure and properties of the bacterial nucleoid. Cell. 1982 Oct;30(3):667–669. doi: 10.1016/0092-8674(82)90269-0. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Pham M. H., Galas D. J. Plasmid permutation vectors to monitor DNA bending. Nucleic Acids Res. 1987 Dec 10;15(23):10060–10060. doi: 10.1093/nar/15.23.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Pham M. H., Gamas P., Chandler M., Galas D. J. Artificial transposable elements in the study of the ends of IS1. Gene. 1987;61(1):91–101. doi: 10.1016/0378-1119(87)90368-4. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ames G. F. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]