Abstract
Chemically defended plant tissues present formidable barriers to herbivores. Although mechanisms to resist plant defenses have been identified in ancient herbivorous lineages, adaptations to overcome plant defenses during transitions to herbivory remain relatively unexplored. The fly genus Scaptomyza is nested within the genus Drosophila and includes species that feed on the living tissue of mustard plants (Brassicaceae), yet this lineage is derived from microbe-feeding ancestors. We found that mustard-feeding Scaptomyza species and microbe-feeding Drosophila melanogaster detoxify mustard oils, the primary chemical defenses in the Brassicaceae, using the widely conserved mercapturic acid pathway. This detoxification strategy differs from other specialist herbivores of mustard plants, which possess derived mechanisms to obviate mustard oil formation. To investigate whether mustard feeding is coupled with evolution in the mercapturic acid pathway, we profiled functional and molecular evolutionary changes in the enzyme glutathione S-transferase D1 (GSTD1), which catalyzes the first step of the mercapturic acid pathway and is induced by mustard defense products in Scaptomyza. GSTD1 acquired elevated activity against mustard oils in one mustard-feeding Scaptomyza species in which GstD1 was duplicated. Structural analysis and mutagenesis revealed that substitutions at conserved residues within and near the substrate-binding cleft account for most of this increase in activity against mustard oils. Functional evolution of GSTD1 was coupled with signatures of episodic positive selection in GstD1 after the evolution of herbivory. Overall, we found that preexisting functions of generalized detoxification systems, and their refinement by natural selection, could play a central role in the evolution of herbivory.
Keywords: plant–herbivore interactions, glutathione S-transferase, detoxification, gene duplication, Drosophila, isothiocyanate
Introduction
Herbivorous insects are remarkably successful: Approximately half of all insect species are herbivorous (Schoonhoven et al. 2005), and herbivorous lineages diversify more rapidly than their nonherbivorous relatives (Mitter et al. 1988). Paradoxically, however, evolutionary transitions to herbivory have occurred in only 9 of 30 extant insect orders (Mitter et al. 1988), despite the evolutionary success of established herbivore lineages. This suggests that feeding on living plant tissue is an evolutionary hurdle that is difficult to overcome.
Plant chemical defenses have been proposed as a major barrier to the evolution of herbivory (Strong et al. 1984; Mitter et al. 1988). Evaluating this hypothesis requires an understanding of the mechanisms enabling nascent herbivores to detoxify plant defensive compounds. Specifically, it is unclear if overcoming plant defensive toxins during evolutionary transitions to herbivory requires metabolic innovations, which may explain why transitions to herbivory are absent in many insect orders, or if newly evolved herbivores can detoxify plant toxins using generalized detoxification systems present in their nonherbivorous ancestors. Our knowledge of adaptations during transitions to herbivory is limited because most herbivorous insect species are within lineages where herbivory arose greater than 150 Ma (Labandeira 1998). Comparative genetic, metabolic, and physiological studies of younger herbivorous lineages and their nonherbivorous relatives therefore hold promise to offer insight into the adaptations facilitating survival exclusively on living plant tissues.
We investigated the mechanism to detoxify plant defensive compounds employed by recently evolved herbivores in the genus Scaptomyza (Scaptomyza flava, S. nigrita, and closely related lineages). Larvae of these species are leaf miners that consume and live within leaves of mustard plants (Brassicaceae), including the model plant Arabidopsis thaliana. Mustard-feeding Scaptomyza are nested phylogenetically within the primarily microbe-feeding Drosophila, and the ancestor of the mustard-feeding clade transitioned to feeding on living plant tissue approximately 6–15 Ma (Whiteman et al. 2012; Lapoint et al. 2013) (fig. 1). Scaptomyza flava feeds on a much broader range of mustard species than S. nigrita, which feeds on only one species of mustard (Wheeler 1952; Wheeler and Takada 1966; Collinge and Louda 1989; Louda and Collinge 1992; Martin 2004; Whiteman et al. 2011) (supplementary note, Supplementary Material online). Differences in the breadth of plant-derived toxins encountered by each species may result in different selective pressures on detoxification pathways and enzymes.
Fig. 1.

An herbivorous lineage nested within the genus Drosophila. (A) Scaptomyza flava larvae and adults complete their life cycle on mustard plants (Brassicaceae), shown on Arabidopsis thaliana. Adult female species pierce leaves with their ovipositors, consume the exudates, and lay eggs (i); larvae form leaf mines after eclosion (ii) and can move between leaves (iii); pupation often occurs in leaf mines (iv); and adults mate on host plants (v). The life cycle of S. nigrita on its host plant, Cardamine cordifolia, is similar. (B) Feeding habit for representative Drosophila and Scaptomyza species indicated on the dated phylogeny of Whiteman et al. (2012).
Although nonherbivorous drosophilids likely encounter toxins constitutively present in feeding substrates, leaf-mining larvae of S. flava and S. nigrita consume defensive compounds that accumulate to high levels in mustard leaves in response to feeding damage (Whiteman et al. 2011, 2012). The primary chemical defense compounds in mustards are derived from glucosinolates, nontoxic thioglucosides that are activated by myrosinases to form toxic isothiocyanates (ITCs or mustard oils) and other hydrolysis products upon tissue damage (Halkier and Gershenzon 2006). ITCs are highly electrophilic, and their insecticidal activity (Winde and Wittstock 2011) is thought to result primarily from disruption of proteins through conjugation to exposed nucleophilic residues (Mi et al. 2011) and cleavage of disulfide bonds (Kawakishi and Kaneko 1985). Although drosophilids utilize a tremendous diversity of feeding substrates (Markow and O'Grady 2005; Lapoint et al. 2013), glucosinolate production is narrowly restricted almost exclusively to mustards and related plant families in the order Brassicales, and glucosinolates are not known to be produced by microbes (Halkier and Gershenzon 2006). Thus, glucosinolate defenses were likely a novel dietary component associated with the transition to mustard feeding within Scaptomyza.
Interactions between herbivores and glucosinolate-bearing host plants have been a fruitful model system for understanding mechanisms of resistance to plant defensive chemicals (Winde and Wittstock 2011). However, the initial adaptations that confer resistance to glucosinolate defenses during the transition to mustard feeding—either from herbivorous or nonherbivorous ancestors—are unknown. On one hand, previously studied insects that specialize on mustards, including representatives from insect orders that span over 300 million years of divergence from a common ancestor, have evolved efficient mechanisms to sequester glucosinolates or otherwise prevent the formation of ITCs (sawfly, Athalia rosae, Muller et al. 2001; harlequin bug, Murgantia histrionica, Aliabadi et al. 2002; diamondback moth, Plutella xylostella, Ratzka et al. 2002; cabbage aphid, Brevicoryne brassicae, Kazana et al. 2007; cabbage butterflies, Pieridae, Wheat et al. 2007). These mustard specialists are relatively insensitive to glucosinolate defenses (Raybould and Moyes 2001; Muller et al. 2010). Accordingly, transitions to mustard feeding might require the evolution of specific adaptations to prevent ITC formation. On the other hand, some insects with broader host ranges that occasionally feed on mustards metabolize ITCs using widely conserved, generalized detoxification mechanisms (Wadleigh and Yu 1988; Schramm et al. 2012) and remain susceptible to glucosinolate defenses (Winde and Wittstock 2011), although these studies have focused on Lepidoptera with broad host plant ranges. Generalized detoxification mechanisms might provide an initial route to resist glucosinolate defenses in insects transitioning to herbivory of mustards.
Glucosinolates slow S. flava larval development (Whiteman et al. 2011, 2012). Therefore, we hypothesized that mustard-feeding Scaptomyza would lack a mechanism to prevent ITC formation, unlike mustard specialists within more ancient herbivorous lineages. To test this hypothesis, we performed metabolic profiling to search for ITCs in larvae feeding on glucosinolate-bearing leaves. Second, we hypothesized that mustard-feeding Scaptomyza would detoxify ITCs using a generalized pathway present in their ancestors. We searched for ITC-derived metabolites known to be produced through generalized detoxification pathways, and for novel ITC metabolites, following glucosinolate or ITC ingestion in mustard-feeding Scaptomyza and in the nonherbivorous relative Drosophila melanogaster. We found that both nonherbivorous and mustard-specialist drosophilid flies use the widely conserved mercapturic acid pathway to detoxify mustard oils.
Given that mustard-feeding Scaptomyza detoxify ITCs using the mercapturic acid pathway, we predicted that an enzyme controlling the pathway’s substrate specificity would evolve enhanced activity against ITCs following the transition to mustard feeding. Glutathione S-transferases (GSTs) are the only enzymes known to catalyze the first step of the mercapturic acid pathway, and thus mediate the pathway’s specificity for different toxins (Habig et al. 1974). GSTs form a large multigene family in metazoans. Up to 45 genes encoding GSTs are found in single Drosophila species with sequenced genomes (Low et al. 2007). Although multiple GSTs, downstream enzymes in the mercapturic acid pathway, and toxin transport enzymes may have been targets of natural selection driven by dietary toxins during the evolution of herbivory, glutathione S-transferase D1 (GSTD1) could play a particularly important role in the detoxification of ITCs. In D. melanogaster, GSTD1 has exceptionally broad substrate specificity, and it is one of the most efficient GSTs catalyzing the conjugation of ITCs to glutathione in vitro (Saisawang et al. 2012). GSTD1 has also been implicated in putatively adaptive responses to environmental toxins in drosophilids, such as resistance to insecticide (Low et al. 2010) and to toxins present in the larval substrates of cactophilic Drosophila species (Matzkin 2008). GstD1 expression in S. flava is induced by ingestion of jasmonate-dependent plant defenses, which include glucosinolates (Whiteman et al. 2011). Because GstD1 evolves under strong purifying selection in Drosophila (Low et al. 2007), characterizing the function and evolution of GstD1 in mustard-feeding Scaptomyza could offer insight into how ecologically important, but conserved, detoxification enzymes evolve during transitions to herbivory.
We conducted in vitro enzyme activity assays to test whether GSTD1 from mustard-feeding Scaptomyza detoxify ITCs more rapidly than orthologous copies from other drosophilids. Then, we solved the crystal structure of a duplicated GSTD1 enzyme from S. nigrita to identify amino acid substitutions in enzyme regions likely to affect substrate specificity, and we tested whether these residues affect the rate of ITC detoxification by GSTD1 in vitro. Finally, we tested whether GstD1 has evolved under neutral, purifying, and/or positive selection in the mustard-feeding Scaptomyza since their divergence from a nonherbivorous Scaptomyza ancestor. Our data suggest that the transition to herbivory, and interactions with plant toxins in particular, can drive the functional evolution of generalized detoxification enzymes. However, additional studies are needed to test the hypothesis that GSTD1 facilitates resistance to mustard oils in Scaptomyza.
Results
Metabolism of Glucosinolates
Previous studies found that chewing herbivores specializing on mustards block formation of toxic ITCs from glucosinolates during feeding (Ratzka et al. 2002; Wheat et al. 2007; Winde and Wittstock 2011). To determine whether a more recently evolved mustard specialist also prevents ITC formation, we profiled glucosinolate metabolites in S. flava larvae reared on glucosinolate-deficient A. thaliana plants (Whiteman et al. 2012) infused with radiolabeled 4-methylsulfinylbutyl glucosinolate, the predominant glucosinolate in A. thaliana accession Col-0 (Brown et al. 2003). Using this unbiased approach, we detected two glucosinolate hydrolysis products in the larvae of S. flava, ITCs and nitriles, and no intact glucosinolates (fig. 2A and supplementary fig. S1A, Supplementary Material online).
Fig. 2.
A common mechanism for ITC detoxification in mustard-feeding Scaptomyza. (A) Distribution of 4msob-glucosinolate-derived products detected in S. flava (Massachusetts) larvae feeding on leaves from A. thaliana glucosinolate-knockout mutants infused with [14C]-4msob-glucosinolate. Mercapturic acid products were the only insect-transformed metabolites detected. NAC: N-acetylcysteine. (B) Inferred route of ITC metabolism in Scaptomyza through conjugation of ITCs to GSH and subsequent transformation using the mercapturic acid pathway. (C) Left: Bayesian 50% majority-rule consensus phylogeny of mustard-feeding Scaptomyza from a partitioned alignment of six genes, rooted with S. pallida. Bayesian posterior probabilities and maximum-likelihood bootstrap values are indicated above and below each node, respectively. Four clades were designated using a maximum divergence cutoff of 3% at the genes COI and COII. Right: ITC-derived metabolites detected in the larvae from four mustard-feeding Scaptomyza lineages using HPLC-MS/MS. The chromatogram for each lineage shows overlaid MS/MS ion traces from compounds 1 to 5. Profiled lineages are indicated by dotted lines. Larvae were fed A. thaliana in the laboratory and profiled for 4-msob-glucosinolate derivatives with the exception of S. nigrita (CO), which could not be reared in the laboratory. Wild-collected S. nigrita (CO) larvae were therefore profiled for derivatives of PE glucosinolate (as well as isopropyl, isobutyl, and sec-butyl glucosinolates, see supplementary fig. S1, Supplementary Material online), which is present in their host plant. Unlike 4msob-ITC, the free ITCs formed from these glucosinolates cannot be detected using the method we employed, and thus only ITC-derived metabolites were profiled in S. nigrita larvae. Numbers above peaks correspond to compounds in panel (B). Collection localities in the United States are indicated following species/lineage names using abbreviated state names; NLD: Netherlands.
In addition to free ITCs, we detected ITC-derived metabolites from the mercapturic acid pathway in the larvae of S. flava (fig. 2A and supplementary fig. S1A, Supplementary Material online). Products in the mercapturic acid pathway are formed through the conjugation of the tripeptide glutathione (GSH) to a toxin and subsequent hydrolytic modification (Boyland and Chasseaud 1969) (fig. 2B). To determine whether ITCs are detoxified using the mercapturic acid pathway across mustard-feeding Scaptomyza, we first inferred the evolutionary relationship among eight mustard-feeding Scaptomyza lineages (fig. 2C). Because the taxonomy of mustard-feeding Scaptomyza is not well resolved, we used a maximum cutoff of 3% sequence divergence at the mitochondrial genes COI and COII to delimit four major clades. We then performed a targeted search for mercapturic acid products in the larvae from one lineage from each clade. Larvae in all four lineages metabolized ITCs using the mercapturic acid pathway (fig. 2C and supplementary fig. S1B and C, Supplementary Material online). We also detected ITC-derived products arising from the mercapturic acid pathway in the adults of D. melanogaster following experimental consumption of ITCs (supplementary fig. S1D, Supplementary Material online).
Duplication of GSTD1, a Candidate ITC Detoxification Enzyme, in Mustard-Feeding Scaptomyza
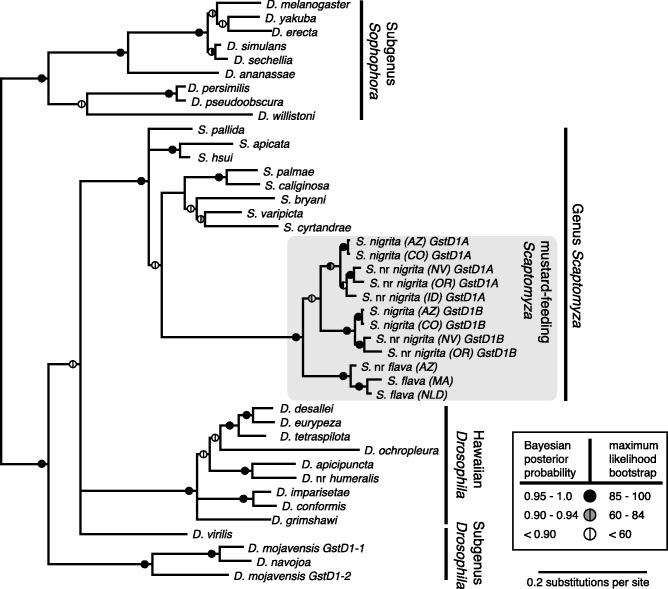
Although GstD1 is present as a single copy in most Drosophila species (Low et al. 2010), we discovered a novel duplication of GstD1 in S. nigrita, and we have named these paralogs GstD1A and GstD1B, respectively (fig. 3). The presence of both paralogs could be verified in many S. nigrita populations in North America (supplementary table S1, Supplementary Material online) and in two of the three lineages in clade II using gene-specific polymerase chain reaction (PCR) primers and sequencing (fig. 3). However, only a single functional copy of GstD1 was found in S. flava from Belmont, MA, through search of a draft genome assembly, and through PCR screens in other clade III and IV lineages (supplementary materials and methods, Supplementary Material online). The most parsimonious reconciliation of the gene and species trees inferred through Bayesian phylogenetic analyses requires only a single duplication event and suggests that the duplication occurred in the ancestor of clades I and II (fig. 2C). A search for GstD1 pseudogenes in the S. flava draft genome assembly does not refute this scenario: Although the assembly contained putative GstD1 pseudogenes, none nested phylogenetically with either GstD1 paralog from S. nigrita (supplementary fig. S2, Supplementary Material online).
Fig. 3.
Phylogeny of GstD1 across Scaptomyza and Drosophila. Analyses were performed using a nucleotide alignment of the full GstD1 coding sequence (630 bp) partitioned by codon position with the GTR + G substitution model. Bayesian posterior probabilities (generated using MrBayes) and maximum-likelihood bootstrap values (generated using RAxML) are shown on the Bayesian 50% majority-rule consensus tree. “GstD1A” and “GstD1B” refer to paralogous copies of GstD1 within the mustard-feeding Scaptomyza. The tree was rooted using the divergence between the subgenera Sophophora and Drosophila.
An estimate of the timing of the duplication giving rise to GstD1A and GstD1B lends further support to the hypothesis that the duplication occurred in the ancestor of clade I and II, after the ancestors of S. flava and S. nigrita diverged. Maximum-likelihood estimates of nucleotide divergence for all pairwise comparisons of GstD1A and GstD1B sequences in our study ranged from 0.027 to 0.074 substitutions per synonymous site. Given that pairwise sequence divergence at putatively neutrally evolving sites is equal to twice the product of the mutation rate and divergence date, we estimated the age of the GstD1 duplication using a long-term mutation rate for the Hawaiian Drosophila (Tamura et al. 2004). The estimated duplication date, 1.2–3.3 Ma, is more recent than the divergence date of S. flava and S. nigrita inferred from a fossil-calibrated phylogeny (Lapoint et al. 2013). Although gene conversion events between GstD1A and GstD1B could bias this estimate toward a younger date, population genetic analyses (Betran et al. 1997) found no evidence for gene conversion among GstD1A and GstD1B in S. nigrita. A phylogenetic analysis (Pond et al. 2006) did not suggest that recombination has occurred between GstD1A and GstD1B since divergence of clades I and II. However, neither analysis is able to detect gene conversion events prior to the divergence of clades I and II.
Functional Evolution of GSTD1 in Mustard-Feeding Scaptomyza
If GSTD1 plays a role in ITC detoxification, GSTD1 from mustard-feeding Scaptomyza might be expected to metabolize ITCs more efficiently than orthologs from species that do not feed on mustard plants. We compared activity profiles of heterologously expressed and purified GSTD1 protein from S. flava (SflaGSTD1; sequenced from the Massachusetts population), S. nigrita (SnigGSTD1A and SnigGSTD1B; sequenced from the Colorado population), and eight nonherbivorous Scaptomyza and Drosophila species that diverged from a common ancestor approximately 70 Ma (Whiteman et al. 2012). In in vitro assays, SnigGSTD1A conjugated seven of the eight structurally diverse ITCs to GSH more rapidly than homologous GSTD1 enzymes from other species, including the ITCs derived from the five most abundant ITC-yielding glucosinolates in the host plant of S. nigrita (Rodman and Louda 1985) (fig. 4 and supplementary table S2, Supplementary Material online). GSTD1A enzymes from two other mustard-feeding lineages, S. nr. nigrita (OR) and S. nr. nigrita (ID), also had higher activity against ITCs than GSTD1 from microbe-feeding Drosophila (supplementary fig. S3, Supplementary Material online). SflaGSTD1 exhibited high specific activity only for phenethyl ITC (PEITC), but not other ITCs (fig. 4).
Fig. 4.
Scaptomyza nigrita GSTD1A is the most efficient GSTD1 ortholog at detoxifying ITCs across the Drosophilidae. Specific activity ± SEM, the velocity at which an enzyme conjugates a substrate to GSH under fixed assay conditions, was determined for eight ITCs (N = 5–7 replicates across 2–3 independent experiments); specific activity values and statistically significant pairwise comparisons are reported in supplementary table S2, Supplementary Material online. Cell shading reflects standard deviations of a measured value from the column mean. Michaelis–Menten kinetic constants ± SEM were determined for BITC by nonlinear regression using variable ITC concentrations at saturating GSH concentration (N ≥ 3 independent replicates). Bolded values are not significantly different than values in the same column for SnigGSTD1A (Mann–Whitney U test). The electrophilic carbon is circled in each ITC structure. kcat, the substrate turnover rate at Vmax; Km, the substrate concentration at which the reaction rate is half of Vmax.
Aromatic ITCs, such as benzyl ITC (BITC), are exceptionally toxic (Wadleigh and Yu 1988; Winde and Wittstock 2011). Their glucosinolate precursors are found across diverse mustard species, including in Cardamine cordifolia, the only known host plant of S. nigrita (Fahey et al. 2001). Therefore, we conducted a more extensive investigation on the efficiency of BITC conjugation to GSH by GSTD1 from S. nigrita and other drosophilids. We measured the activity of each enzyme across a range of BITC concentrations under a saturating GSH concentration. Two kinetic constants can be determined from this data: kcat, the maximum BITC turnover rate per unit of enzyme, and Km, the BITC concentration at which reaction velocity is half of the maximum velocity for that enzyme. In some cases, SnigGSTD1A did not have more efficient values of kcat or Km than GSTD1 enzymes from other species (one-sided Mann–Whitney U test, P < 0.05 for seven of the ten pairwise comparisons of kcat and eight of the ten comparisons of Km after correcting for multiple tests; fig. 4). However, the specificity constant, kcat/Km, which predicts the rate at which BITC is conjugated to GSH under the low BITC and high GSH concentrations likely predominating in cells (Zhang 2001), was 200–450% higher for SnigGSTD1A than orthologous GSTD1 enzymes (Mann–Whitney U test, P < 0.01 for all comparisons between SnigGSTD1A and other enzymes; fig. 4). Results for PEITC, another aromatic ITC, were similar (supplementary fig. S4, Supplementary Material online). Our activity measurements suggest that GSTD1 could increase the rate of BITC detoxification by 2–3 orders of magnitude relative to the uncatalyzed reaction under physiologically relevant conditions, even in drosophilid species that do not naturally encounter ITCs (supplementary table S3, Supplementary Material online). However, GSTs other than GSTD1 may be important for ITC detoxification in vivo.
Activity assays using three structurally diverse non-ITC substrates of GSTs did not reveal a loss of substrate breadth or a change in affinity for GSH (reflected in Km) by SnigGSTD1A (supplementary fig. S4, Supplementary Material online). On the other hand, SnigGSTD1B exhibited little activity against ITCs or these other substrates, and also had a lower affinity (higher Km) for GSH (fig. 4 and supplementary fig. S4, Supplementary Material online).
Molecular Basis of Elevated Activity Against ITCs in a Paralogous GSTD1 Enzyme from Mustard-Feeding Scaptomyza
To determine how substitutions in SnigGSTD1A affect enzyme topology, we used X-ray crystallography to solve the crystal structure of SnigGSTD1A in complex with GSH (fig. 5A and B and supplementary table S4, Supplementary Material online). As is typical for GSTs, GSH was bound adjacent to a hydrophobic toxin-binding pocket (fig. 5B). Comparison of SnigGSTD1A with the structure of D. melanogaster GSTD1 (DmelGSTD1, PDB ID: 3EIN) (Low et al. 2010) revealed no dramatic changes to the structure of the protein (RMSD = 0.45 Å, Cα atoms).
Fig. 5.
A single amino acid in the GSTD1 hydrophobic binding cleft, and additional residues outside the cleft, affect the rate of ITC detoxification by SnigGSTD1A and DmelGSTD1 in vitro. (A) Novel structure of the SnigGSTD1A homodimer cocrystallized with GSH. (B) Surface residues in the GSH binding site (purple) and hydrophobic toxin-binding site (orange) are represented as stick models. (C) Amino acid differences between SnigGSTD1A and other drosophilid GSTD1 enzymes at sites incorporated into site-directed mutagenesis experiments. (D–F) Location of amino acid residues from panel (C) in SnigGSTD1A (yellow) and DmelGSTD1 (blue). In SnigGSTD1A, these residues form a novel hydrogen bond network (D), rearrange packing in a hydrophobic zipper motif (E), and form a novel hydrogen bond in a α-helix near the binding cleft (F). Site 166 is shown in all panels. (G) Kinetic constants of purified enzymes with candidate amino acids (panel C) switched between SnigGSTD1A and DmelGSTD1. For each purified enzyme, Michaelis–Menten kinetic constants ± SEM were determined for BITC by nonlinear regression using variable ITC concentrations at saturating GSH concentration (N = 4 independent replicates). Groupings of enzymes with statistically indistinguishable specificity constants (kcat/Km), determined using a one-way analysis of variance and Tukey’s post hoc test, are indicated with letters a–e. kcat, substrate turnover rate at Vmax; Km, substrate concentration at which reaction rate is half of Vmax.
SnigGSTD1A differed from orthologous GSTD1 enzymes in other drosophilids at 20–29 (of 209) amino acids. To identify candidates among these amino acid differences that may confer enhanced activity of SnigGSTD1 against ITCs, we identified amino acid substitutions in SnigGSTD1A at conserved sites in regions known to impact substrate specificity and catalytic efficiency in related Delta-class GSTs (fig 5C), described in detail below. Because the activity of GSTD1 in the ancestor of S. nigrita prior to the duplication of GstD1 is unknown, we considered unique substitutions in SnigGSTD1A and substitutions shared by SnigGSTD1A and SnigGSTD1B as potential candidates underlying elevated activity of SnigGSTD1A against ITCs. Although resurrecting and performing site-directed mutagenesis on inferred ancestral proteins could offer insight into whether GSTD1 acquired higher activity against ITCs before or after the duplication in ancestral Scaptomyza, a marginal reconstruction approach (Yang 2007) could not infer the ancestral states at four amino acids (posterior probability ranging from 0.5 to 0.7). Therefore, we instead performed reciprocal replacements of candidate amino acids between SnigGSTD1A and DmelGSTD1—either singly or in combination with other, nearby amino acids—and we measured the rate at which these modified enzymes metabolize ITCs.
Hydrophobic Binding Cleft
Changes in substrate specificity and catalytic efficiency in insect Delta-class GSTs often arise from substitutions to residues contributing to a hydrophobic toxin-binding cleft, termed the H-site (Vararattanavech et al. 2006; Wongsantichon et al. 2010). Residues forming the H-site wall (Low et al. 2010) are highly conserved in GSTD1 across drosophilids: We observed only a single substitution among 30 nonmustard-feeding species of Drosophila and Scaptomyza, with the exception of an evolutionarily labile aromatic amino acid at the periphery of the H-site. Interestingly, one amino acid substitution in the hydrophobic H-site wall was fixed in SnigGSTD1A following the GstD1 duplication event (T166S). The same site is highly conserved across GSTD1 in schizophoran flies, including in microbe-feeding species of Drosophila and Scaptomyza (fig. 5C). Adjacent H-site residues are shifted in SnigGSTD1A relative to DmelGSTD1 (F204 and Y207 in fig. 5D and F), putatively due to the loss of a methyl group contributing to the H-site wall due to the T166S substitution. Reversing the T166S substitution lowered the specificity constant (kcat/Km) for BITC in SnigGSTD1A, and introducing an S166T substitution into DmelGSTD1 increased the specificity constant (P < 0.05 for both comparisons, one-way analysis of variance with Tukey’s HSD; fig. 5G). In both cases, specificity constants for BITC were altered by approximately 50%. A similar trend was observed for specific activity measured at high concentrations of BITC, PEITC, and an aliphatic ITC derived from glucosinolates in the host plant of S. nigrita (supplementary fig. S5, Supplementary Material online). However, these changes in activity were more subtle than the changes to the specificity constant, kcat/Km, suggesting that the effect of this substitution on enzyme efficiency is magnified at low, physiologically relevant ITC concentrations.
Residues Not Exposed to the Binding Cleft
Two amino acid substitutions unique to SnigGSTD1A reorganize packing of buried residues in an aromatic zipper motif near residue 166 (fig. 5C and E). Additionally, unlike most drosophilid GSTD1 enzymes, SnigGSTD1A has a polar amino acid residue at site 202, which protrudes from α-helix 8 near the hydrophobic binding cleft (fig. 5C and F). Site 202 was identified as one of the two sites putatively evolving under positive selection across Scaptomyza and the subgenus Drosophila as a whole (supplementary table S6, Supplementary Material online). After substituting all four candidate amino acids in the zipper motif, α-helix 8, and H-site from SnigGSTD1A into DmelGSTD1, the modified DmelGSTD1 enzyme had a specificity constant (kcat/Km) for BITC that was greater than wild-type DmelGSTD1 and not different from SnigGSTD1A (fig. 5G).
Interface of Toxin and GSH Binding Clefts
GSTD1 enzymes across mustard-feeding Scaptomyza have undergone multiple substitutions at highly conserved amino acids (sites 33, 35–36) in a loop region between the toxin-binding H-site and GSH binding cleft. In SnigGSTD1A, these residues form a novel hydrogen bond network absent in the structure of DmelGSTD1 (fig. 5C and D). However, exchanging amino acids at these positions between DmelGSTD1 and SnigGSTD1A—either alone (fig. 5G) or in conjunction with the other four substitutions discussed previously (supplementary table S5, Supplementary Material online)—did not affect the activity against ITCs.
Rapid Molecular Evolution of GstD1 in Mustard-Feeding Scaptomyza
To illuminate patterns of selection driving the evolution of GstD1 across drosophilids, we used phylogenetic models of evolution that rely on the rates of nonsynonymous and synonymous substitutions expected under different selection pressures. Episodic positive selection results in an excess of normalized nonsynonymous substitutions compared with synonymous substitutions (ω > 1), whereas synonymous substitutions are fixed much more frequently than nonsynonymous substitutions under purifying selection (ω < 1).
Across the genus Scaptomyza and the subgenus Drosophila, in which Scaptomyza and the Hawaiian Drosophila are nested phylogenetically, we found a strong signal of gene-wide purifying selection in GstD1 (one-ratio model, gene-wide ω = 0.1100). A model allowing some codons to evolve under positive selection was a better fit than a model constraining all codons to evolve under purifying selection or neutral evolution (models M7 versus M8, likelihood-ratio test, P = 0.006; supplementary table S6, Supplementary Material online). Although identification of single codons under positive selection should be interpreted cautiously, there was strong support (posterior probability > 0.95) that codons at position 133 and 202 of GstD1 were evolving under positive selection across drosophilids. However, the model allowing positive selection was not a significantly better fit than a conservative null model less prone to false positives when some codons are evolving neutrally (models M8a vs. M8, P > 0.1; supplementary table S6, Supplementary Material online). Thus, conservative model comparisons did not find strong evidence for episodic positive selection (ω > 1) acting on specific codons in GstD1 across the subgenus Drosophila and genus Scaptomyza generally.
Next, we tested for an elevated rate of nonsynonymous substitutions in GstD1 in individual lineages of mustard-feeding Scaptomyza (Yang 1998). Although evolution of GstD1 was dominated by purifying selection across microbe-feeding drosophilids, ω was elevated after the evolution of herbivory in Scaptomyza (branches indicated as -A,B; -A; -B; and -F in fig. 6). Interestingly, however, ω along more recent branches of the phylogeny for SnigGstD1A, SnigGstD1B, or SflaGstD1 was not statistically different than in Drosophila and Scaptomyza as a whole. This is striking because the presence of locally adaptive or slightly deleterious nonsynonymous polymorphism, which contributes proportionally less than synonymous polymorphism to among-species divergence, would be expected to inflate ω in terminal branches for closely related lineages relative to deeper branches of the phylogeny (Peterson and Masel 2009). Although ω was elevated in GstD1 following the evolution of herbivory, only two branches showed ω > 1, and the models allowing ω > 1 along either branch did not fit significantly better than a model fixing ω = 1 (ln L = −4009.958; for -A, ln L = −4013.494 for -A,B; likelihood-ratio test P > 0.05 in both cases). More powerful tests to detect a subset of codons evolving under positive selection (ω > 1) (Zhang et al. 2005) could not be applied to individual branches because these tests may be prone to false positives when the total number of substitutions is small (Nozawa et al. 2009). Therefore, although elevated ω in mustard-feeding Scaptomyza could result from episodic positive selection affecting only a subset of codons in GstD1, phylogenetic models of variation in ω cannot rule out past relaxation of purifying selection as the cause of this pattern.
Fig. 6.

GstD1 evolved rapidly in ancestral Scaptomyza lineages following the evolution of herbivory. (A) Branches or clades tested for an elevated normalized ratio of nonsynonymous to synonymous substitutions (ω) are indicated on the gene tree of GstD1 from mustard-feeding Scaptomyza. Background branches (not shown) included all subgenus Drosophila, Hawaiian Drosophila, and genus Scaptomyza species in figure 3. (B) Branch-specific ω values, likelihood scores, and significance values for likelihood-ratio tests comparing a model allowing elevated ω in the specified foreground branch or clade (CodeML two-ratio branch model) with a model estimating a single ω across all species in the subgenus Drosophila, including the Hawaiian Drosophila, and genus Scaptomyza (CodeML one-ratio model M0, ω = 0.1100, ln L = −4015.596). Phylogenetic studies (Lapoint et al. 2013) suggest herbivory evolved along the branch labeled “-A,B,F.” Multiple test correction used the method of Benjamini and Hochberg. Filled circle indicates 0.1 < P < 0.05; *P < 0.05; **P < 0.01. ω = ∞ reflects that no synonymous changes were inferred along the branch of interest.
The McDonald–Kreitman test detects an excess of nonsynonymous divergence relative to expectations based on current patterns of polymorphism. Polarized McDonald–Kreitman tests revealed an excess of nonsynonymous substitutions in SflaGstD1, SnigGstD1A, and SnigGstD1B (table 1 and supplementary table S9, Supplementary Material online). The proportion of amino acid substitutions predicted to have been fixed by positive selection (α; Smith and Eyre-Walker 2002) exceeded 85% for each gene (table 1), although this statistic fluctuates stochastically and should be interpreted cautiously (Nei et al. 2010). Genes without roles in detoxification did not yield statistically significant McDonald–Kreitman tests or show elevated ω in S. flava or S. nigrita (supplementary tables S7 and S8, Supplementary Material online). This suggests that evidence for positive selection on SnigGSTD1A, SnigGSTD1B, and SflaGSTD1 was not due to demographic processes, such as a population bottleneck, which can leave genome-wide molecular signatures resembling positive selection. Although statistical tests for positive selection may yield false inferences when assumptions are violated, the results of our independent tests are consistent with a scenario in which the transition to herbivory in Scaptomyza is associated with positive selection driving evolution of GstD1.
Table 1.
Polarized McDonald–Kreitman Tests for GstD1 in Mustard-Feeding Scaptomyza.
| Species | Gene | Methoda | Nchrb | Psc | Dsd | Pne | Dnf | αg | Ph |
|---|---|---|---|---|---|---|---|---|---|
| S. flava | GstD1 | 1 | 20 | 8 | 7 | 0 | 11 | 1 | 0.0074* |
| S. nigrita | GstD1A | 1 | 48 | 9 | 0 | 2 | 9 | 1 | 0.0003** |
| S. nigrita | GstD1B | 1 | 36 | 8 | 4 | 2 | 8 | 0.88 | 0.0427* |
| S. flava | GstD1 | 2 | 20 | 8 | 6 | 0 | 8 | 1 | 0.0177* |
| S. nigrita | GstD1A | 2 | 48 | 9 | 0 | 2 | 5 | 1 | 0.0048* |
| S. nigrita | GstD1B | 2 | 36 | 8 | 3 | 2 | 8 | 0.91 | 0.0300* |
aMethod 1 included all sites (full 209 codons for S. nigrita; 182 codon partial gene sequence for S. flava). Method 2 excluded sites for which the codon identity in the ancestor of S. flava and S. nigrita (for S. flava) or S. nigrita GstD1A and GstD1B (for both S. nigrita paralogs) could not be inferred with >0.90 probability using maximum-likelihood reconstruction (see supplementary materials and methods, Supplementary Material online).
bNumber of chromosomes sampled.
cNumber of synonymous polymorphisms within a lineage.
dNumber of synonymous substitutions within a lineage.
eNumber of nonsynonymous polymorphisms within a lineage.
fNumber of nonsynonymous substitutions within a lineage
gEstimated proportion of nonsynonymous substitutions fixed by positive selection (α = 1 − DsPn/DnPs).
hP value from Fisher’s exact test. P values are presented without correction for multiple tests. Significance after correcting for multiple tests using the method of Benjamini and Hochberg is indicated.
*P < 0.05; **P < 0.01.
Discussion
An Ancient Metabolic Pathway Detoxifies Plant-Derived Chemicals in a Recently Evolved Herbivorous Insect Lineage
Most mustard-specialist herbivores are highly resistant to glucosinolate defenses (Raybould and Moyes 2001; Muller et al. 2010). Notably, chewing insect herbivores that specialize on mustard plants have evolved mechanisms to prevent the activation of glucosinolates into toxic ITCs (Muller et al. 2001; Ratzka et al. 2002; Wheat et al. 2007; Winde and Wittstock 2011), which otherwise occurs in mustards when leaf tissue is damaged during feeding. However, studies have focused on mustard specialist species within lineages for which the timing of the transition to herbivory is ancient or unknown. Thus, it is unclear whether highly efficient mechanisms to prevent ITC formation are necessary for evolutionary transitions to herbivory of mustards.
ITCs are highly toxic to D. melanogaster (Lichtenstein et al. 1962), a nonherbivorous relative of Scaptomyza, and may have presented a barrier to the evolution of mustard feeding. The evolutionary transition to mustard herbivory in Scaptomyza occurred without novel, highly efficient mechanisms to prevent ITC formation. Instead, lineages spanning the diversity of mustard-feeding Scaptomyza, in which herbivory evolved relatively recently (Whiteman et al. 2012; Lapoint et al. 2013), detoxify ITCs through the mercapturic acid pathway. This is also the primary mode of ITC metabolism in generalist feeders, including humans (Mennicke et al. 1988), some lepidopteran larvae (Schramm et al. 2012), and mollusks (Falk et al. 2013). In an unbiased quantitative analysis, no ITC-derived metabolites other than mercapturic acid products could be detected as metabolites produced by S. flava larvae. We found that D. melanogaster, a nonherbivorous relative of mustard-feeding Scaptomyza, also detoxifies ITCs using the mercapturic acid pathway. The ability to detoxify ITCs through the mercapturic acid pathway was therefore likely present in the earliest ancestral drosophilids and predated the evolution of mustard herbivory in Scaptomyza.
Scaptomyza flava feeds and develops more slowly when encountering glucosinolate defenses (Whiteman et al. 2012), unlike mustard specialists that prevent ITC formation (Raybould and Moyes 2001; Muller et al. 2010). Two aspects of the glucosinolate and ITC metabolism in S. flava may underlie this observation. First, detoxification of ITCs through the mercapturic acid pathway in S. flava may be nutritionally costly. The GST-catalyzed step in the mercapturic acid pathway involves the addition of a nitrogen-containing compound (GSH) to ITCs, and herbivores are thought to be nitrogen limited (Mattson 1980). Furthermore, GSH synthesis requires cysteine, which insects can obtain directly from the diet or synthesize from methionine. Cysteine and methionine are among the least abundant amino acids in plant leaves and may be growth limiting for chewing herbivores, and a large fraction (∼20%) of total cysteine content is used to produce GSH (Barbehenn, Kochmanski, et al. 2013; Barbehenn, Niewiadomski, et al. 2013). Second, ITCs have direct chemotoxic effects. Free ITCs bind to nucleophilic residues of cellular proteins (Mi et al. 2011; Zhang 2012), disrupt disulfide bridges important for protein structure (Kawakishi and Kaneko 1985), and can induce cell death and apoptosis (Brown and Hampton 2011). These chemotoxic effects of ITCs likely contribute to the induction of stress response genes following ingestion of dietary glucosinolates in S. flava larvae (Whiteman et al. 2012).
Evolution of an Increased Rate of ITC Detoxification in a GST from a Mustard-Feeding Drosophilid
GSTs, a family of enzymes that catalyze the mercapturic acid pathway, have broad substrate specificity (Hayes et al. 2005). However, if ITCs imposed strong selection on GSTs following the transition to mustard herbivory in Scaptomyza, these enzymes could evolve increased activity against ITCs. We tested this hypothesis by comparing activity against ITCs by the enzyme GSDT1, an abundant and ecologically important detoxification enzyme in drosophilids. We found that SnigGSTD1A, one of the two copies of GSTD1 in S. nigrita resulting from a gene duplication event in the mustard-feeding Scaptomyza lineage, initiates the detoxification of diverse ITCs at a rate unmatched by orthologous GSTD1 enzymes we profiled in other Drosophila. However, more radical kinetic changes have been inferred in other detoxification enzymes—such as cytochrome P450s—following host shifts or exposure to pesticides (Li et al. 2003, 2007; Joußen et al. 2012). Larger activity increases often involve enzymes that undergo rapid duplication and loss (Li et al. 2003) or form novel chimeric enzymes through recombination (Joußen et al. 2012). Purifying selection—which we found to be pervasive for GstD1 across microbe-feeding species in the genus Scaptomyza and subgenus Drosophila—might prevent similarly large changes in GSTD1 function. Furthermore, radical increases in activity against ITCs may be difficult to achieve. The specificity constants (kcat/Km) of drosophilid GSTD1 enzymes against two ITCs are within an order of magnitude of the largest known for these substrates among any GST (Kolm et al. 1995; Wiktelius and Stenberg 2007), and random engineering of a human GST with high activity against BITC and PEITC resulted in no more than 2-fold increases in activity (Runarsdottir and Mannervik 2010). On the other hand, because Drosophila GSTs other than GSTD1 are also able to metabolize ITCs (Saisawang et al. 2012), the absence of a larger increase in activity against ITCs may reflect a limited role for GSTD1 in ITC detoxification. Nonetheless, kinetic changes similar in magnitude to those observed in SnigGSTD1A can have large effects on organismal fitness (Walkiewicz et al. 2012), and functional evolution of SnigGSTD1A is consistent with an adaptive evolutionary response to dietary ITCs.
Although molecular evolutionary analyses suggested that GstD1 evolved under positive selection in the ancestor of S. flava, we did not observe an increase in GSTD1 activity against most ITCs in this species. SflaGSTD1 did, however, exhibit high specific activity and catalytic efficiency (kcat/Km) against PEITC. Phenethyl glucosinolate (which yields PEITC) and hydroxylated phenethyl glucosinolate dominate the glucosinolate profile of B. vulgaris (van Leur et al. 2006), the host plant from which the S. flava individuals used in this study were collected in Belmont, MA. Thus, a role for ITCs in driving evolution of SflaGSTD1 cannot be rejected.
Structural Basis of Changes in GSTD1 Function Following Gene Duplication
Evolutionary genetic analyses suggest that many amino acid substitutions in SnigGSTD1A, which acquired elevated activity against ITCs, were likely fixed by positive selection following gene duplication, although forces other than glucosinolate defenses may have been responsible. Therefore, linking structural and functional changes in SnigGSTD1A could reveal new insight into adaptive structural changes during the divergence of paralogous GSTs coupled with exposure to novel selective pressures. To identify residues in SnigGSTD1A that enhanced activity against ITCs, we assayed enzyme activity after swapping candidate amino acid residues between SnigGSTD1A and DmelGSTD1, which has a specificity constant (kcat/Km) for BITC that is less than 40% of the specificity constant of SnigGSTD1A.
Only one substitution occurred in the hydrophobic toxin-binding pocket of SnigGSTD1A (T166S), and site-directed mutagenesis revealed that this substitution accounts for approximately 50% of the difference in specificity for ITCs in SnigGSTD1A versus DmelGSTD1. This suggests that elevated activity against ITCs by SnigGSTD1A evolved, at least partly, following gene duplication. Although the substituted site is highly conserved in drosophilid GSTD1 enzymes, a parallel substitution occurred in a GstD1 paralog resulting from gene duplication in D. repleta group, which contains cactophilic (though nonherbivorous sensu stricto) species that are also tightly linked to their host plants. The substituted (Ser) and ancestral (Thr) amino acids do not differ in charge and are of a similar size. Despite this, substitutions between Thr and Ser have independently evolved at a positively selected site in the binding pockets of mammalian Mu-class GSTs, and these substitutions alter substrate specificity (Norrgård et al. 2006). Our results are consistent with previous findings that nonradical substitutions in the hydrophobic toxin-binding pocket can drive functional divergence of GSTs.
We also observed a cluster of substitutions unique to SnigGSTD1A or shared by mustard-feeding Scaptomyza species at the interface of the hydrophobic and GSH binding pockets, which form a novel hydrogen bond network. This region affects substrate specificity in another dipteran Delta-class GST (Vararattanavech et al. 2006). Furthermore, specialization for metabolism of dietary furanocoumarins by papilionid cytochrome P450 enzymes appears to result, in part, from novel stabilizing networks in the catalytic pocket that reduce flexibility and maintain an enzymatic conformation favorable for furanocoumarin metabolism (Li et al. 2003, 2004). However, this region did not affect specificity for ITCs in GSTD1. Given that this region differs between SnigGSTD1A, SnigGSTD1B, and SflaGSTD1—each of which have likely undergone episodic positive selection—its functional importance in these enzymes warrants further investigation.
Because substitutions of exposed amino acids in the binding cleft did not fully explain differences in activity against ITCs between SnigGSTD1A and DmelGSTD1, we investigated substitutions unique to SnigGSTD1A or both SnigGSTD1 paralogs that occurred outside the binding cleft. Two substitutions rearranged packing of an aromatic zipper buried near the hydrophobic binding cleft, which affects flexibility, and in turn substrate specificity, in another dipteran Delta-class GST (Wongsantichon et al. 2010). Additionally, a charged amino acid substitution formed a novel hydrogen bond with the backbone of the mobile α-8 helix, which partially occludes the hydrophobic binding cleft (Low et al. 2010). Although this may affect the stability of the α-8 helix (Scholtz and Baldwin 1992), its functional importance is not clear. Swapping these three candidate substitutions in the α-8 helix and hydrophobic zipper between DmelGSTD1 and SnigGSTD1A, in conjunction with the T166S substitution to the wall of hydrophobic binding cleft, nearly completely reversed specificity constants of SnigGSTD1A and DmelGSTD1 for ITCs. The direction of these changes in activity suggests that one or more of these substitutions may have been adaptive in S. nigrita.
Although we observed only very small topological changes in regions affecting SnigGSTD1A activity against ITCs, GSTs undergo conformational shifts upon substrate binding (Wongsantichon et al. 2012), and our attempts to cocrystallize SnigGSTD1A with ITCs were unsuccessful. Therefore, our structural analysis may underestimate the extent of topological changes relevant to ITC detoxification.
A Potential Role for Relaxation of Functional Constraint on GstD1 Following a Lineage-Specific Duplication in S. nigrita
Strong purifying selection we and others (Low et al. 2007) inferred for GstD1 in drosophilids may result from pressure to maintain both broad substrate specificity and an essential developmental function. GSTD1 exhibits the broadest known substrate specificity among GSTs in D. melanogaster (Saisawang et al. 2012), and we found that the ability to metabolize a broad range of toxins is maintained across the drosophilid radiation. Furthermore, RNAi-mediated knockdown of GstD1 in D. melanogaster results in pupal-stage lethality, suggesting that GSTD1 also plays a developmental role (RNAi stock 10045R-4, National Institute of Genetics, Japan). The retention of at least one GstD1 copy across the Drosophila and Scaptomzya radiations, despite the dynamic turnover of many GSTs observed in fully sequenced Drosophila genomes (Low et al. 2007), further suggests that one or more functions performed by GSTD1 are essential in drosophilids.
When a single gene performs multiple functions, pleiotropy may constrain the simultaneous optimization of both functions, termed adaptive conflict (Piatigorsky and Wistow 1991; Des Marais and Rausher 2008). This model is particularly relevant to promiscuous detoxification enzymes that metabolize a diversity of toxic substrates, such as GSTD1 (Storz 2008). Gene duplication allows escape from adaptive conflict. Different functions are then refined in each paralog via positive selection. Both GstD1 paralogs in S. nigrita show signatures of episodic positive selection, and phylogenetic models revealed that nonsynonymous substitutions were fixed at an accelerated rate in both paralogs following gene duplication but not in the more recent history of either paralog. This pattern would be expected if different ancestral functions were refined in each paralog following duplication. Interestingly, however, SnigGSTD1B had minimal activity against all ITC and non-ITC substrates. One evolutionary scenario is that refinement of a developmental role by SnigGSTD1B enabled SnigGSTD1A to acquire increased activity against ITCs without negative pleiotropic effects. Similarly, the loss of high activity against many substrates observed in SnigGSTD1B may have been enabled by the retention of these activities in SnigGSTD1A. Characterizing differences in the timing and location of SnigGstD1A and SnigGstD1B expression is likely to give further insight into their functional divergence. Future studies could test for roles of both enzymes in generalized detoxification, mustard oil resistance, and development by expressing either SnigGstD1A or SnigGstD1B in D. melanogaster.
Differences in host breadth between S. flava and S. nigrita may also influence the strength of purifying selection on GstD1. Although S. flava feeds on >40 mustard species and a few unrelated host plant species (Whiteman et al. 2011), S. nigrita specializes on a single mustard species (Collinge and Louda 1989) (supplementary note, Supplementary Material online) and is likely to be exposed to a more restricted set of plant-derived toxins. The narrow host breadth of S. nigrita could have permitted the evolution of increased efficiency and specificity for ITCs at the expense of activity against other toxins not present in its only known host plant species. In an analogous situation, narrow dietary specialization may have enabled functional specialization for furanocoumarins by promiscuous cytochrome P450 enzymes in papilionid larvae (Li et al. 2003, 2004; Wen et al. 2006).
Generalized Detoxification Systems May Facilitate the Evolution of Herbivory
The larvae of most Scaptomyza species feed on microbes living in rotting vegetation (Lapoint et al. 2013). We have successfully reared S. pallida and S. apicata, which last shared a common ancestor with herbivorous Scaptomyza >15 Ma (Whiteman et al. 2012; Lapoint et al. 2013), in rotting tissue from mustard plants (supplementary note, Supplementary Material online). The ability to detoxify mustard oils using the mercapturic acid pathway—which is shared by herbivorous mustard-feeding Scaptomyza and microbe-feeding D. melanogaster—could have facilitated feeding on rotting and then living mustard tissues.
Interestingly, herbivory has evolved more times in flies (Diptera) than in other insect orders, including multiple times in the Drosophila radiation (Mitter et al. 1988; Whiteman et al. 2012). Comparative functional and genomic studies of these species and their closest relatives, which are not herbivores, will shed further light on the role of GSTs and other detoxification enzymes in facilitating transitions that are among the most important in the history of terrestrial life.
Conclusion
Rapid species radiations follow evolutionary transitions to herbivory, such that herbivorous insects account for approximately half of insect species diversity. However, these transitions are absent from most insect orders. Understanding the behavioral, morphological, and physiological underpinnings of the transition to herbivory is important to addressing this paradox. From a physiological perspective, plant toxins present in living plant tissue have long been hypothesized to render living plant tissues recalcitrant to invasion by insects. Although specialized herbivorous species possess derived and efficient detoxification mechanisms, it is unclear whether more generalized detoxification mechanisms play an important role in the transition to herbivory. Here, we tested the hypothesis that generalized detoxification mechanisms may play important roles in evolutionary transitions to herbivory. We found that a recently derived lineage of drosophilid herbivore uses generalized, ancestral detoxification pathway to metabolize ITCs, a major defensive toxin in their host plants. After the transition to herbivory, an otherwise highly conserved GST, catalyzing the initiation of this pathway, may have evolved under positive selection in the herbivorous lineage. Signatures of positive selection are coupled with the evolution of higher activity against ITCs in one paralog following gene duplication. We found that a single amino acid substitution in this enzyme is critical for the increased specificity against ITCs. Given these results, and the fact that GSTs arose before the divergence of plants and animals (Sheehan et al. 2001), the generalized detoxification pathway catalyzed by GSTs could have helped to facilitate the very first transitions to herbivory in insects >350 Ma (Labandeira 1998). We anticipate that studies in other lineages will provide insight into the extent to which recently derived herbivores use generalized detoxification pathways, rather than entirely novel mechanisms, to metabolize dietary plant toxins. This could help illuminate the evolutionary origins of herbivory, one of the most widespread, complex, and ecologically important phenotypes known in metazoans.
Materials and Methods
Detailed Materials and Methods with references are described in the Supporting Information (Supplementary Material online).
Analysis of Glucosinolate and 4msob-ITC Metabolism
To enable quantitative analysis in larval S. flava, [14C]-4msob-glucosinolate was prepared biosynthetically from [U-14C]-Met, HPLC purified, and introduced into detached leaves of glucosinolate-deficient A. thaliana (cyp79b2 cyp79b3 myb28 myb29 in the Col-0 background) (Schramm et al. 2012). After feeding, S. flava larvae were homogenized, extracts were fractionated by HPLC, and metabolites were quantified by scintillation counting (Schramm et al. 2012). Metabolic analyses in other species were semiquantitative. Mustard-feeding Scaptomyza larvae were fed A. thaliana in the laboratory or wild-collected from mustard plants (see supplementary materials and methods, Supplementary Material online). Adult D. melanogaster were fed with 1 mM 4msob-ITC in 5% sucrose. Compounds were analyzed by HPLC-mass spectrometry (MS)/MS and identified based on comparison of retention times to previously isolated compounds or synthetic standards (Schramm et al. 2012).
PCR and Sequencing of GstD1
Degenerate PCR primers (supplementary table S10, Supplementary Material online) were used to amplify the GstD1 coding sequences (CDS) (630 bp) from S. flava, S. nigrita, additional mustard-feeding lineages that could not be confidently identified as described species, and 16 species of Scaptomyza and Hawaiian Drosophila. Searches for new paralogs in S. flava and S. nigrita used high-efficiency thermal asymmetric interlaced PCR (Liu and Chen 2007) and, for S. flava, a preliminary genome assembly. Specific primers were designed to amplify GstD1 from individuals from single populations of S. flava and S. nigrita for population genetic analysis. Sequencing of purified PCR products or plasmids generated overlapping reads across the CDS. DNA sequences have been deposited in GenBank under accession codes listed in supplementary table S1, Supplementary Material online.
Construction of GSTD1 Expression Vectors and Enzyme Purification
The coding sequence of GstD1 (or, for S. nigrita, GstD1A and GstD1B) was amplified from each of the ten Drosophila and Scaptomyza species and inserted into the pT7-MAT-Tag-2 expression vector using restriction digest and ligation. Constructs were sequence verified before and after transformation into Rosetta 2 E. coli. Enzymes were overexpressed and purified using affinity for Glutathione Sepharose (supplementary fig. S6, Supplementary Material online).
Specific Activity and Kinetic Assays
Initial reaction rates of GSH:substrate conjugation were determined using spectrophotometry (supplementary table S11, Supplementary Material online), with published or newly determined extinction coefficients (supplementary materials and methods, Supplementary Material online). Specific activities reported are the mean of four to eight independent replicates. For kinetic assays, initial reaction rates were measured (N = 3–7) at seven to nine concentrations of the rate-limiting substrate, and Michaelis–Menten kinetic parameters were determined using nonlinear regression.
Structure Determination and Refinement
Successful crystallization conditions for SnigGSTD1A with a GSH ligand were identified by robotic screening trials and optimized using a sitting-drop vapor diffusion method. The structure was determined from X-ray diffraction data by molecular replacement using D. melanogaster GSTD1 (PDB ID: 3EIN) (Low et al. 2010), manually rebuilt, and refined. Coordinates and structure factors for the final model have been deposited in the Protein Data Bank under accession code 4I97.
Phylogenetic and Molecular Evolutionary Analysis
Phylogenetic analyses of GstD1 in drosophilids and a multigene phylogeny for mustard-feeding Scaptomyza were implemented under maximum likelihood (Stamatakis 2006) and Bayesian (Ronquist and Huelsenbeck 2003) frameworks with the general time reversible (GTR)+G substitution model. The tree topology for GstD1 in mustard-feeding Scaptomyza inferred through Bayesian analyses, presented in figure 3, was used for phylogenetic analysis of codon evolution rates (ω). Because many internal nodes in the GstD1 genealogy could not be confidently inferred, previously described species relationships were used to specify the topology for nonherbivorous taxa in this analysis. Models implemented in PAML 4.5 (Yang 2007) tested for positive selection across the genus Scaptomyza and subgenus Drosophila (including the Hawaiian Drosophila) generally, and for accelerated evolution and positive selection in specific mustard-feeding lineages. McDonald–Kreitman tests (McDonald and Kreitman 1991), polarized using marginal ancestral reconstruction conducted using PAML 4.5 (Yang 2007), were also used to test for lineage-specific positive selection in S. flava and S. nigrita. Similar analyses were conducted for additional genes to control for demographic effects.
Supplementary Material
Supplementary note, materials and methods, figures S1–S6, and tables S1–S11 are available at Molecular Biology and Evolution online (http://mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the editor and two anonymous reviewers for helpful comments on the manuscript; Richard Lapoint for collection and identification of many fly specimens used in this study; Noel Kitchen for assistance with experiments; and Vahe Bandarian, Mark Beilstein, Matthew Cordes, and Megan McEvoy for use of equipment at the University of Arizona. Specimen collection permits were provided by the United States Forest Service. This work was supported by the National Institutes of Health (R01 HL062969 to W.R.M.; Shared Instrumentation Grant S10 RR025485 to A.W.; PERT fellowship 5K12GM000708-13 to A.N.D.), the National Science Foundation (DEB-1256758 to N.K.W.; DEB-1405966, DGE-1143953, and University of Arizona IGERT Genomics Fellowship DGE-0654435 to A.D.G.), the John Templeton Foundation (Grant ID #41855 to N.K.W.), the National Geographic Society (9097-12 to N.K.W.), the University of Arizona (Faculty Seed Grant, Center for Insect Science Seed Grant, and laboratory set-up grant to N.K.W.), the Rocky Mountain Biological Laboratory (fellowships to A.D.G. and N.K.W.), and the Max Planck Society (funds to J.G.). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory, and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393), and the National Center for Research Resources (P41RR001209).
References
- Aliabadi A, Renwick JA, Whitman DW. Sequestration of glucosinolates by harlequin bug Murgantia histrionica. J Chem Ecol. 2002;28:1749–1762. doi: 10.1023/a:1020505016637. [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Kochmanski J, Menachem B, Poirier LM. Allocation of cysteine for glutathione production in caterpillars with different antioxidant defense strategies: a comparison of Lymantria dispar and Malacosoma disstria. Arch Insect Biochem Physiol. 2013;84:90–103. doi: 10.1002/arch.21116. [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Niewiadomski J, Kochmanski J. Importance of protein quality versus quantity in alternative host plants for a leaf-feeding insect. Oecologia. 2013;173:1–12. doi: 10.1007/s00442-012-2574-7. [DOI] [PubMed] [Google Scholar]
- Betran E, Rozas J, Navarro A, Barbadilla A. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics. 1997;146:89–99. doi: 10.1093/genetics/146.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyland E, Chasseaud LF. The role of glutathione and glutathione S-transferases in mercapturic acid biosynthesis. Adv Enzymol Relat Areas Mol Biol. 1969;32:173–219. doi: 10.1002/9780470122778.ch5. [DOI] [PubMed] [Google Scholar]
- Brown KK, Hampton MB. Biological targets of isothiocyanates. Biochim Biophys Acta. 2011;1810:888–894. doi: 10.1016/j.bbagen.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry. 2003;62:471–481. doi: 10.1016/s0031-9422(02)00549-6. [DOI] [PubMed] [Google Scholar]
- Collinge SK, Louda SM. Scaptomyza nigrita Wheeler (Diptera: Drosophilidae), a leaf miner of the native crucifer, Cardamine cordifolia A. Gray (Bittercress) J Kans Entomol Soc. 1989;62:1–10. [Google Scholar]
- Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–765. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56:5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- Falk KL, Kästner J, Bodenhausen N, Schramm K, Paetz C, Vassão DG, Reichelt M, Knorre D, Bergelson J, Erb M. The role of glucosinolates and the jasmonic acid pathway in resistance of Arabidopsis thaliana against molluskan herbivores. Mol Ecol. 2013;23:1188–1203. doi: 10.1111/mec.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Joußen N, Agnolet S, Lorenz S, Schöne SE, Ellinger R, Schneider B, Heckel DG. Resistance of Australian Helicoverpa armigera to fenvalerate is due to the chimeric P450 enzyme CYP337B3. Proc Natl Acad Sci U S A. 2012;109:15206–15211. doi: 10.1073/pnas.1202047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakishi S, Kaneko T. Interaction of oxidized glutathione with allyl isothiocyanate. Phytochemistry. 1985;24:715–718. [Google Scholar]
- Kazana E, Pope TW, Tibbles L, Bridges M, Pickett JA, Bones AM, Powell G, Rossiter JT. The cabbage aphid: a walking mustard oil bomb. Proc Biol Sci. 2007;274:2271–2277. doi: 10.1098/rspb.2007.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolm RH, Danielson UH, Zhang Y, Talalay P, Mannervik B. Isothiocyanates as substrates for human glutathione transferases: structure-activity studies. Biochem J. 1995;311:453–459. doi: 10.1042/bj3110453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira CC. Early history of arthropod and vascular plant associations. Annu Rev Earth Planet Sci. 1998;26:329–377. [Google Scholar]
- Lapoint RT, Whiteman NK, O'Grady PM. Diversification and dispersal of the Hawaiian Drosophilidae: the evolution of Scaptomyza. Mol Phylogenet Evol. 2013;69:95–108. doi: 10.1016/j.ympev.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Schuler MA, Berenbaum MR. Diversification of furanocoumarin-metabolizing cytochrome P450 monooxygenases in two papilionids: specificity and substrate encounter rate. Proc Natl Acad Sci U S A. 2003;100:14593–14598. doi: 10.1073/pnas.1934643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Baudry J, Berenbaum MR, Schuler MA. Structural and functional divergence of insect CYP6B proteins: from specialist to generalist cytochrome P450. Proc Natl Acad Sci U S A. 2004;101:2939–2944. doi: 10.1073/pnas.0308691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- Lichtenstein E, Strong F, Morgan D. Naturally occurring insecticides, identification of 2-phenylethylisothiocyanate as an insecticide occurring naturally in the edible part of turnips. J Agric Food Chem. 1962;10:30–33. [Google Scholar]
- Liu YG, Chen Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques. 2007;43:649–654. doi: 10.2144/000112601. [DOI] [PubMed] [Google Scholar]
- Louda SM, Collinge SK. Plant resistance to insect herbivores: a field test of the environmental stress hypothesis. Ecology. 1992;73:153–169. [Google Scholar]
- Low WY, Feil SC, Ng HL, Gorman MA, Morton CJ, Pyke J, McConville MJ, Bieri M, Mok YF, Robin C, et al. Recognition and detoxification of the insecticide DDT by Drosophila melanogaster glutathione S-transferase D1. J Mol Biol. 2010;399:358–366. doi: 10.1016/j.jmb.2010.04.020. [DOI] [PubMed] [Google Scholar]
- Low WY, Ng HL, Morton CJ, Parker MW, Batterham P, Robin C. Molecular evolution of glutathione S-transferases in the genus Drosophila. Genetics. 2007;177:1363–1375. doi: 10.1534/genetics.107.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA, O'Grady PM. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu Rev Genet. 2005;39:263–291. doi: 10.1146/annurev.genet.39.073003.112454. [DOI] [PubMed] [Google Scholar]
- Martin NA. History of an invader, Scaptomyza flava (Fallen, 1823) (Diptera: Drosophilidae) New Zeal J Zool. 2004;31:27–32. [Google Scholar]
- Mattson WJ. Herbivory in relation to plant nitrogen content. Ann Rev Ecol Syst. 1980;11:119–161. [Google Scholar]
- Matzkin LM. The molecular basis of host adaptation in cactophilic Drosophila: molecular evolution of a glutathione S-transferase gene (GstD1) in Drosophila mojavensis. Genetics. 2008;178:1073–1083. doi: 10.1534/genetics.107.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mennicke WH, Gorler K, Krumbiegel G, Lorenz D, Rittmann N. Studies on the metabolism and excretion of benzyl isothiocyanate in man. Xenobiotica. 1988;18:441–447. doi: 10.3109/00498258809041680. [DOI] [PubMed] [Google Scholar]
- Mi L, Hood BL, Stewart NA, Xiao Z, Govind S, Wang X, Conrads TP, Veenstra TD, Chung FL. Identification of potential protein targets of isothiocyanates by proteomics. Chem Res Toxicol. 2011;24:1735–1743. doi: 10.1021/tx2002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter C, Farrell F, Wiegmann B. The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification? Am Nat. 1988;132:107–128. [Google Scholar]
- Muller C, Agerbirk N, Olsen CE, Boeve JL, Schaffner U, Brakefield PM. Sequestration of host plant glucosinolates in the defensive hemolymph of the sawfly Athalia rosae. J Chem Ecol. 2001;27:2505–2516. doi: 10.1023/a:1013631616141. [DOI] [PubMed] [Google Scholar]
- Muller R, de Vos M, Sun JY, Sonderby IE, Halkier BA, Wittstock U, Jander G. Differential effects of indole and aliphatic glucosinolates on lepidopteran herbivores. J Chem Ecol. 2010;36:905–913. doi: 10.1007/s10886-010-9825-z. [DOI] [PubMed] [Google Scholar]
- Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 2010;11:265–289. doi: 10.1146/annurev-genom-082908-150129. [DOI] [PubMed] [Google Scholar]
- Norrgård MA, Ivarsson Y, Tars K, Mannervik B. Alternative mutations of a positively selected residue elicit gain or loss of functionalities in enzyme evolution. Proc Natl Acad Sci U S A. 2006;103:4876–4881. doi: 10.1073/pnas.0600849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Suzuki Y, Nei M. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci U S A. 2009;106:6700–6705. doi: 10.1073/pnas.0901855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson GI, Masel J. Quantitative prediction of molecular clock and ka/ks at short timescales. Mol Biol Evol. 2009;26:2595–2603. doi: 10.1093/molbev/msp175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J, Wistow G. The recruitment of crystallins: new functions precede gene duplication. Science. 1991;252:1078–1079. doi: 10.1126/science.252.5009.1078. [DOI] [PubMed] [Google Scholar]
- Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- Ratzka A, Vogel H, Kliebenstein DJ, Mitchell-Olds T, Kroymann J. Disarming the mustard oil bomb. Proc Natl Acad Sci U S A. 2002;99:11223–11228. doi: 10.1073/pnas.172112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould A, Moyes C. The ecological genetics of aliphatic glucosinolates. Heredity. 2001;87:383–391. doi: 10.1046/j.1365-2540.2001.00954.x. [DOI] [PubMed] [Google Scholar]
- Rodman JE, Louda SM. Seasonal flux of isothiocyanate-yielding glucosinolates in roots, stems and leaves of Cardamine cordifolia. Biochem Syst Ecol. 1985;13:405–412. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Runarsdottir A, Mannervik B. A novel quasi-species of glutathione transferase with high activity towards naturally occurring isothiocyanates evolves from promiscuous low-activity variants. J Mol Biol. 2010;401:451–464. doi: 10.1016/j.jmb.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Saisawang C, Wongsantichon J, Ketterman AJ. A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster. Biochem J. 2012;442:181–190. doi: 10.1042/BJ20111747. [DOI] [PubMed] [Google Scholar]
- Scholtz JM, Baldwin RL. The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct. 1992;21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- Schoonhoven LM, van Loon JJA, Dicke M. Insect-plant biology. New York: Oxford University Press; 2005. [Google Scholar]
- Schramm K, Vassao DG, Reichelt M, Gershenzon J, Wittstock U. Metabolism of glucosinolate-derived isothiocyanates to glutathione conjugates in generalist lepidopteran herbivores. Insect Biochem Mol Biol. 2012;42:174–182. doi: 10.1016/j.ibmb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NG, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Storz JF. Genome evolution: gene duplication and the resolution of adaptive conflict. Heredity. 2008;102:99–100. doi: 10.1038/hdy.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong DR, Lawton JH, Southwood TRE. Insect on plants: community patterns and mechanisms. Cambridge: Harvard University Press; 1984. [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- van Leur H, Raaijmakers CE, van Dam NM. A heritable glucosinolate polymorphism within natural populations of Barbarea vulgaris. Phytochemistry. 2006;67:1214–1223. doi: 10.1016/j.phytochem.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Vararattanavech A, Prommeenate P, Ketterman AJ. The structural roles of a conserved small hydrophobic core in the active site and an ionic bridge in domain I of delta class glutathione S-transferase. Biochem J. 2006;393:89–95. doi: 10.1042/BJ20050555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadleigh RW, Yu SJ. Detoxification of isothiocyanate allelochemicals by glutathione transferase in three lepidopterous species. J Chem Ecol. 1988;14:1279–1288. doi: 10.1007/BF01019352. [DOI] [PubMed] [Google Scholar]
- Walkiewicz K, Benitez Cardenas AS, Sun C, Bacorn C, Saxer G, Shamoo Y. Small changes in enzyme function can lead to surprisingly large fitness effects during adaptive evolution of antibiotic resistance. Proc Natl Acad Sci U S A. 2012;109:21408–21413. doi: 10.1073/pnas.1209335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Rupasinghe S, Niu G, Berenbaum MR, Schuler MA. CYP6B1 and CYP6B3 of the black swallowtail (Papilio polyxenes): adaptive evolution through subfunctionalization. Mol Biol Evol. 2006;23:2434–2443. doi: 10.1093/molbev/msl118. [DOI] [PubMed] [Google Scholar]
- Wheat CW, Vogel H, Wittstock U, Braby MF, Underwood D, Mitchell-Olds T. The genetic basis of a plant-insect coevolutionary key innovation. Proc Natl Acad Sci U S A. 2007;104:20427–20431. doi: 10.1073/pnas.0706229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MR. The Drosophilidae of the nearctic region, exclusive of the genus Drosophila. Univ Texas Publ. 1952;5204:162–218. [Google Scholar]
- Wheeler MR, Takada H. The nearctic and neotropical species of Scaptomyza Hardy (Diptera; Drosophilidae) Univ Texas Publ. 1966;6615:3–38. [Google Scholar]
- Whiteman NK, Gloss AD, Sackton TB, Groen SC, Humphrey PT, Lapoint RT, Sonderby IE, Halkier BA, Kocks C, Ausubel FM, et al. Genes involved in the evolution of herbivory by a leaf-mining, Drosophilid fly. Genome Biol Evol. 2012;4:788–804. doi: 10.1093/gbe/evs063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman NK, Groen SC, Chevasco D, Bear A, Beckwith N, Gregory TR, Denoux C, Mammarella N, Ausubel FM, Pierce NE. Mining the plant-herbivore interface with a leafmining Drosophila of Arabidopsis. Mol Ecol. 2011;20:995–1014. doi: 10.1111/j.1365-294X.2010.04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiktelius E, Stenberg G. Novel class of glutathione transferases from cyanobacteria exhibit high catalytic activities towards naturally occurring isothiocyanates. Biochem J. 2007;406:115–123. doi: 10.1042/BJ20070328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winde I, Wittstock U. Insect herbivore counteradaptations to the plant glucosinolate-myrosinase system. Phytochemistry. 2011;72:1566–1575. doi: 10.1016/j.phytochem.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Wongsantichon J, Robinson RC, Ketterman AJ. Structural contributions of delta class glutathione transferase active-site residues to catalysis. Biochem J. 2010;428:25–32. doi: 10.1042/BJ20091939. [DOI] [PubMed] [Google Scholar]
- Wongsantichon J, Robinson RC, Ketterman AJ. Structural evidence for conformational changes of delta class glutathione transferases after ligand binding. Arch Biochem Biophys. 2012;521:77–83. doi: 10.1016/j.abb.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Molecular mechanism of rapid cellular accumulation of anticarcinogenic isothiocyanates. Carcinogenesis. 2001;22:425–431. doi: 10.1093/carcin/22.3.425. [DOI] [PubMed] [Google Scholar]
- Zhang Y. The molecular basis that unifies the metabolism, cellular uptake and chemopreventive activities of dietary isothiocyanates. Carcinogenesis. 2012;33:2–9. doi: 10.1093/carcin/bgr255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.