Abstract
Uncovering how a new gene acquires its function and understanding how the function of a new gene influences existing genetic networks are important topics in evolutionary biology. Here, we demonstrate nonconservation for the embryonic functions of Drosophila Bonus and its newest vertebrate relative TIF1-γ/TRIM33. We showed previously that TIF1-γ/TRIM33 functions as an ubiquitin ligase for the Smad4 signal transducer and antagonizes the Bone Morphogenetic Protein (BMP) signaling network underlying vertebrate dorsal–ventral axis formation. Here, we show that Bonus functions as an agonist of the Decapentaplegic (Dpp) signaling network underlying dorsal–ventral axis formation in flies. The absence of conservation for the roles of Bonus and TIF1-γ/TRIM33 reveals a shift in the dorsal–ventral patterning networks of flies and mice, systems that were previously considered wholly conserved. The shift occurred when the new gene TIF1-γ/TRIM33 replaced the function of the ubiquitin ligase Nedd4L in the lineage leading to vertebrates. Evidence of this replacement is our demonstration that Nedd4 performs the function of TIF1-γ/TRIM33 in flies during dorsal–ventral axis formation. The replacement allowed vertebrate Nedd4L to acquire novel functions as a ubiquitin ligase of vertebrate-specific Smad proteins. Overall our data reveal that the architecture of the Dpp/BMP dorsal–ventral patterning network continued to evolve in the vertebrate lineage, after separation from flies, via the incorporation of new genes.
Keywords: Bonus/TIF1/TRIM, Dorsal/NF-κB, Dpp/BMP/TGF-β, dorsal–ventral axis, embryonic development, Drosophila
Introduction
The classic model for the origin of new genes and their acquisition of novel functions is based on gene duplication; one copy of a pair of newly duplicated genes maintains the original function whereas the other copy accumulates mutations. Accumulation of deleterious mutations leads to a loss of function whereas accumulation of mutations conferring a selective advantage leads to a novel function and phenotypic evolution (Haldane 1933). The first empirical evidence for this model of new gene origination came from studies revealing that a duplication of the Bar locus effected eye phenotypes in Drosophila (Muller 1936).
Several recent studies have advanced our understanding of new gene origins, their influence on genetic systems (including developmental networks), and the resulting effects on phenotypic evolution. Regarding origins, a study in mouse embryonic stem cells revealing pervasive bidirectional transcription suggested to the authors that opposite strand transcription could provide a robust source for new genes (Almada et al. 2013; Wu and Sharp 2013). Regarding genetic systems, studies of new genes in Drosophila (those present in just a few species) showed that they can quickly become essential for viability, male fertility, and foraging behavior (Chen et al. 2010; Chen, Ni, et al. 2012; Chen, Spletter, et al. 2012). Indispensability for fertility and behavior was shown to result from the incorporation of new genes into existing networks and the reshaping of those networks to take maximum advantage of the new gene’s novel function (Chen et al. 2013; Long et al. 2013). Detailed analysis of a new gene required for centromere integrity in Drosophila revealed that the rapid acquisition of essentiality is facilitated by a surprisingly small number of mutations (Ross et al. 2013).
Regarding development, two recent studies have expanded our understanding of the multifaceted nature of the evolution of development, a process for which a major focus of investigation was evolutionary conservation (e.g., Hox genes). A comparative analysis of the Hippo growth control pathway in mice and flies revealed that differences in pathway regulation were achieved via the gain and loss of function by pre-existing genes. This report demonstrates that the architecture of a vertebrate signaling pathway is not static but instead was impacted by the acquisition of new functions by existing genes after divergence from the lineage leading to flies (Bossuyt et al. 2014). A second study identified an example of a new gene-driven developmental switch leading to phenotypic evolution in the nematode genus Pristionchus (Ragsdale et al. 2013). Here, alternative forms of adult mouth morphology were found to be dependent upon the dosage of an often duplicated sulfatase that functions downstream of a pheromone sensing pathway. This indicated that gene duplication and the subsequent incorporation of the duplicates into a developmental network, even without neofunctionalization, can facilitate phenotypic diversity.
Developmental networks often employ morphogen gradient systems as a mechanism for regulating gene expression. In a morphogen gradient, secreted signaling molecules orchestrate distinct differentiation programs in target cells via concentration-dependent responses (Wolpert 1969). Among the best-understood gradients are the two that pattern the dorsal–ventral axis of the Drosophila embryo (Anderson 1998). First, activation of the transmembrane receptor Toll on the ventral side leads to a maternal ventralizing gradient of the transcription factor Dorsal with the highest level of Dorsal activity in the ventral-most region. Dorsal activates ventral-specific genes and modulates the expression of two signaling proteins; it represses decapentaplegic (dpp) and activates short gastrulation (sog). Second, extracellular interactions between Dpp and Sog then create a zygotic dorsalizing gradient of Dpp. The highest level of Dpp activity is in the dorsal-most region. Cells interpret the local concentration of Dpp along the dorsal–ventral axis via a pair of Smad signal transducers Mad and Medea and then adopt one of the five cell fates (O’Connor et al. 2006).
The reversal of dorsal–ventral polarity between insect and vertebrate embryos (insect “nerve cords” develop on the ventral side) is due to a zygotic ventralizing gradient of BMP that employs homologous proteins in same architecture and plays the same role as the Dpp dorsalizing gradient (Holley et al. 1995, 1996; De Robertis 2008). Recent studies identified a new component of the developmental network supporting the Dpp/BMP gradients that play a conserved role in flies and vertebrates. In both networks, the activities of the homologous signal transducers Medea and Smad4 are activated by the homologous deubiquitylases Fat facets (Faf) and USP9X (Dupont et al. 2009; Stinchfield et al. 2012). In vertebrates, an additional new participant was recently revealed; TIF1-γ/TRIM33 is a Ring class E-3 ubiquitin ligase that functions opposite USP9X by deactivating Smad4 and antagonizing BMP signaling (Dupont et al. 2005, 2012). The conservation of roles for Medea/Smad4 and Faf/USP9X formally requires an ubiquitin ligase for Medea; without a ligase there is no need for the Faf deubiquitylase. We wondered which Drosophila ubiquitin ligase performs the functions of TIF1-γ/TRIM33 in the Dpp dorsal–ventral signaling network?
Bonus is the Drosophila protein most closely related to TIF-γ/TRIM33. More precisely, Bonus is most closely related to four vertebrate members of a tightly linked subfamily within the TIF1/TRIM family: TIF1-α/TRIM24, TIF1-β/TRIM28, TIF-γ/TRIM33, and TIF-δ/TRIM66. The TIF1/TRIM family is a large and ancient one whose origins predate the diversification of metazoan animals, as shown by the presence of a TRIM37-like protein in a variety of protozoan species. The TIF-γ/TRIM33 subfamily is among the older subfamilies with a representative in all Bilaterian species (Marin 2012). To confirm subfamily age, our reciprocal Blast searches easily identified and confirmed a TIF-γ/TRIM33 family member in Nematostalla vectensis.
The connection between Bonus and the TIF-γ/TRIM33 subfamily is supported by experimental data. Studies of bonus mutants revealed activities as a nuclear receptor cofactor and as an inhibitor of βFTZ-F1-dependent transcription during metamorphosis (Beckstead et al. 2001, 2005). TIF1-α/TRIM24 interacts with retinoic acid and estrogen nuclear receptors, via the same LxxLL motif that mediates Bonus and βFTZ-F1 binding (Le Douarin et al. 1996). Side-by-side assays of Bonus and TIF1-β/TRIM28 in cultured cells identified a common role as a nuclear corepressor of c-Myb-dependent transcription (Nomura et al. 2004). A further parallel between Bonus and TIF1-β/TRIM28 is the ability to recruit the chromodomain protein HP1a to specific sites where together they alter chromatin configuration and modulate transcription (Nielsen et al. 1999; Ito et al. 2012).
We tested the hypothesis that Bonus has a conserved function as an ubiquitin ligase for Medea that serves as an antagonist of the Dpp dorsal–ventral network. We found that the role of Bonus is not the same as that of TIF1-γ/TRIM33 and instead the ubiquitin ligase Nedd4 does the job of TIF-γ/TRIM33 in Dpp signaling. Taken together our phylogenetic and developmental genetics data demonstrate that the architecture of the Dpp/BMP dorsal–ventral patterning network continued to evolve in the vertebrate lineage, after the separation from flies, via the incorporation of new genes.
Results
Birth Order of Vertebrate TIF1/TRIM Subfamily Members Most Closely Related to Bonus
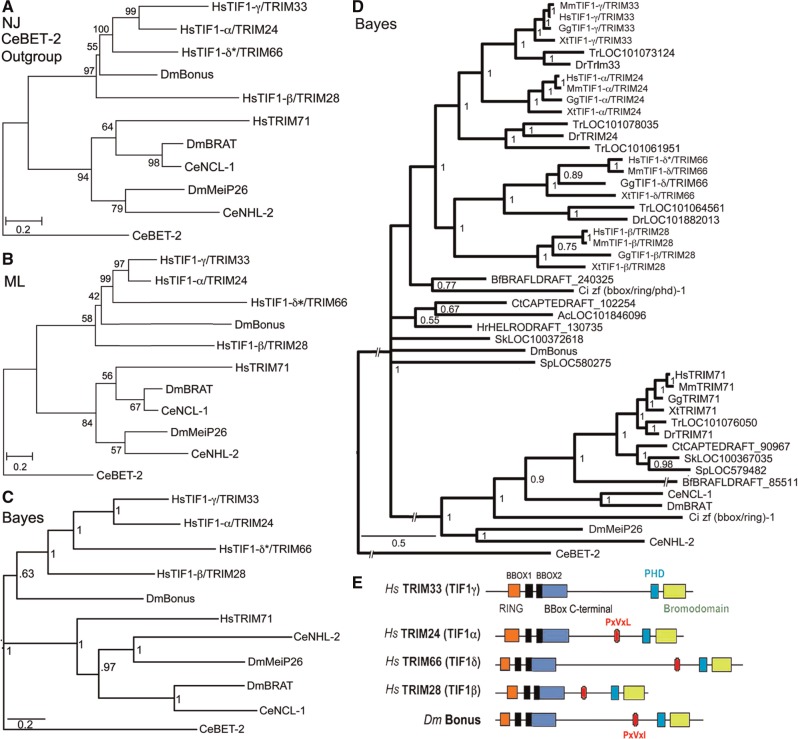
To better characterize the relationship between Bonus, TIF1-γ/TRIM33 and other vertebrate subfamily members we conducted a series of phylogenetic studies. First, we created a set of six small trees that utilized 11 sequences from humans, flies, and nematodes that share multiple structural domains with Bonus (see supplementary table S1, Supplementary Material online, for accession numbers and supplementary table S2, Supplementary Material online, for domain comparisons). In our view employing just these sequences has three benefits: 1) the alignments contain a large number of informative sites, 2) the resulting trees can be easily compared with trees from the TGF-β, Wnt, and Hippo pathways (e.g., Newfeld et al. 1999; Konikoff et al. 2010; Wisotzkey et al. 2012) and 3) flies and nematodes provide the ability to experimentally evaluate the functional consequences of sequence divergence.
We then created a set of six larger trees employing 47 sequences. These included an additional 36 that we identified in species that lie taxonomically between nematodes and humans. We added sequences from four additional vertebrate species (mouse, chicken, zebrafish, and pufferfish) as well as four species from groups that are regarded as vertebrates’ closest relatives (cephalochordates, echinoderms, urochordates, and hemichordates). We also included three lophotrocozoan species (two annelids and a sea slug) to fill the large evolutionary gap between nematodes/flies and vertebrates/chordates. The alignments underlying these trees had fewer informative sites leading to lower resolution but nevertheless revealed important insights on the origin of the Bonus-TIF1-γ/TRIM33 subfamily.
Initial alignments with the 11 sequences revealed that the current sequence of human TIF1-δ/TRIM66 corresponds to the shorter mouse TIF1-δ/TRIM66 isoform2 that does not contain a RING domain. Given that the RING domain is a canonical feature of the TIF1/TRIM family (Boudinot et al. 2011), we predicted and then identified a human TIF1-δ/TRIM66 isoform1 that contains this domain (supplementary figs. S1–S4, Supplementary Material online). We employed this extended sequence, that we named TIF1-δ*/TRIM66, in our analysis.
We included vertebrate TRIM71 proteins as they belong to a distinct TIF1/TRIM subfamily (SubgroupD; Sardiello et al. 2008). SubgroupD also contains two fly proteins (DmMei-P26 and DmBrat) and two nematode proteins (CeNHL-2 and CeNCL-1). The SubgroupD sequences provided an important second subfamily for our trees that aided in interpreting the placement of nonmodel organism sequences. We utilized three distinct algorithms (Neighbor Joining, Maximum Likelihood, and Bayesian) to generate trees and each was rooted with two carefully chosen outgroups. We employed CeBET-2 as an outgroup in half of our trees because it contains only two Bromo domains (one of the five types of structural domain found in TRIM family members; supplementary table S2, Supplementary Material online). We utilized HsTRIM37 as an outgroup in the other half because this protein never clustered with any other human TRIM protein (Sardiello et al. 2008). Features common to many trees engender the most confidence.
All trees (fig. 1 and supplementary fig. S5, Supplementary Material online) support the previously reported subfamily of Bonus with four vertebrate TIF1/TRIM sequences (SubgroupC; TIF1-α/TRIM24, TIF1-β/TRIM28, TIF1-γTRIM33, and TIF1-δ/TRIM66; Sardiello et al. 2008). Examination of branch topology and length as well as node confidence in our trees supports the hypotheses that human TIF1-α/TRIM24 and TIF1-γ/TRIM33 resulted from the most recent duplication in this cluster. These two sequences cluster together in all trees and always have the shortest branches. Eleven of our 13 trees provide strong statistical support for this cluster with bootstrap or posterior probabilities above 90%. In addition, two trees in Sardiello et al. (2008) display 100% bootstrap confidence in this cluster (their figs. 4C and 6B). Taking a step back, the data suggest that TIF1-β/TRIM28 is the progenitor of this pair via an older duplication; it is linked to them in seven trees whereas TIF1-δ*/TRIM66 is linked to them in three (two of our trees have TIF1-β/TRIM28 and TIF1-δ*/TRIM66 clustered).
Fig. 1.
Phylogenetic relationships and structure of Bonus-Tif1-γ/TRIM33 subfamily members. (A) Neighbor Joining (NJ), (B) Maximum Likelihood (ML) and (C) Bayesian trees of ten human (Hs), fly (Dm), and nematode (Ce) TRIM family sequences related to Bonus. Accession numbers are in supplementary table S1, Supplementary Material online. HsTRIM71 was chosen to anchor the non-Bonus fly and nematode sequences into a single subfamily based on Sardiello et al. (2008). CeBET-2 was chosen as the outgroup because it does not belong to the TRIM family and shares only the BROMO of the five structural domains shared by the others. The alignment contained 551 informative positions. A scale bar showing amino acid substitutions per site is present. Bootstrap values (NJ and ML trees) above 40% and posterior probabilities above 0.5 (Bayesian tree) are shown. Bootstrap values above 70% and posterior probabilities above 0.95 are considered statistically significant. Each tree contains the same two distinct subfamilies. Within one subfamily, Bonus is either the most distant sequence or between TIF1-β/TRIM28 and TIF1-δ/TRIM61, whereas TIF1-γ/TRIM33 and TIF1-α/TRIM24 consistently group together. (D) Expanded Bayesian tree containing all 47 sequences in supplementary table S1, Supplementary Material online, derived from an alignment with 173 informative positions. The same two statistically supported subfamilies are visible. The topology of the Bonus-TIF1-γ/TRIM33 subfamily is a slightly different from the other trees. The four vertebrate sequences resolve into two clusters with Bonus a distant outlier. (E) Schematic comparison of the domain structure of Bonus-TIF1-γ/TRIM33 subfamily proteins. The locations of seven distinct domains are shown: RING, orange; BBOX, black (2); BBox C-terminal, blue; PxVxL (or the biochemically similar PxVxI in Bonus), red; PHD, aqua; and BROMO, green.
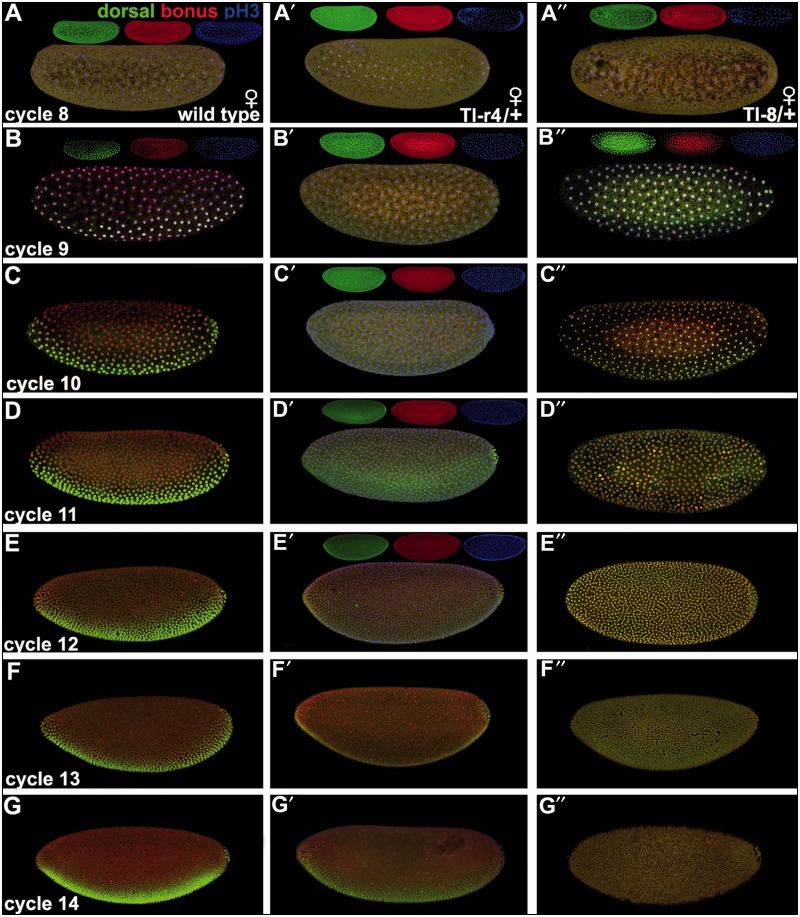
Fig. 4.
Synchronous nuclear translocation of Bonus and Dorsal. Embryos in lateral view in mitotic cycles 8–14. Mothers were wild type (A–G), Tollr4/+ (A′–G′), and Toll8/+ (A″–G″) mated to wild type fathers. The molecular lesion in each Toll mutant is described in the Materials and Methods. Embryos display Bonus (red), Dorsal (green), and the mitotic marker pH3 (blue) for cycles 8 and 9 (wild type and Toll8) or cycles 8–12 (Tollr4). For three-color images, each channel is also shown as an inset. The remaining images display only Bonus (red) and Dorsal (green). Left column: Wild type. (A) Cycle 8: pH3 is ubiquitously nuclear with Bonus and Dorsal ubiquitously cytoplasmic. (B–G) Cycles 9–14: Bonus is ubiquitously nuclear with Dorsal visible within the ventral-most 40% of nuclei while remaining cytoplasmic elsewhere. High magnification views of nuclei in cycles 8–10 embryos are shown in supplementary figure S9, Supplementary Material online. Middle column: Tollr4/+. (A′–C′) Cycles 8–10: Bonus and Dorsal are ubiquitously cytoplasmic. (D′) Cycle 11: Bonus shows a mixture of ubiquitiously cytoplasmic and weak nuclear expression. Dorsal is also mixed with primarily cytoplasmic and weak nuclear expression in the ventral-most 40%. (E′) Cycle 12: Increasing ubiquitous nuclear and diminished cytoplasmic Bonus. Dorsal shows increasing nuclear expression in the ventral-most 40% with cytoplasmic expression elsewhere. (F′, G′) Cycles 13 and 14: Roughly wild type expression of Bonus and Dorsal. Right column: Toll8/+. (A″) Cycle 8: Bonus and Dorsal are ubiquitously cytoplasmic. (B″–G″) Cycles 9–14: Bonus and Dorsal are ubiquitously nuclear.
Identifying the oldest vertebrate subfamily member proved more complicated as TIF1-β/TRIM28 and TIF1-δ*/TRIM66 show a variety of relationships to Bonus. TIF1-δ*/TRIM66 maps closest in eight trees and TIF1-β/TRIM28 closest in four trees. However, TIF1-β/TRIM28 has the longest branch in eight trees and TIF1-δ*/TRIM66 the longest in four trees. In this regard, we noted that the paper reporting the cloning and characterization of mouse TIF1-δ/TRIM66, the only paper describing its expression in any species, states that this gene is testis-specific (Khetchoumian et al. 2004). Thus, it seems likely that distinct selective pressures on germline-exclusive genes led this protein to accumulate amino acid changes at a rate exceeding those of its somatically expressed siblings. As a result, it moved from its expected position to one of the greater divergence and branch length in a subset of trees. Taken together one hypothesis we favor is that TIF1-β/TRIM28 is the oldest human family member and the most similar to Bonus; TIF1-β/TRIM28 gave rise first to TIF1-δ*/TRIM66 and then to the original member of the TIF1-α/TRIM24 and TIF1-γTRIM33 pair.
Completing the larger picture of this subfamily in vertebrates, we noted that TIF1-β/TRIM28 was lost in the lineage leading to fish whereas TIF1-α/TRIM24 experienced a duplication in pufferfish. In contrast to the four subfamily members present in all vertebrates except fish, all nonvertebrate species contain a single family member. Thus, the three rounds of duplication that led from TIF1-β/TRIM28 to TIF1-γ/TRIM33 occurred in the common ancestor of all vertebrates but after the divergence from their closest nonvertebrate relatives. This scenario of frequent duplication and subsequent lineage-specific deletion is visible throughout the TRIM family (Marin 2012). As a result, there are 66 members in humans (Sardiello et al. 2008) with a mix of common and unique family members among the 55 zebrafish and 44 pufferfish family members (Boudinot et al. 2011).
To identify the newest vertebrate subfamily member, either TIF1-α/TRIM24 or TIF1-γ/TRIM33, we relied on three lines of evidence. Each of these supports the hypothesis that TIF1-γ/TRIM33 is the newest. First, TIF1-γ/TRIM33 has the shortest branch in 9 of 12 trees. Second, TIF1-γ/TRIM33 does not contain an HP1-interacting PxVxL sequence (fig. 1E). This interaction is conserved in Bonus via a similar PxVxI motif (Nielsen et al. 1999; Ito et al. 2012). Third, domain comparisons indicate that TIF1-γ/TRIM33 never shows the greatest identity or similarity to Bonus (supplementary table S3 and fig. S6, Supplementary Material online). Taken together these criteria support the hypothesis that TIF1-γ/TRIM33 is the most evolutionarily distant from Bonus and thus the newest vertebrate subfamily member. This designation is consistent with reports that TIF1-γ/TRIM33 functions primarily as a monoubiquitin ligase, a role not frequently associated with its siblings (Deshaies and Joazeiro 2009). The best-characterized role of TIF1-γ/TRIM33 is as an antagonist of the BMP signaling network during dorsal–ventral axis formation (Dupont et al. 2005, 2009; Morsut et al. 2010; Agricola et al. 2011).
Notwithstanding our hypothesis that TIF1-β/TRIM28 is the oldest vertebrate member of the subfamily, the pattern of conservation between Bonus and its four vertebrate relatives is uneven. None of the vertebrate proteins was the most identical or the most similar to Bonus in more than two of the six structural domains. Further, in two domains, the vertebrate protein with the highest identity to Bonus did not have the highest similarity (e.g., greatest identity in the BBC domain is TIF1-β/TRIM28 but greatest similarity is TIF1-δ*/TRIM66; supplementary table S3, Supplementary Material online). Thus, it was conceivable that Bonus, as the only subfamily member in flies, was capable of functioning similarly to any of the vertebrate proteins including TIF1-γ/TRIM33. We then tested the hypothesis that Bonus serves as an ubiquitin ligase for Medea that antagonizes the Dpp signaling network during dorsal–ventral axis formation in Drosophila.
bonus Zygotic Nuclear Requirement for Dpp Responsiveness
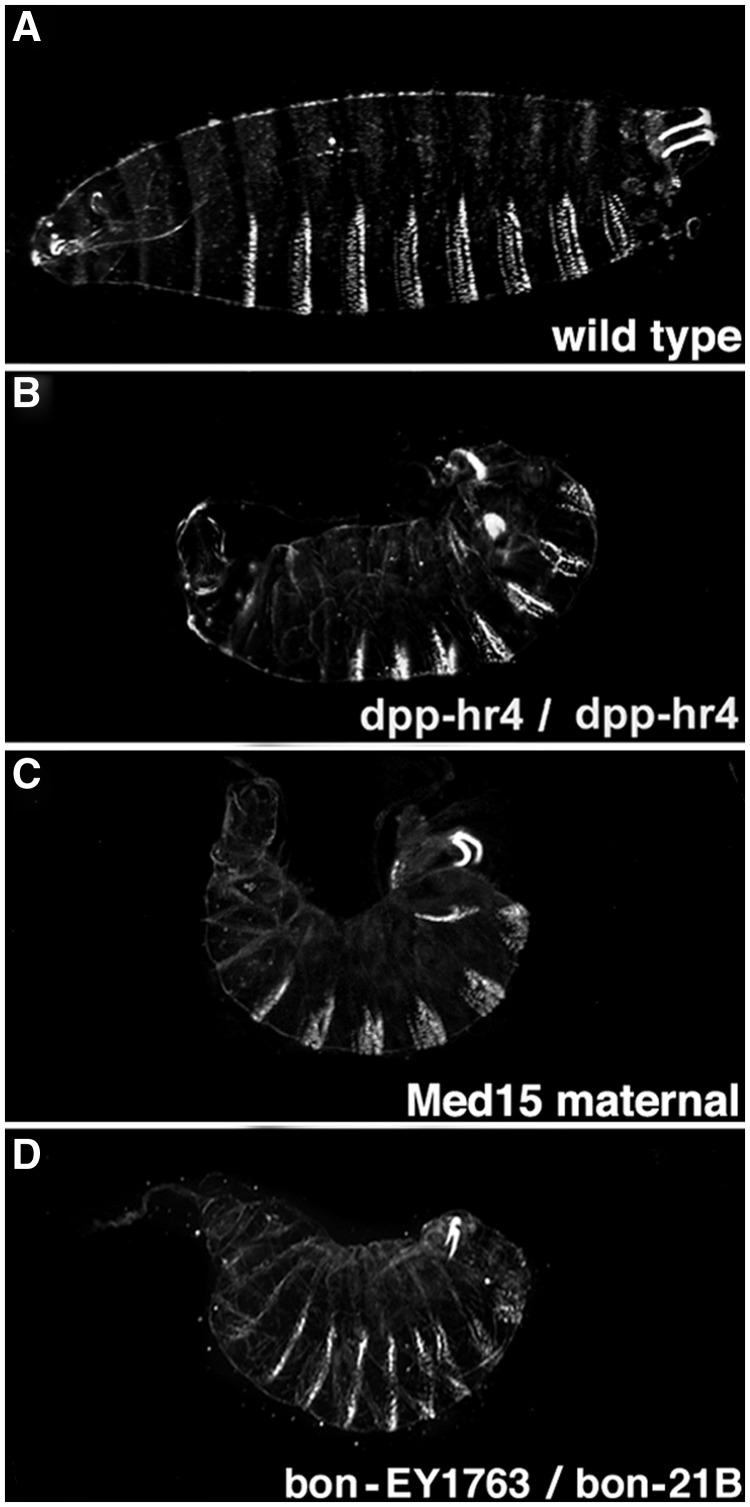
Initial studies of embryonic cuticles revealed that two bonus zygotic mutant genotypes displayed defects in dorsal–ventral patterning. Eighteen percent of bonus487/bonus21B and 13% of bonus21B/bonusEY1763 cuticles were partially ventralized (see Materials and Methods for descriptions of alleles). The most severely affected individuals resembled dpphr4 cuticles and cuticles generated by Medea15 females (fig. 2 and supplementary table S4, Supplementary Material online). Further, heterozygosity for bonus21B partially rescued dorsalization defects due to hemizygosity for sogyl26 (supplementary table S5 and fig. S7, Supplementary Material online). This effect is also seen with dpphr4, Medea15, and fafB6 (Stinchfield et al. 2012). These data suggest that Bonus plays an instructive role in dorsal–ventral patterning, a role not predicted by analogy to TIF1-γ/TRIM33. If Bonus were a ubiquitin ligase for Medea, then bonus zygotic mutants would be dorsalized due to reduced Medea ubiquitylation and hyperactive Dpp signaling.
Fig. 2.
bonus zygotic mutants phenocopy dpp mutants. Cuticles in lateral view with anterior to the left and dorsal up. The maternally contributed allele is listed first. The molecular lesion in each mutant is described in the Materials and Methods. (A) Wild type. The broad white denticle belts on the ventral surface (bottom), narrow white Filzkorper in the posterior spiracles (upper right corner) and internal head skeleton (left side) are visible. (B) Homozygous dpphr4 ventralized cuticle with a short curved body, dorsally extended denticles, herniated head, and defective Filzkorper. (C) Homozygous maternal Medea15 ventralized cuticle is similar to dpphr4. (D) bonusEY1763/bonus21B transheterozygous ventralized cuticle is also similar to dpphr4.
We then adjusted our model for Bonus function by incorporating a different target. In the new hypothesis, Bonus serves as a Toll pathway ubiquitin ligase. Here, the prediction is that loss of Bonus ubiquitin ligase activity will lead to Toll hyperactivation, expansion of Dorsal, increased repression of Dpp, and ventralization of the embryo. We examined this hypothesis in bonus21B/bonusEY1763 embryos stained with Bonus antibody.
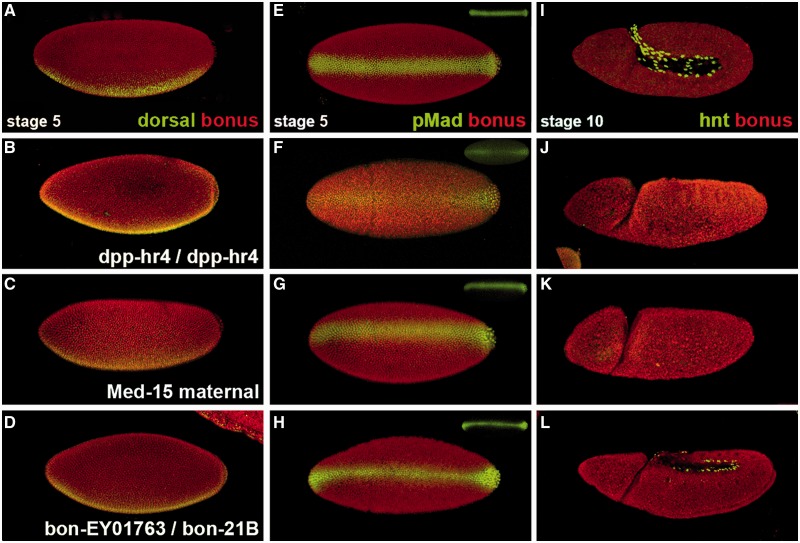
Dorsal protein shows normal ventral nuclear accumulation at stage 5 in bonus mutants (fig. 3A–D). To confirm this result, we examined pMad expression as a marker of Dpp pathway activity. In wild type, at stage 5, pMad is present as a narrow stripe in the dorsal-most region reflecting the concentration of Dpp there via extracellular transport by Sog. dpphr4 mutants have a point mutation that interferes with transport and thus display reduced dorsal pMad accumulation in a background of diffuse staining. Medea15 maternal mutants show normal pMad due to the loss of signaling downstream of Mad phosphorylation. bonus zygotic mutants have normal pMad as well (fig. 3E–H). Thus, Bonus does not antagonize either of these pathways.
Fig. 3.
bonus zygotic requirement for Dpp responsiveness. (A–D) Stage 5 embryos (nuclear cycle 14) in lateral view displaying Bonus (red) and Dorsal (green). (E–H) Stage 5 embryos in dorsal view displaying Bonus (red) and pMad (green) with pMad alone as an inset. (I–L) Stage 10 embryos in lateral view with Bonus (red) and Hindsight (green). (A, E, I) Wild type embryos with ubiquitous nuclear Bonus, ventral Dorsal nuclear stripe, sharp dorsal pMad nuclear stripe, and nuclear Hindsight in amnioserosa cells. (B, F, J) dpphr4 homozygous embryo with normal nuclear Bonus and Dorsal. The pMad dorsal stripe is expanded with diffuse pMad throughout the embryo. Hindsight is absent. (C, G, K) Embryo from a homozygous Medea15 female with normal Bonus, Dorsal, and pMad but no Hindsight. (D, H, L) bonusEY1763/bonus21B embryo has normal nuclear Bonus, Dorsal, and pMad but significantly reduced Hindsight.
These experiments also showed that at stage 5, Bonus is a ubiquitous nuclear protein. Thus, we then hypothesized that bonus’s positive role in dorsal–ventral patterning was associated with the regulation of Dpp responsiveness. To test this, we analyzed Hindsight expression in stage 10 embryos. Hindsight is a marker for the amnioserosa, the dorsal-most tissue and the one that requires the highest level of Dpp signaling (Ray et al. 1991). Roughly 17% of bonus21B/bonusEY1763 embryos displayed severely reduced Dpp-dependent Hindsight expression in the amnioserosa (fig. 3I–L) consistent with the frequency of cuticle defects in this genotype. Individuals of this genotype contained normal Dpp-independent Hindsight expression in the foregut and hindgut (supplementary fig. S8, Supplementary Material online). Data from bonus mutants, together with Bonus nuclear localization after stage 5 and documented roles in chromatin remodeling (Beckstead et al. 2001), are consistent with a zygotic nuclear requirement for Bonus in dorsal–ventral patterning as a modulator of Dpp responsiveness.
Bonus and Dorsal Nuclear Translocation Is Synchronous in Multiple Genotypes
The nonconservation of zygotic bonus function in the Dpp dorsal–ventral signaling network with that of TIF1-γ/TRIM33 in the BMP dorsal–ventral signaling network did not preclude the possibility that a maternal Bonus function could be conserved. Examination of unfertilized eggs showed that Bonus is maternally loaded and cytoplasmic (supplementary fig. S9A′–A″′, Supplementary Material online). As Bonus is ubiquitously nuclear by stage 5 (e.g., fig. 3A), we tested the hypothesis that Bonus nuclear translocation occurs between mitotic cycles 9 and 12, the stages when Dorsal (cycle 9) and pMad (cycle 12) enter the nucleus in response to their respective signals (Roth et al. 1989; Steward 1989; Shimmi et al. 2005). We examined wild type as well as Toll maternal gain and loss of function embryos to test the hypothesis that this pathway influences Bonus nuclear translocation.
In wild type at cycle 8, the last cycle of strictly maternal expression (Pritchard and Schubiger 1996), Bonus and Dorsal are wholly cytoplasmic. The maternal to zygotic transition begins during cycle 9 and in wild type embryos Bonus becomes ubiquitously nuclear. At the same time, Dorsal becomes nuclear within the ventral-most 40% of nuclei whereas remaining cytoplasmic elsewhere. The maternal-to-zygotic transition is complete during stage 10 and in wild type embryos Bonus and Dorsal maintain their respective subcellular locations through cycle 14 (fig. 4A–G; left column with high magnification views in supplementary fig. S9B–D, Supplementary Material online).
Tollr4 is a loss of function allele with modest haploinsufficiency (Anderson, Bokla, et al. 1985; Anderson, Jürgens, et al. 1985). In embryos from Tollr4 heterozygous mothers, Bonus and Dorsal are wholly cytoplasmic at stage 8 and then their nuclear translocation is delayed. During cycles 9 and 10, ubiquitous cytoplasmic expression of both proteins persists. In cycle 11, they begin nuclear translocation with normal spatial parameters. During cycles 12 and 13, they become increasingly nuclear until the wild type pattern appears at cycle 14, a delay of four nuclear cycles (fig. 4A′–G′; middle column). Toll8 is strong gain of function allele (Anderson, Bokla, et al. 1985; Anderson, Jürgens, et al. 1985). In embryos from Toll8 heterozygous mothers, Bonus and Dorsal are wholly cytoplasmic at stage 8. They then become ubiquitously nuclear during cycle 9 and maintain this pattern through cycle 14 (fig. 4A″–G″; right column). In all genotypes, Bonus and Dorsal translocate to the nucleus synchronously.
bonus Maternal Cytoplasmic Requirement for Dorsal Nuclear Translocation
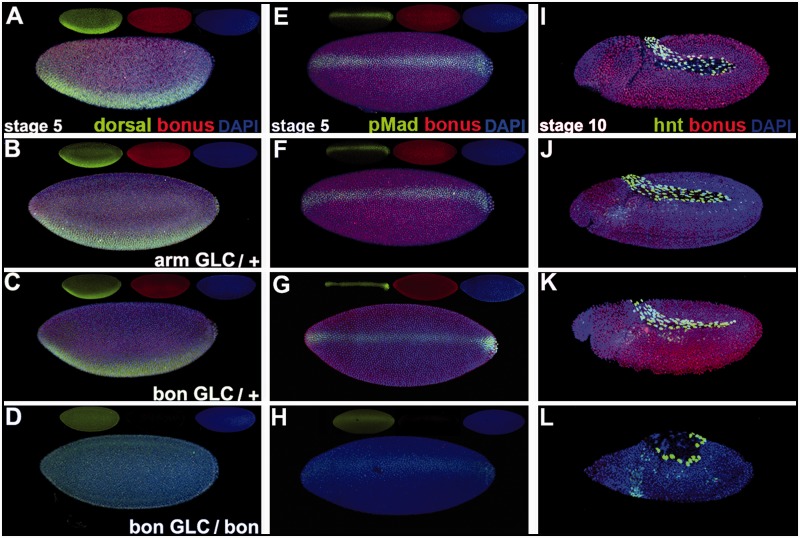
Given this synchrony, we then examined Dorsal nuclear localization in embryos without Bonus derived from bonus21B germline clone bearing females. We mated these females to heterozygous bonus21B males. Germline clone eggs are maternally hemizygous for the bonus mutation. Germline clone embryos can be either zygotically homozygous or zygotically heterozygous for bonus21B depending upon the paternally contributed chromosome. bonus21B deletes the normal initiator methionine (Beckstead et al. 2001) and thus a reduction of wild type Bonus expression identified the zygotically homozygous bonus21B embryos. In this experiment, we can distinguish maternal from zygotic requirements for Bonus in embryonic development.
At stage 5, wild type, arm-lacZ germline clone embryos (a control for the method) and zygotically heterozygous bonus21B germline clone embryos display normal Dorsal expression. Zygotically homozygous bonus21B germline clone embryos show no visible nuclear Dorsal (fig. 5A–D). This reveals a maternal cytoplasmic requirement for bonus in Dorsal nuclear translocation. Without any nuclear Dorsal bonus21B germline clone embryos should be fully dorsalized due to loss of Dorsal repression of dpp ventrally and the absence of Dorsal activation of Sog. Consistent with this prediction, homozygous bonus21B germline clone embryos display ventrally expanded pMad nuclear accumulation barely above a background of ubiquitous expression (fig. 5E–H). This observation is supported by dpp in situ hybridization studies in these embryos revealing ventral expansion of Dpp transcription. The presence of any pMad nuclear accumulation, in embryos without visible Dorsal nuclear translocation, may be explained by the presence of a cryptic methionine in the portion of bonus exon1 that is not deleted in bonus21B (supplementary fig. S10, Supplementary Material online). bonus21B may not be a protein null allowing invisible but functional amounts of Dorsal into the nucleus and the slight dorsal–ventral pMad polarity seen in homozygous bonus21B germline clone embryos.
Fig. 5.
bonus maternal requirement for Dorsal nuclear translocation. (A–D) Stage 5 embryos in lateral view displaying Bonus (red), Dorsal (green), and the DNA dye DAPI (blue) plus individual channels as insets. (E–H) Stage 5 embryos in dorsal view with Bonus (red), pMad (green), and DAPI (blue) plus insets. (I–L) Stage 10 embryos in lateral view with Bonus (red), Hindsight (green), and DAPI (blue). (A, E, I) Wild type embryos show ubiquitiously nuclear Bonus, ventral Dorsal nuclear stripe, sharp dorsal pMad nuclear stripe, and Hindsight in amnioserosa cells. (B, F, J) arm-lacZ germline clone embryos display normal Bonus, Dorsal, pMad, and Hindsight. (C, G, K) Heterozygous bonus21B germline clone embryos with paternal rescue display normal Bonus, Dorsal, pMad, and Hindsight. (D, H, L) Homozygous bonus21B germline clone embryos with no paternal rescue were identified via reduction of Bonus expression. These embryos display no visible nuclear Dorsal, faint ventrally expanded dorsal pMad stripe with diffuse pMad background, and significantly reduced Hindsight in amnioserosa cells.
The absence of Dorsal nuclear translocation and subsequent reduction in pMad nuclear accumulation are inconsistent with the loss of Hindsight expression in the amnioserosa at stage 10 observed in homozygous bonus21B germline clone embryos (fig. 5I–L). If bonus has no effect on dorsal–ventral patterning beyond the maternal cytoplasmic requirement for Dorsal nuclear translocation, then bonus21B germline clone embryos should be almost completely dorsalized. Dorsalized embryos display expanded Hindsight. To confirm this prediction, we examined the effect of reduced Dorsal nuclear accumulation on Hindsight expression in embryos derived from Tollr3/Tollr4 females. This temperature-sensitive genotype has severely, but not completely, compromised Toll signaling (Schneider et al. 1991). In embryos from these females, pMad nuclear accumulation and Hindsight amnioserosa expression are both ventrally expanded (supplementary fig. S11, Supplementary Material online). Homozygous bonus21B germline clone embryos display roughly the same effect on pMad as Tollr3/Tollr4 (supplementary fig. S12, Supplementary Material online) but they display the opposite effect on Hindsight. In homozygous bonus21B germline clone embryos Hindsight expression is instead consistent with that seen in bonus zygotic mutant embryos, reflecting a distinct zygotic nuclear requirement in Dpp responsiveness.
Discussion
Bonus Contributes to Two Embryonic Dorsal–Ventral Patterning Networks
From a developmental perspective, the data reveal that bonus plays two distinct roles in dorsal–ventral axis formation. There is a maternal cytoplasmic role in Toll signaling required for Dorsal nuclear accumulation and there is a zygotic nuclear role downstream of pMad required for Dpp responsiveness. The latter fits squarely within the existing paradigm for TIF1/TRIM proteins in chromatin remodeling and transcription regulation. TIF1/TRIM proteins, including Bonus, interact directly via their PxVxL/I domains with members of the HP1 family to promote the modulation of gene expression (Le Douarin et al. 1996; Nielsen et al. 1999; Beckstead et al. 2001). Perhaps the PxVxI motif in Bonus facilitates Dpp responsiveness via interactions with its signal transducers Mad, Medea, or Schnurri.
Regarding the maternal role, the presence of a near-perfect match in Dorsal to the AF2-AD consensus sequence (supplementary fig. S13, Supplementary Material online) suggests that this function may also be mediated by a TIF1/TRIM family paradigmatic mechanism. In this case, Bonus and TIF1/TRIM proteins employ their LxxLL motif to bind the AF2-AD consensus sequence in βFTZ-F1 (Beckstead et al. 2001) or nuclear hormone receptors (Le Douarin et al. 1996). We determined that there is no AF2-AD sequence in Cactus, the protein that sequesters Dorsal in the cytoplasm further suggesting that Bonus regulation of Dorsal nuclear translocation may be direct. Perhaps this interaction is necessary because several reports suggest that release from Cactus is insufficient for Dorsal nuclear translocation. For example, the phosphorylation of Dorsal is also required (Whalen and Steward 1993; Drier et al. 1999). Thus, one potential function for Bonus may be to facilitate Dorsal phosphorylation. Alternatively, Bonus may act as a chaperone to help Dorsal efficiently locate its targets during the rapid nuclear cycles 8, 9, and 10. We propose that the ability of Bonus to translocate to nuclei throughout the embryo, rather than only ventrally like Dorsal, is due to its ability to respond to lower doses of Toll signaling than Dorsal. This scenario is similar to the differential responses of spalt (narrow response to high concentrations) and optomotor blind (wide response to high and low concentrations) to the Dpp gradient in third instar wing imaginal discs (O’Connor et al. 2006).
Acquisition of Nedd4 Function by TIF1-γ/TRIM33 during Vertebrate Evolution
From an evolutionary perspective, the analyses showed that vertebrates’ closest relatives have only a single Bonus-TIF1-γ/TRIM33 subfamily sequence. A preponderance of the phylogenetic data suggests that TIF1-β/TRIM28 is most similar to the common ancestor of the four vertebrate subfamily members. Then during the vertebrate whole-genome duplication TIF1-δ/TRIM66 was created and diverged significantly. Subsequently, but prior to the divergence of fish, TIF1-β/TRIM28 generated TIF1-α/TRIM24 and then TIF1-α/TRIM24 generated TIF1-γ/TRIM33. After the divergence of fish but before the split between zebrafish and pufferfish the gene for TIF1-β/TRIM28 was lost in the fish lineage.
Thus, our demonstration that the role of the newest subfamily member TIF1-γ/TRIM33 as an ubiquitin ligase for Smad4 is not conserved for Bonus and Medea begs the question, who does this job in flies? There must be an ubiquitin ligase for Medea that complements the role of the conserved deubiquitylase Faf in Dpp dorsal–ventral signaling. We eliminated the ubiquitin ligase dSmurf (known as Lack in Flybase) even though it is a well-known antagonist of Dpp dorsal–ventral signaling (Podos et al. 2001). dSmurf was explicitly shown to target Mad but not Medea (Liang et al. 2003). Another candidate is Highwire (Hiw), an ubiquitin ligase for Medea at the larval neuro-muscular junction (McCabe et al. 2004). A third candidate is Nedd4, the only fly member of a HECT class E-3 ubiquitin ligase family and homolog of Nedd4L that antagonizes Smad activity during TGF-β signaling in mammals (Alarcón et al. 2009; Gao et al. 2009; Aragón et al. 2011). To date Nedd4 has not been connected to TGF-β signaling in Drosophila.
We examined the ability of the latter candidates to suppress the roughly 40% haploinsufficiency associated with dpphr27 (Wharton et al. 1993). The logic was that an excess of maternally provided, nonubiquitylated Medea will amplify the weak Dpp signal associated with this allele. Maternal homozygosity for two hiw alleles had no effect, consistent with reports that hiw mRNA is not visible in unfertilized eggs (Wan et al. 2000). Maternal heterozygosity for either nedd4DG05310 or nedd4T119FS (Huet et al. 2002; Sakata et al. 2004) dominantly suppressed dpphr27 haploinsufficiency at roughly the same rate as smurfKG07014 (supplementary table S6, Supplementary Material online). This is consistent with reports that nedd4 mRNA is maternally loaded (Tadros et al. 2007) and suggests that Nedd4 is the Medea ubiquitin ligase functioning opposite Faf in Dpp dorsal–ventral patterning.
One explanation for Bonus nonconservation is that the ubiquitylation role for TIF1-γ/TRIM33 is derived. After the amplification of the TIF1/TRIM family in the lineage leading to vertebrates, the newest member TIF1-γ/TRIM33 accumulated sufficient mutations to become an effective Ring class ubiquitin ligase. Supporting this hypothesis, expression of Xenopus TIF1-γ/TRIM33 in fly wings generates vein defects consistent with a role as a Medea ubiquitin ligase and these defects are suppressed by coexpression of Faf (Dupont et al. 2009). Then after becoming an ubiquitin ligase, TIF1-γ/TRIM33 did not assume a novel role that would confer a selective advantage as predicted by the classical model of new gene evolution by gene duplication. Instead TIF1-γ/TRIM33 assumed the role in dorsal–ventral patterning performed by the existing gene Nedd4L as a Smad4 ubiquitin ligase. Perhaps the selective advantage of this replacement is that the RING class TIF1-γ/TRIM33 is more efficient then the HECT class Nedd4L as a Smad4 ubiquitin ligase. Thus, Nedd4L became free to assume a novel function.
Phylogenetic analyses of the TGF-β family (Kahlem and Newfeld 2009) and the Nedd4 family (supplementary fig. S14, Supplementary Material online) support this view. The TGF-β/Activin/Nodal subfamily that employs Smad2/Smad3 signal transducers expanded from 4 in flies to 14 during vertebrate evolution. Alternatively, there is just a pair of Nedd4 proteins in vertebrates and sequence similarity to Drosophila Nedd4 indicates that Nedd4L is the ancestral form (supplementary table S7, Supplementary Material online). Notwithstanding its close relationship to fly Nedd4, Nedd4L specifically targets Smad3 and Smad7, two vertebrate-specific Smads (Aragón et al. 2011, 2012). Evidence for a vertebrate origin of Smad3 rather than Smad2, and for Smad7 rather than Smad6 derives from analyses of conservation and expression in fly wings (supplementary table S8, Supplementary Material online; Marquez et al. 2001). Thus, in the vertebrate lineage Nedd4L was freed from the responsibility of ubiquitylating Smad4 by TIF1-γ/TRIM33 at the same time that duplications were creating new TGF-β pathways. Nedd4L then adopted the novel function of regulating the vertebrate-specific signal transducers Smad3 and Smad7.
Multi-Step Model of New Gene Evolution and the Dynamic Nature of Developmental Networks
Taken together the results suggest an expansion of the classic model for the origin of new genes and their acquisition of novel functions based on gene duplication. What is necessary is the inclusion of a potential intermediate step in the process. In this step, new genes resulting from duplication do not immediately assume a novel function themselves, but instead they assume the function of an existing gene. This substitution then allows the gene whose function was replaced to acquire a novel function.
Studies in Drosophila revealed that the rapid incorporation of new genes into existing genetic networks underlies their requirement for fertility and foraging behavior (Chen et al. 2013; Long et al. 2013). Thus, a prediction is that the incorporation of new genes into developmental networks underlies their requirement for viability. Our data that the new gene TIF1-γ/TRIM33 replaced the function of the existing gene Nedd4L in the vertebrate dorsal–ventral patterning network validate this prediction. The replacement of the Nedd4 HECT domain ubiquitin ligase by the TIF1-γ/TRIM33 RING domain ligase as an antagonist of the vertebrate BMP gradient is consistent with a study of the cave crustacean Asellus aquaticus (Protas et al. 2011). That study also showed that developmental phenotypes (in their case eye loss) can be achieved by multiple underlying genetic mechanisms.
As was demonstrated for the Hippo signaling network (Bossuyt et al. 2014), our data indicate that the architecture of the vertebrate dorsal–ventral patterning network was not simply conserved since the divergence from their common ancestor with flies. Instead these two networks were impacted by the acquisition of new functions by existing genes or by the incorporation of new genes after divergence from each other. Thus, the vertebrate developmental program contains highly conserved features such as Hox genes and dynamic features such as Hippo signaling and the dorsal–ventral patterning network.
In summary, our data extend our understanding of developmental evolution and the architecture of two developmental networks. For developmental evolution, the nonconservation of Bonus and TIF1-γ/TRIM33 functions in the conserved Dpp/BMP dorsal–ventral patterning pathway reveals that new genes may displace an existing gene, allowing the displaced gene to assume a novel function. For developmental networks, our data reveal that Bonus is a common signal transducer in the Toll and Dpp pathways whose function is likely mediated by distinct mechanisms. We conclude that the architecture of the Dpp/BMP dorsal–ventral patterning network continued to evolve in the vertebrate lineage, after the separation from arthropods, via the incorporation of new genes.
Materials and Methods
Identification of Human TIF1-δ/TRIM66 Isoform1
Alignment of human and mouse TIF1-δ/TPIM66 isoforms showed that the human initiator methionine is homologous to the methionine at position103 in mouse isoform1 and to the initiator methionine of mouse isoforms2 and 3. TBLASTN was employed to query the human RefSeq Genomic database with the N-terminal region of mouse isoform1. This identified two regions in the human genome upstream of the current TIF1-δ/TRIM66 start site that displayed significant similarity when translated. One region is 20-kb distant and the other is immediately adjacent to the current initiator methionine. The distantly upstream genomic region contains an in-frame methionine that was identified as a potential new start codon. However, there are stop codons in all three frames roughly 100–120 amino acids downstream of this new initiator methionine. Further analysis identified potential splice sites deleting the stop codons and allowing an open-reading frame containing the new initiator methionine and a RING domain to connect in-frame with the immediately upstream region. The immediately upstream region then continues in-frame into the current TIF1-δ/TRIM66 sequence. TBLASTN revealed that sequences encoding peptides with 98% identity to the 5′-extension of human TIF1-δ/TRIM66 are present in the Pan troglodytes chromosome 11 genomic scaffold. TBLASTN with the proposed upstream extension of human Tif1-δ/TRIM66 was utilized to query the human expressed sequence tag (EST) database. This analysis identified an EST (TESTI4024751) containing both the donor and acceptor sites confirming their existence. However, the EST does not directly join the two splice sites together. This suggests human TIF1-δ/TRIM66 isoform3 contains three additional exons between the start of isoform1 and the current TIF1-δ/TRIM66. GenBank recently added a new prediction for a HsTIF1-δ/TRIM66 isoform that contains a nearly identical 5′-extension (GI 530396083 and Protein XP_005253327.1).
Phylogenetics
An alignment of 47 sequences (supplementary table S1, Supplementary Material online) was created with default settings in the Clustal Omega server at the EMBL-EBI website (www.ebi.ac.uk/Tools/msa/clustalo/) and visually inspected. Neighbor Joining and Maximum Likelihood trees were generated in MEGA5 (Tamura et al. 2011). For both methods, bootstrap consensus trees are inferred from 1,000 replicates and evolutionary distances computed using the Poisson correction method. Bootstrap values above 70 are considered statistically significant (Sitnikova 1996). Bayesian trees were created in MrBayes 3.2 (Ronquist et al. 2011). The prior amino acid model was set to Poisson (assumes equal stationary state frequencies and equal substitution rates; mrbayes.sourceforge.net/). The number of generations was set to 100,000 with a sample frequency of 100 and burn-in of 0.25. Posterior probabilities above 0.95 are considered significant. Other parameters were set to default.
Domains and Comparisons
Known structural or functional domains were predicted via SMART at EMBL (http://smart.embl-heidelberg.de/). The E value for a given region represents the number of sequences with a score greater than, or equal to, the score of the query sequence that can be expected absolutely by chance as calculated by Hidden Markov Models. Pairwise alignments of individual domains were generated with EMBOSS Needle at EMBL-EBI (http://www.ebi.ac.uk/Tools/psa/), a program employing the Needleman–Wunsch algorithm to produce an optimal global alignment of two sequences. The score was determined using the Blosum62 matrix with a gap penalty of −10.
Mutants
Fly stocks are as described: bonusS048706 and bonus21B (Beckstead et al. 2001); bonusEY1763 (Bellen et al. 2004), dpphr4, and dpphr27 (Spencer et al. 1982); fafB6 (Fischer-Vize et al. 1992), hiwBG02015, and hiwEP1308 (Wan et al. 2000); Medea15 and Medea17 (Hudson et al. 1998); nedd4DG05310 (Huet et al. 2002); nedd4T119FS (Sakata et al. 2004), sogy506 (Ferguson and Anderson 1992); smurfKG07014 (Tyler et al. 2007); nos.Gal4:VP16-MVD1 (van Doren et al. 1998); and UASp.GFP-αTub84B (Grieder et al. 2000). Tollr3, Tollr4, and Toll8 were originally known as Tollr632, Tollrm9/rm10, and Toll10B, respectively (Anderson, Bokla, et al. 1985; Anderson, Jürgens, et al. 1985). bonus21B is a small deletion resulting from an imprecise excision of a P-element in the 5′-untranslated region of exon1. This deletion does not affect the promoter. It eliminates all but the first 12 nt of exon1 including the initiator methionine as well as the splice acceptor and 324 nt of intron1. However, low level expression of nearly full-length Bonus is visible on Westerns and in embryos (Beckstead et al. 2001). Within the remaining 12 nt of exon1 is a methionine that if connected to the splice donor at the 5′-end of exon2 would result in a protein missing only the coding portion of exon1 (roughly 5% of the protein). Such a protein would contain all of the functional domains of full-length Bonus. bonus487 has a P element insertion in intron1 that acts as a strong loss of function allele. bonusEY1763 has a P element insertion in the 5′-untranslated region of exon1 that acts as a weak loss of function allele (Bellen et al. 2004). Tollr3 is recessive loss of function allele with moderate effect and no identifiable mutations (Anderson, Bokla, et al. 1985; Anderson, Jürgens, et al. 1985; Schneider et al. 1991). Tollr4 is a recessive loss of function allele associated with modest haploinsufficiency with two missense mutations in the extracellular domain (Anderson, Bokla, et al. 1985; Anderson, Jürgens, et al. 1985; Schneider et al. 1991). Temperature sensitivity of Tollr3/Tollr4 is as described (Wang et al. 2005). Toll8 is a dominant gain of function allele with a missense mutation in the extracellular domain (Anderson, Bokla, et al. 1985; Anderson, Jürgens, et al. 1985; Erdelyi and Szabad 1989; Schneider et al. 1991). Standard blue balancers and germline clone-related strains are described in Flybase (Marygold et al. 2013).
Genetics
Toll maternal effect mutations were analyzed in two ways. First, in embryos derived from matings of Tollr4/+ or Toll8/+ females with wild type males. Second, egg-lays containing temperature-sensitive, transheterozygous Tollr3/Tollr4 females were collected at the permissive temperature (18 °C) and maintained at that temperature through virgin collection. Tollr3/Tollr4 females were mated to wild type males at the restrictive temperature (25 °C) and embryos aged 4 h at that temperature before fixation. Germline clone bearing females were generated employing a FRT82B bonus21B chromosome (Beckstead et al. 2001). FRT82B arm-lacZ was employed as a control for germline clone induction as described (Chou and Perrimon 1996). In brief, germline clone bearing females were mated to bonus21B heterozygous males to assay bonus21B homozygous embryos and paternally rescued heterozygous embryos. Germline clone embryos containing a paternal bonus21B chromosome were identified via wild type Bonus antibody staining. The only difference from the published germline clone method was that heat shocks were employed on Day 8 (pupal ages 144–192 AEL) and Day 9 (168–216 AEL).
Embryos
Cuticle preparations, antibody staining, and RNA in situ hybridization were as described (Stinchfield et al. 2012). Primary antibodies were: Hindsight (DSHB-1G9), Dorsal (DSHB-7A4), lacZ (DSHB-401A and Organon Teknika), Bonus-GP37 (Beckstead et al. 2001), pSmad (Epitomics), and pH3 (Abcam). Primary antibodies were detected with Alexa Fluor 488, 546, or 633 goat α-rabbit, α-mouse, α-sheep, or α-guinea pig (Molecular Probes) or with VectaStain Elite (Vector Labs). eGFP and DAPI (Sigma) were visualized directly. Labeled Dpp-HI cDNAs were detected with α-DIG according to Takaesu et al. (2008). Pixel intensity plots reflecting pMad expression were created from single channel images in ImageJ. A single perpendicular line was drawn at the midpoint of the embryonic A/P axis. Pixel intensity along the line was analyzed with Plot Profile and the pixel intensity values were imported into Excel and graphed.
Supplementary Material
Supplementary tables S1–S8 and figures S1–S14 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the following: Estela Arciniega, Hugo Bellen, Ela Serpe, Osamu Shimmi, Nancy Tran, Bloomington Stock Center, and Iowa Hybridoma Bank. This work was supported by National Institute of Health grants to S.J.N. (HG002516 and NS072128). The authors declare no competing financial interests. S.J.N. designed research and wrote paper; R.G.W., J.C.Q., M.J.S., and S.J.N. performed research; R.G.W., J.C.Q., and S.J.N. analyzed data.
References
- Agricola E, Randall R, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1-γ to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell. 2011;43:85–96. doi: 10.1016/j.molcel.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Alarcón C, Zaromytidou A, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad activation and turnover in BMP and TGF-β pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almada A, Wu X, Kriz A, Burge C, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–366. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KV. Pinning down positional information: dorsal–ventral polarity in the Drosophila embryo. Cell. 1998;95:439–442. doi: 10.1016/s0092-8674(00)81610-4. [DOI] [PubMed] [Google Scholar]
- Anderson KV, Bokla L, Nüsslein-Volhard C. Establishment of dorsal–ventral polarity in the Drosophila embryo: induction of polarity by Toll. Cell. 1985;42:791–798. doi: 10.1016/0092-8674(85)90275-2. [DOI] [PubMed] [Google Scholar]
- Anderson KV, Jürgens G, Nüsslein-Volhard C. Establishment of dorsal–ventral polarity in the Drosophila embryo: genetic studies of Toll. Cell. 1985;42:779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- Aragón E, Goerner N, Xi Q, Gomes T, Gao S, Massagué J, Macias M. Structural basis for the interactions of Smad7 with WW domains in TGF-β pathways. Structure. 2012;20:1726–1736. doi: 10.1016/j.str.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón E, Goerner N, Zaromytidou A, Xi Q, Escobedo A, Massagué J, Macias M. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead R, Ner S, Hales K, Grigliatti T, Baker BS, Bellen H. Bonus, a Drosophila TIF1 homolog, is a chromatin-associated protein that acts as a modifier of position-effect variegation. Genetics. 2005;169:783–794. doi: 10.1534/genetics.104.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead R, Ortiz J, Sanchez C, Prokopenko S, Chambon P, Losson R, Bellen H. Bonus, a Drosophila homolog of TIF1 proteins, interacts with nuclear receptors and can inhibit FTZ-F1-dependent transcription. Mol Cell. 2001;7:753–765. doi: 10.1016/s1097-2765(01)00220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H, Levis R, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. BDGP gene disruption project: transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt W, Chen C, Chen Q, Sudol M, McNeill H, Pan D, Kopp A, Halder G. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene. 2014;33:1218–1228. doi: 10.1038/onc.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudinot P, van der Aa L, Jouneau L, Du Pasquier L, Pontarotti P, Briolat V, Benmansour A, Levraud J. Origin and evolution of TRIM proteins: new insights from the complete TRIM repertoire of zebrafish and pufferfish. PLoS One. 2011;6:e22022. doi: 10.1371/journal.pone.0022022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky B, Long M. New genes as drivers of phenotypic evolution. Nat Rev Genet. 2013;14:645–660. doi: 10.1038/nrg3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ni X, Krinsky B, Zhang Y, Vibranovski M, White KP, Long M. Reshaping of global gene expression networks and sex-biased gene expression by integration of a young gene. EMBO J. 2012;31:2798–2809. doi: 10.1038/emboj.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Spletter M, Ni X, White KP, Luo L, Long M. Frequent recent origination of brain genes shaped the evolution of foraging behavior in Drosophila. Cell Rep. 2012;1:118–132. doi: 10.1016/j.celrep.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang Y, Long M. New genes in Drosophila quickly become essential. Science. 2010;330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM. Evo-devo: variations on ancestral themes. Cell. 2008;132:185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R, Joazeiro C. Ring domain E3 ubiquitin ligases. Ann Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Drier E, Huang L, Steward R. Nuclear import of the Drosophila Rel protein Dorsal is regulated by phosphorylation. Genes Dev. 1999;13:556–568. doi: 10.1101/gad.13.5.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Inui M, Newfeld S. Regulation of TGF-β signal transduction by mono- and deubiquitylation of Smads. FEBS Lett. 2012;586:1913–1920. doi: 10.1016/j.febslet.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. FAM/USP9X a deubiquitinating enzyme in TGF-β signaling controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Erdelyi M, Szabad J. Isolation and characterization of dominant female sterile mutations of Drosophila. I. Mutations on the third chromosome. Genetics. 1989;122:111–127. doi: 10.1093/genetics/122.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Anderson KV. Dpp acts as a morphogen to organize dorsal–ventral pattern in the Drosophila embryo. Cell. 1992;71:451–461. doi: 10.1016/0092-8674(92)90514-d. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize J, Rubin G, Lehmann R. The fat facets gene is required for Drosophila eye and embryo development. Development. 1992;116:985–1000. doi: 10.1242/dev.116.4.985. [DOI] [PubMed] [Google Scholar]
- Gao S, Alarcón C, Sapkota G, Rahman S, Chen P, Goerner N, Macias M, Erdjument-Bromage H, Tempst P, Massagué J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder N, de Cuevas M, Spradling A. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 2000;127:4253–4264. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. Part played by recurrent mutation in evolution. Am Nat. 1933;67:5–19. [Google Scholar]
- Holley S, Jackson P, Sasaim Y, Lum B, De Robertis EM, Hoffmann FM, Ferguson EL. A conserved system for dorsal–ventral patterning in insects and vertebrates involving sog and chordin. Nature. 1995;376:249–253. doi: 10.1038/376249a0. [DOI] [PubMed] [Google Scholar]
- Holley S, Neul J, Attisano L, Wrana J, Sasai Y, O’Connor MB, De Robertis EM, Ferguson EL. The Xenopus dorsalizing factor Noggin ventralizes Drosophila embryos by preventing Dpp from activating its receptor. Cell. 1996;86:607–617. doi: 10.1016/s0092-8674(00)80134-8. [DOI] [PubMed] [Google Scholar]
- Hudson J, Podos S, Keith K, Simpson S, Ferguson EL. The Drosophila Medea gene is required downstream of Dpp and encodes a functional homolog of human Smad4. Development. 1998;125:1407–1420. doi: 10.1242/dev.125.8.1407. [DOI] [PubMed] [Google Scholar]
- Huet F, Lu J, Myrick K, Baugh L, Crosby M, Gelbart W. A deletion-generator compound element allows saturation analysis for genomewide phenotypic annotation. Proc Natl Acad Sci U S A. 2002;99:9948–9953. doi: 10.1073/pnas.142310099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Sato K, Koganezawa M, Ote M, Matsumoto K, Hama C, Yamamoto D. Fruitless recruits two antagonistic chromatin factors to establish single-neuron sexual dimorphism. Cell. 2012;149:1327–1338. doi: 10.1016/j.cell.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Kahlem P, Newfeld S. Informatics approaches to understanding TGF-β pathway regulation. Development. 2009;136:3729–3740. doi: 10.1242/dev.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetchoumian K, Teletin M, Mark M, Lerouge T, Cerviño M, Oulad-Abdelghani M, Chambon P, Losson R. TIF1-δ a novel HP1-interacting member of the TIF1 family expressed by elongating spermatids. J Biol Chem. 2004;279:48329–48341. doi: 10.1074/jbc.M404779200. [DOI] [PubMed] [Google Scholar]
- Konikoff C, Wisotzkey R, Stinchfield M, Newfeld SJ. Distinct molecular evolutionary mechanisms underlie the functional diversification of the Wnt and TGF-β pathways. J Mol Evol. 2010;70:303–312. doi: 10.1007/s00239-010-9337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Nielsen A, Garnier J, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1-α and TIF1-β in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Lin X, Liang M, Brunicardi F, ten Dijke P, Chen Z, Choi K, Feng XH. dSmurf selectively degrades Dpp-activated Mad and its overexpression disrupts imaginal disc development. J Biol Chem. 2003;278:26307–26310. doi: 10.1074/jbc.C300028200. [DOI] [PubMed] [Google Scholar]
- Long M, VanKuren N, Chen S, Vibranovski M. New gene evolution: little did we know. Annu Rev Genet. 2013;47:325–351. doi: 10.1146/annurev-genet-111212-133301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I. Origin and diversification of TRIM ubiquitin ligases. PLoS One. 2012;7:e50030. doi: 10.1371/journal.pone.0050030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez R, Singer M, Takaesu N, Waldrip W, Kraytsberg Y, Newfeld SJ. Transgenic analysis of the Smad family of TGF-β signal transducers suggests new roles and new interactions between family members. Genetics. 2001;157:1639–1648. doi: 10.1093/genetics/157.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold S, Leyland P, Seal R, Goodman J, Thurmond J, Strelets V, Wilson R, FlyBase Consortium FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751–D757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe B, Hom S, Aberle H, Fetter R, Marques G, Haerry T, Wan H, O’Connor MB, Goodman CS, Haghighi AP. Highwire regulates presynaptic BMP signaling essential for synaptic growth. Neuron. 2004;41:891–905. doi: 10.1016/s0896-6273(04)00073-x. [DOI] [PubMed] [Google Scholar]
- Morsut L, Yan K, Enzo E, Aragona M, Soligo SM, Wendling O, Mark M, Khetchoumian K, Bressan G, Chambon P, et al. Negative control of Smad activity by Ecto/Tif1-γ patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Bar duplication. Science. 1936;83:528–530. doi: 10.1126/science.83.2161.528-a. [DOI] [PubMed] [Google Scholar]
- Newfeld SJ, Wisotzkey R, Kumar S. Molecular evolution of a developmental pathway: phylogenetic analyses of TGF-β family ligands, receptors and Smad signal transducers. Genetics. 1999;152:783–795. doi: 10.1093/genetics/152.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A, Ortiz J, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with HP1 family members and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Tanikawa J, Akimaru H, Kanei-Ishii C, Ichikawa-Iwata E, Khan M, Ito H, Ishii S. Oncogenic activation of c-Myb correlates with a loss of negative regulation by TIF1-β and Ski. J Biol Chem. 2004;279:16715–16726. doi: 10.1074/jbc.M313069200. [DOI] [PubMed] [Google Scholar]
- O’Connor MB, Umulis D, Othmer H, Blair S. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podos S, Hanson K, Wang Y, Ferguson EL. The dSmurf ubiquitin ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev Cell. 2001;1:567–578. doi: 10.1016/s1534-5807(01)00057-0. [DOI] [PubMed] [Google Scholar]
- Pritchard D, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 1996;10:1131–1142. doi: 10.1101/gad.10.9.1131. [DOI] [PubMed] [Google Scholar]
- Protas M, Trontelj P, Patel NH. Genetic basis of eye and pigment loss in the cave crustacean Asellus aquaticus. Proc Natl Acad Sci U S A. 2011;108:5702–5707. doi: 10.1073/pnas.1013850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale E, Müller M, Rödelsperger C, Sommer RJ. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell. 2013;155:922–933. doi: 10.1016/j.cell.2013.09.054. [DOI] [PubMed] [Google Scholar]
- Ray R, Arora K, Nusslein-Volhard C, Gelbart W. The control of cell fate along the dorsal–ventral axis of the Drosophila embryo. Development. 1991;113:35–54. doi: 10.1242/dev.113.1.35. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, vanderMark P, Ayres D, Darling A, Hohna S, Larget B, Liu L, Suchard M, Huelsenbeck J. MrBayes3.2: efficient phylogenetic inference and model choice across a large model space. Syst Biol. 2011;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross B, Rosin L, Thomae A, Hiatt M, Vermaak D, de la Cruz A, Imhof A, Mellone B, Malik HS. Stepwise evolution of essential centromere function in a Drosophila neogene. Science. 2013;340:1211–1214. doi: 10.1126/science.1234393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Stein D, Nusslein-Volhard C. A gradient of nuclear localization of Dorsal determines dorsoventral pattern in the Drosophila embryo. Cell. 1989;59:1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. Drosophila Nedd4 regulates endocytosis of Notch and suppresses its ligand-independent activation. Curr Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups with distinct evolutionary properties. BMC Evol Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Hudson K, Lin T, Anderson KV. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal–ventral polarity in the Drosophila embryo. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- Shimmi O, Umulis D, Othmer H, O’Connor MB. Facilitated transport of Dpp/Scw by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnikova T. Bootstrap method of interior-branch test for phylogenetic trees. Mol Biol Evol. 1996;13:605–611. doi: 10.1093/oxfordjournals.molbev.a025620. [DOI] [PubMed] [Google Scholar]
- Spencer F, Hoffmann FM, Gelbart W. Decapentaplegic: a gene complex affecting morphogenesis in Drosophila. Cell. 1982;28:451–461. doi: 10.1016/0092-8674(82)90199-4. [DOI] [PubMed] [Google Scholar]
- Steward R. Relocalization of the Dorsal protein from the cytoplasm to the nucleus correlates with its function. Cell. 1989;59:1179–1188. doi: 10.1016/0092-8674(89)90773-3. [DOI] [PubMed] [Google Scholar]
- Stinchfield M, Takaesu NT, Quijano JC, Castillo A, Tiusanen N, Shimmi O, Enzo E, Dupont S, Piccolo S, Newfeld SJ. Fat facets deubiquitylation of Medea/Smad4 modulates interpretation of a Dpp morphogen gradient. Development. 2012;139:2721–2729. doi: 10.1242/dev.077206. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman A, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes T, Westwood J, Smibert C, Lipshitz H. Smaug is a regulator of maternal mRNA destabilization and its translation is activated by Pangu. Dev Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Takaesu NT, Bulanin D, Johnson A, Orenic T, Newfeld SJ. A combinatorial enhancer recognized by Mad, TCF and Brinker first activates then represses dpp expression in the posterior spiracles of Drosophila. Dev Biol. 2008;313:829–843. doi: 10.1016/j.ydbio.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler D, Li W, Zhuo N, Pellock B, Baker NE. Genes affecting cell competition in Drosophila. Genetics. 2007;175:643–657. doi: 10.1534/genetics.106.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doren M, Williamson A, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Wan H, DiAntonio A, Fetter R, Bergstrom K, Strauss R, Goodman CS. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Tao Y, Reim I, Gajewski K, Frasch M, Schulz R. Expression, regulation and requirement of Toll in dorsal vessel formation in Drosophila. Mol Cell Biol. 2005;25:4200–4210. doi: 10.1128/MCB.25.10.4200-4210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen A, Steward R. Dissociation of Dorsal–Cactus complex and phosphorylation of Dorsal protein correlates with nuclear localization of Dorsal. J Cell Biol. 1993;123:523–534. doi: 10.1083/jcb.123.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K, Ray R, Gelbart W. An activity gradient of dpp is necessary for the specification of dorsal pattern in the Drosophila embryo. Development. 1993;117:807–822. doi: 10.1242/dev.117.2.807. [DOI] [PubMed] [Google Scholar]
- Wisotzkey R, Konikoff C, Newfeld SJ. Hippo pathway phylogenetics predicts monoubiquitylation of Salvador and Merlin/Nf2. PLoS One. 2012;7:e51599. doi: 10.1371/journal.pone.0051599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- Wu X, Sharp PA. Divergent transcription: a driving force for new gene origination? Cell. 2013;155:990–996. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.