Abstract
Background:
Uropathogenic Escherichia coli (UPEC) can cause urinary tract infection (UTI). To prevent urine flow lavage, UPEC has acquired several virulence factors called adhesins. These adhesins are expressed and controlled by different genes.
Aim:
This study was aimed to determine some of the most important genes that control virulence factors of UPEC (pyelonephritis associated pili [pap], S fimbrial adhesion [sfa] and A fimbrial adhesion [afa] genes), which code for adhesins and phenotypic factors.
Materials and Methods:
In total, 205 UPEC isolates from in- and out-patients with UTI were obtained. Polymerase chain reaction was used for gene amplification. One drop of bacterial suspension, one of red blood cells and one of peripheral blood smear were mixed for hemagglutination (HA). Formation of a clump was considered to be positive. Bacteria were grown on blood agar to determine hemolysis. Surface hydrophobicity was determined using the SAT test.
Result:
Frequencies of pap, afa and sfa were 42 (20.5%), 17 (8.3%) and 44 (21.5%), respectively. Frequencies of HA, hemolysis and hydrophobicity were 138 (67.3%), 56 (27.3%) and 39 (19%), respectively. Among HA-positive bacteria, 103 (74.6%) were mannose resistant. Our results highlight higher frequency of HA than that of other virulence factors, indicating a crucial role of this virulence factor in UPEC.
Discussion:
We concluded that major differences exist in the prevalence of virulence factors among different UPEC isolated from different countries. The association observed between pathogenicity and virulence factors may promote UPEC survival and growth within the urinary tract. Detecting these genes as the primary controllers of UPEC virulence factors may aid in better management of related infections.
Keywords: Hemagglutination, hemolysin, hydrophobicity, uropathogenic Escherichia coli, virulence factors
INTRODUCTION
Urinary tract infection (UTI) is a prevalent infectious condition with potentially severe complications, thus representing a major cause of human discomfort.[1] UTIs are one of the most frequently encountered infections in developing countries as well as one of the most common medical problems. It is estimated that 150 million patients are diagnosed with UTI every year, which result in at least $6 billion in health care expenditures.[2] Clinical manifestations of UTI are highly variable in extent and severity, with cystitis generally resolving without sequela, whereas pyelonephritis can cause serious morbidity and fatality.[3]
A broad spectrum of bacteria can cause UTI, such as Klebsiella, Proteus, Enterobacter, Pseudomonas, Serratia and Citrobacter species. Escherichia coli is by far the most common cause of UTI, accounting for 80-90% of all UTIs among ambulatory populations.[4] E. coli isolates are divided to intestinal, extra intestinal and commensal pathogens[5] and the varitypes causing UTI are called uropathogenic E. coli (UPEC). UPEC is the most common etiological agent of community-acquired UTIs, accounting for more than 80% of these infections.[6] The severity of the infection depends on both the virulence of the infecting bacteria and the susceptibility of the host. UPEC adhesions to the uroepithelium may protect the bacteria from urinary lavage, thus increasing their ability to multiply and invade the renal tissue. UPEC strains encode several virulence factors, which enable the bacteria to colonize the urinary tract and persist in the presence of highly effective host defense. UPEC possess adherence factors called pilus or fimbriae, which allow them to successfully initiate infections. The 2 primary pili in UPEC are type 1 and P. Moreover, specific adhesion is mediated by bacterial proteins called adhesions, which may or may not be associated with fimbriae, e.g., pyelonephritis associated pili (pap), S fimbrial adhesion (sfa) and A fimbrial adhesion (afa).[7] In the late 1970s, it was recognized for the first time that UTI-causing E. coli strains typically agglutinate human erythrocytes despite the presence of mannose (mannose-resistant hemagglutination [MRHA]) and adhere to human uroepithelial cells. Moreover, adherence to uroepithelial cells is usually unaffected by mannose (mannose-resistant adherence) and is more common among strains exhibiting MRHA than among those exhibiting only mannose-sensitive hemagglutination (HA)[8] HA preparations are cytotoxic toward tissue culture cells.[9] The expected correlation between hemolysin and necrotoxin production is associated with the production of a “liquid” hemolysin, which is cytotoxic.[10] A close association has been observed between hemolysin production and cytotoxicity in 1 strain of E. coli (serotype 06, the standard hemolytic strain). The absence of strains that produce a “liquid” but not a “solid” hemolysin in UTIs indicates the importance of the production of non-diffusible hemolysins.[10] Recent studies have shown that HA and erythrocyte hemolysis can help bacteria to adhere to host cells and colonize them.[11] Cell surface hydrophobicity is one of the unspecific factors for bacteria to adhere to cell surfaces.[12] It is an important factor that helps E. coli adhere to various surfaces for colonization. Bacteria are lysed by normal serum due to the activity of the complement system. The alternate pathway of complement activation is potentially more important than the classical pathway. Bacterial resistance to killing by serum results from individual or combined effects of capsular polysaccharides, lipopolysaccharides and surface proteins.[13] Genes encoding urovirulence factors usually form gene clusters (also called as pathogenicity islands).[14]
Prevalence of virulent genes in UPEC vary according to the phylogenetic group, clinical conditions of host and geographical localization.[2] Given this generic virulence “arsenal” of UPEC, strains from different geographical regions show different disease severity and should be genetically different. Data on the genetic characteristics of UPEC are limited and UPEC are studied less in the west of Iran. Therefore, the present study was performed to determine some of the most important genes that control virulence factors in UPEC (afa, pap and sfa genes), with hemolysin, HA and hydrophobicity as the important phenotypic factors.
MATERIALS AND METHODS
Study population and strains
E. coli strains were isolated from urine samples of 71 outpatients and 134 inpatients referred to different hospitals in west Iran (Kermanshah city). Clinical presentation of patients having lower urinary tract syndrome, including chills, fever, dysuria, frequency and sometimes urgency and cost vertebral angle tenderness, were noted. Urine samples were collected from clean-catch midstream urine. E. coli isolates were then identified using routine biochemical reactions. Suspicious isolates were confirmed using API20E kit.[15]
Deoxyribonucleic acid extraction and polymerase chain reaction assay
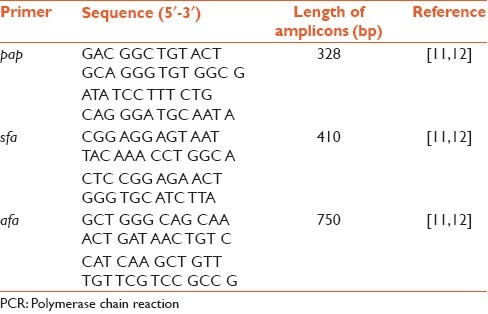
E. coli isolates were grown on Luria Bertani (LB) broth at 37°C overnight. DNA extraction was performed using the boiling method. To evaluate the purity of the extracted DNA, a spectrophotometer in ratio OD of 260/280 ≥ 1.7 was used. The quality of extracted DNA was controlled on 2% agarose gel. Amplification were performed in a thermal cycle C1000 (BioRad, Hercules, USA) under the following conditions. Multiplex PCR assays were performed in a final volume of 25 μL, containing DNA (5 μL boiling lysate), 1.5 mM MgCl2, 200 μM dNTP, 1.2 U Taq DNA polymerase (Fermentas), ×10 Taq DNA polymerase buffer and 0.45 μM primers. The reaction cycle included heating to 94°C for 10 min; 5 cycles of denaturation (94°C for 1 min), annealing (56°C for 1 min) and extension cycle (72°C for 4 min); and 25 cycles of denaturation (94°C for 40 s), annealing (61°C for 40 s) and extension (72°C for 3 min), followed by final extension (72°C for 5 min).[16,17] Expected lengths of the amplicons were ascertained by electrophoresis in 1% agarose gel. Bands were visualized in a GelDoc system (BioRad) under ultra violet light after ethidium bromide staining. Random samples of positive reaction were delivered to Fazabiotech Company for sequencing. Sequence analysis of PCR products was performed using a basic local alignment search tool search on the Genbank database [Table 1].
Table 1.
Primer sequence and predicted length of amplified PCR products
Hemolysin
Hemolysin test was performed according to the method described by Santo et al. In brief, bacteria were first cultured on LB broth and incubated for 16 h at 37°C. Next, a volume of 50 μl of bacterial suspension was inoculated in sheep blood agar and reincubated for 16 h at 37°C. Hemolysis production was verified by formation of a transparent halo around the spot.[18]
Surface hydrophobicity
In brief for this test, 50 μl of bacterial suspension was mixed with dilutions of 0.25, 0.5, 1, 1.4 and 2 M of ammonium sulfate. Isolates that formed colonies in 1.4 M or lower dilutions were considered as hydrophobic isolates. Sterile 96-well microplates were also used in this text.[19]
HA
Direct bacterial HA was performed for this test. In brief, bacteria were cultured on 1% nutrient broth and incubated at 37°C for 3-5 days to obtain the maximum production of fimbriae.[20]
Statistical analysis
Data entered in the Microsoft Access XP software were exported into the SPSS statistical software (version 16.0, SPSS, Inc., Chicago, IL), which was used for data analyzes. Categorical data were compared using a Chi-squared test. All P values were 2-sided, with a P < 0.05 considered to be statistically significant.
RESULTS
In total, 205 strains of E. coli were isolated from 42 (20.4%) male and 163 (79.5%) female with UTI. The age of patients ranged from 1 month to 107 years old (mean age, 31.88 ± 22.69 years) with the mean age for male 29.7 ± 40.9 and for female 19.9 ± 29.5. Of all the patients, 71 (34.6%) were out-patients and 134 (65.3%) were in-patients. Patients aged more than 40 years had more frequencies in both hospitals.
Virulence genotypes
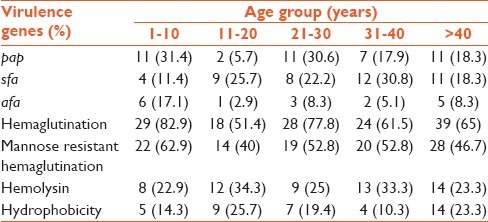
Virulence factors such as pap, afa and sfa were analyzed by the PCR assay. Products of the PCR assay are shown in Figure 1. Frequencies of pap, afa and sfa were 42 (20.5%), 17 (8.3%) and 44 (21.5%), respectively. The results showed that 110 (53.7%) isolates carried just 1 virulence factor, with 23 (11.2%) carrying pap, 10 (4.9%) carrying afa and 29 (14.1%) carrying sfa. 1 (0.5%) isolate carried all the 3 genes. However, no virulence factor was found in 123 (60%) isolates. There was a significant correlation between pap and the age of patients; the prevalence of pap was more common in patients aged 1-10 years (P < 0.048) [Table 2].
Figure 1.
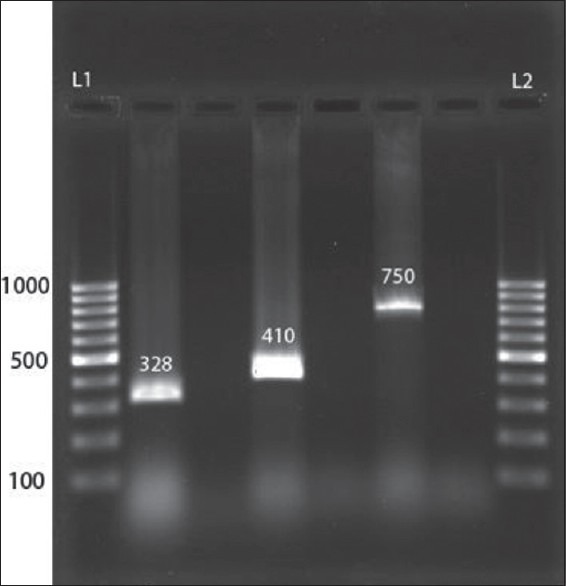
Polymerase chain reaction products shown on agarose gel. L1, deoxyribonucleic acid (DNA) size marker 100-bp; lane 2, pyelonephritis associated pili (328 bp), lane 4, S fimbrial adhesion (410 bp), lane 6, A fimbrial adhesion (750 bp); L2, DNA size marker 100-bp; and lanes 3, 5, 7, control negatives
Table 2.
Prevalence of virulence genes according to age groups
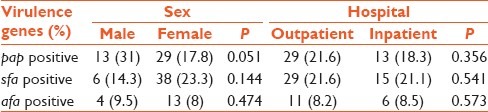
The result also showed correlation between the sex of patients and the pap gene, with the gene being more prevalent in male patients (P < 0.051) [Table 3].
Table 3.
Prevalence of virulence genes and their relation with sex and source of bacteria
Virulence phenotyping
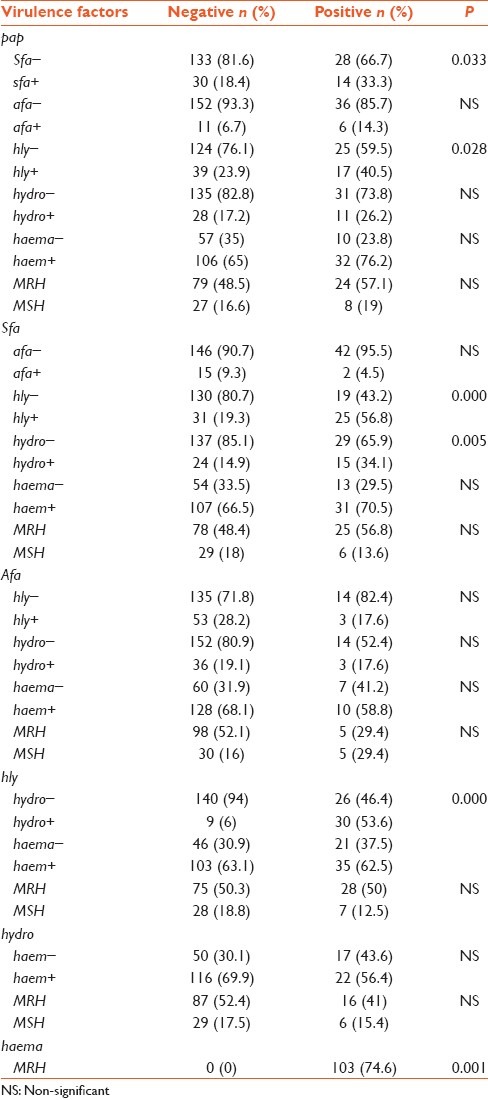
Frequencies of HA, hemolysin and hydrophobicity were 138 (67.3%), 56 (27.3%) and 39 (19%), respectively. Among isolates showing HA, 103 (74.6%) were mannose resistant. 98 (47.8%) isolates were positive for HA, 8 (3.9%) for hemolysin and 4 (2%) for hydrophobicity. Our data indicated that there was a significant correlation between hemolysin and outpatients (P < 0.024) [Table 4] and between HA and the age of patients; it was more common in patients aged 1-10 years and minimum in patients aged 10-20. Frequencies of isolates with genes pap, afa and sfa and phenotypic factors such as HA, hemolysin and hydrophobicity were 1 (0.5%) and 30 (14.6%) isolates carried none of the genes or phenotypic factors [Table 2]. Statistical significance of co-occurrence of 2 virulence factors was assessed using χ2 and Fisher's exact test. A significant difference was found for pap-sfa (P < 0.033), pap-hly (P < 0.028), sfa-hly (P < 0.000), sfa-hydro (P < 0.005), hly-hydro (P < 0.000) and hemo-MRH (P < 0.001); co-occurrence was tested for the same sample [Table 5].
Table 4.
Prevalence of virulence factors according to sex and hospital
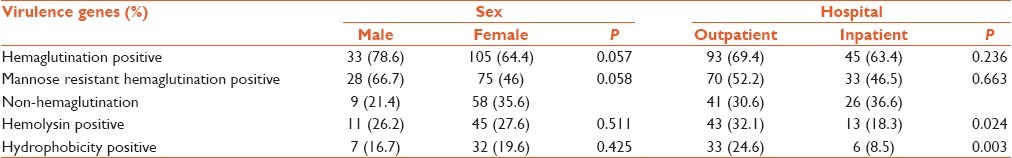
Table 5.
significant relations between six virulence factors
DISCUSSION
UPEC are a major cause of UTIs and may be responsible for approximately 90% of UTIs. In the present study, we found that 20.5%, 8.3% and 21.5% E. coli isolates from urine samples of in- and out-patients with UTI were positive for the pap, afa and sfa genes, respectively. Frequencies of isolates with HA, hemolysin and hydrophobicity were 67.3%, 27.3% and 19%, respectively. Various studies investigated the prevalence of virulence factors in E. coli isolates from patients with UTI. A study from Ankara, similar to our study, showed that almost 23% and 9.94% E. coli isolates were positive for pap and afa genes, respectively. However, there was a significant difference between the frequencies of sfa gene (6.21% versus 21.5%)[21] In both studies, a significant correlation was observed between pap and sfa genes. Bogylova in Slovaki, in the same investigation, reported that 8.3% extraintestinal E. coli isolates were positive for the afa gene, which is exactly same as our result. However, there is a significant difference between 2 other gene frequencies (74% and 65% for pap and sfa genes, respectively).[22] These differences may be due to the differences in geographical regions. In Iran (Shiraz), Farshad reported 30.2% and 18.75% frequencies for pap and sfa genes, respectively. In another study in Shiraz, Emamghoreishi reported a frequency of 14.6% for the sfa gene.[23] In our study, a significant difference was detected between age groups infected with pap-positive bacteria (P < 0.043). Patients aged 1-10 and 21-30 years were more infected with pap-positive E. coli (31.4% and 30.6%, respectively) [Table 2]. The pap gene encodes for P-fimbriae that allows bacteria to adhere to epithelial surfaces and protect them against urine lavage, thus allowing them to ascend further to even cause a serious disease. In the first decade of life, hygiene problems are the most important reason for bacterial infection, particularly in girls because of a short urethra (approximately 4 cm). The third decade of life is a time of sexual activity.
Previous studies have shown that UPEC are transmitted during sexual activity.[16,17] Our results highlight higher frequency of HA (67.3%) than other virulence factors, indicating a crucial role of virulence factors in UTI-causing E. coli. Therefore, it appears that virulence factors are the most important armament to adhere to epithelial surface and to break host defense system. The ability of bacteria to adhere to urinary epithelium can protect them from urinary lavage. HA is significantly correlated with MRH (P < 0.001). Prevalence of isolates that have HA was 67.3% either alone (47.8%) or in association with 2 other phenotypic factors (11.2%).
In total, 3.9% isolates were only positive for hemolysin and 2% were positive for hydrophobicity. Only 1 (0.5%) of the UPEC isolates was positive for all 6 virulence factors. These observations confirm the higher prevalence of HA among our isolates and probably the more significant role in pathogenicity in this geographical region. In Raksha's study on phenotypic virulence factors in E. coli in India, with prevalence of HA was 30.9% and hemolysin with 41.36%, have had the most frequency among phenotypic pathogenic factors.[24] Although HA is the most important factor in our isolates, it does not cause a serious disease leading to hospital admission. We found a significant difference between in- and out-patients infected with HA-positive isolates (18.3% in-patients versus 32.1% outpatients; P < 0.024). Therefore, it appears that in our area, HA has a significantly higher prevalence and perhaps plays a crucial role in the pathogenicity of our isolates.
Due to differences in social, economic, health, hygiene and environmental conditions between developed and developing countries, more studies are needed from developing countries to detect the virulence factors of UPEC to determine the pattern of pathogenicity in these bacteria. Furthermore, studies are needed to determine the relationship between the expression of virulence factors and antibiotic resistance. These studies will be important in understanding the role of these factors in causing UTIs, which in turn may lead to the development of universal vaccines to prevent such infections.
ACKNOWLEDGMENTS
This work was performed in partial fulfillment of the requirements for MD thesis in medicine (Hosna Khademi and Roya Ebrahimi). The authors would like to acknowledge Kermanshah University of Medical Sciences. The study was financially supported by the Kermanshah University of Medical Sciences for grant 88090. The authors also thank members of department of microbiology in Kermanshah University of Medical Sciences, Particularly Baharak Norozi and Zhila Rostami for their assistance with the PCR assessment in this study.
Footnotes
Source of Support: The study was financially supported by the Kermanshah University of Medical Sciences for grant 88090.
Conflict of Interest: None declared.
REFERENCES
- 1.Tanagho EA, McAninch JW. Smith's General Urology. 17th ed. New York: Lange Medical Books/McGraw-Hill; c2008. p. 193. [Google Scholar]
- 2.Oliveira FA, Paludo KS, Arend LN, Farah SM, Pedrosa FO, Souza EM, et al. Virulence characteristics and antimicrobial susceptibility of uropathogenic Escherichia coli strains. Genet Mol Res. 2011;10:4114–25. doi: 10.4238/2011.October.31.5. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Foxman B. Molecular epidemiology of Escherichia coli mediated urinary tract infections. Front Biosci. 2003;8:e235–44. doi: 10.2741/1007. [DOI] [PubMed] [Google Scholar]
- 4.Abe CM, Salvador FA, Falsetti IN, Vieira MA, Blanco J, Blanco JE, et al. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol Med Microbiol. 2008;52:397–406. doi: 10.1111/j.1574-695X.2008.00388.x. [DOI] [PubMed] [Google Scholar]
- 5.Mobley HL, Warren JW, editors. Urinary Tract Infections: molecular Pathogenesis and Clinical Management. Washington, DC: ASM Press; 1996. [Google Scholar]
- 6.Cheng CH, Tsau YK, Kuo CY, Su LH, Lin TY. Comparison of extended virulence genotypes for bacteria isolated from pediatric patients with urosepsis, acute pyelonephritis, and acute lobar nephronia. Pediatr Infect Dis J. 2010;29:736–40. doi: 10.1097/INF.0b013e3181dab249. [DOI] [PubMed] [Google Scholar]
- 7.Mulvey MA. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol. 2002;4:257–71. doi: 10.1046/j.1462-5822.2002.00193.x. [DOI] [PubMed] [Google Scholar]
- 8.Arthur M, Arbeit RD, Kim C, Beltran P, Crowe H, Steinbach S, et al. Restriction fragment length polymorphisms among uropathogenic Escherichia coli isolates: Pap-related sequences compared with rrn operons. Infect Immun. 1990;58:471–9. doi: 10.1128/iai.58.2.471-479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokurenko EV, Hasty DL, Dykhuizen DE. Pathoadaptive mutations: Gene loss and variation in bacterial pathogens. Trends Microbiol. 1999;7:191–5. doi: 10.1016/s0966-842x(99)01493-6. [DOI] [PubMed] [Google Scholar]
- 10.Struve C, Krogfelt KA. In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiology. 1999;145(Pt 10):2683–90. doi: 10.1099/00221287-145-10-2683. [DOI] [PubMed] [Google Scholar]
- 11.Starcic Erjavec M, Rijavec M, Krizan-Hergouth V, Fruth A, Zgur-Bertok D. Chloramphenicol- and tetracycline-resistant uropathogenic Escherichia coli (UPEC) exhibit reduced virulence potential. Int J Antimicrob Agents. 2007;30:436–42. doi: 10.1016/j.ijantimicag.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 12.Donnenberg MS, Welch RA. Urinary Tract Infections: molecular Pathogenesis and Clinical Management. Washington, DC: ASM Press; 1996. Virulence determinant of uropathogenic Escherichia coli; pp. 135–74. [Google Scholar]
- 13.Leffler H, Svanborg-Edén C. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect Immun. 1981;34:920–9. doi: 10.1128/iai.34.3.920-929.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–79. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 15.Beauchamp CJ, Simao-Beaunoir AM, Beaulieu C, Chalifour FP. Confirmation of E. coli among other thermotolerant coliform bacteria in paper mill effluents, wood chips screening rejects and paper sludges. Water Res. 2006;40:2452–62. doi: 10.1016/j.watres.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Foxman B, Manning SD, Tallman P, Bauer R, Zhang L, Koopman JS, et al. Uropathogenic Escherichia coli are more likely than commensal E. coli to be shared between heterosexual sex partners. Am J Epidemiol. 2002;156:1133–40. doi: 10.1093/aje/kwf159. [DOI] [PubMed] [Google Scholar]
- 17.Fergusson DM, McLeod GF, Horwood LJ. Childhood sexual abuse and adult developmental outcomes: Findings from a 30-year longitudinal study in New Zealand. Child Abuse Negl. 2013;37(9):664–74. doi: 10.1016/j.chiabu.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Santo E, Macedo C, Marin JM. Virulence factors of uropathogenic Escherichia coli from a university hospital in Ribeirão Preto, São Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2006;48:185–8. doi: 10.1590/s0036-46652006000400002. [DOI] [PubMed] [Google Scholar]
- 19.Wojnicz D, Jankowski S. Effects of subinhibitory concentrations of amikacin and ciprofloxacin on the hydrophobicity and adherence to epithelial cells of uropathogenic Escherichia coli strains. Int J Antimicrob Agents. 2007;29:700–4. doi: 10.1016/j.ijantimicag.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Vagarali MA, Karadesai SG, Patil CS, Metgud SC, Mutnal MB. Haemagglutination and siderophore production as the urovirulence markers of uropathogenic Escherichia coli. Indian J Med Microbiol. 2008;26:68–70. doi: 10.4103/0255-0857.38863. [DOI] [PubMed] [Google Scholar]
- 21.Arisoy M, Aysev D, Ekim M, Ozel D, Köse SK, Ozsoy ED, et al. Detection of virulence factors of Escherichia coli from children by multiplex polymerase chain reaction. Int J Clin Pract. 2006;60:170–3. doi: 10.1111/j.1742-1241.2005.00668.x. [DOI] [PubMed] [Google Scholar]
- 22.Bogyiová E, Kmetová M, Biros E, Siegfried L. Detection of pap-, sfa- and afa-specific DNA sequences in Escherichia coli strains isolated from extraintestinal material. Folia Microbiol (Praha) 2002;47:723–6. doi: 10.1007/BF02818678. [DOI] [PubMed] [Google Scholar]
- 23.Anvarinejad M, Farshad Sh, Ranjbar R, Giammanco GM, Alborzi A, Japoni A. Genotypic analysis of E. coli strains isolated from patients with cystitis and pyelonephritis. Iran Red Crescent Med J. 2012;14:408–16. [PMC free article] [PubMed] [Google Scholar]
- 24.Raksha R, Srinivasa H, Macaden RS. Occurrence and characterisation of uropathogenic Escherichia coli in urinary tract infections. Indian J Med Microbiol. 2003;21:102–7. [PubMed] [Google Scholar]


