Abstract
Mutations in the lysosomal enzyme, N-sulfoglucosamine sulfohydrolase (SGSH), also called sulfamidase, cause accumulation of lysosomal inclusion bodies in the brain of children born with mucopolysaccharidosis type IIIA, also called Sanfilippo type A syndrome. Enzyme replacement therapy with recombinant SGSH does not treat the brain because the enzyme is a large molecule drug that does not cross the blood–brain barrier (BBB). A BBB-penetrating form of SGSH was produced by re-engineering the enzyme as an IgG fusion protein, where the IgG domain is a monoclonal antibody (mAb) against the human insulin receptor (HIR). The HIRMAb domain of the HIRMAb–SGSH fusion protein acts as a molecular Trojan horse to ferry the fused enzyme across the BBB. The HIRMAb–SGSH was produced in stably transfected host cells and purified to homogeneity by protein A chromatography. The fusion protein reacted with antibodies against either human IgG or SGSH on Western blotting. High affinity binding to the HIR was retained following SGSH fusion to the HIRMAb, with an EC50 of 0.33 ± 0.05 nM in an HIR binding ELISA. The SGSH enzyme activity of the HIRMAb–SGSH fusion protein was 4712 ± 388 units/mg protein based on a two-step fluorometric enzyme assay. The HIRMAb–SGSH was taken up by lysosomes in MPSIIIA fibroblasts, and treatment of these cells with the fusion protein caused an 83% reduction in sulfate incorporation into lysosomal glycosoaminoglycans. The HIRMAb–SGSH fusion protein was radiolabeled with the [125I]-Bolton–Hunter reagent and injected intravenously in the Rhesus monkey. The brain uptake of the fusion protein was high, ∼1% injected dose/brain. Calculations, based on this level of brain uptake, suggest normalization of brain SGSH enzyme activity is possible following administration of therapeutic doses of the fusion protein. These studies describe a novel IgG–SGSH fusion protein that is a new noninvasive treatment of the brain in MPS type IIIA.
Keywords: blood−brain barrier, lysosomal storage, sulfamidase, fusion protein, drug delivery
Introduction
Mucopolysaccharidosis (MPS) type IIIA, also called MPSIIIA, or Sanfilippo A syndrome, is an inborn failure of metabolism caused by mutations in the gene encoding the lysosomal enzyme, N-sulfoglucosamine sulfohydrolase (SGSH), also called sulfamidase, which degrades heparan sulfate type glycosoaminoglycans or GAGs.1 SGSH is the only human enzyme that causes the hydrolysis of N-linked sulfate groups from the nonreducing terminal glucosaminide residues of heparan sulfate.1 An insufficient level of SGSH causes a pathological buildup of heparan sulfate, a GAG, in tissues, including the central nervous system (CNS). However, unlike other MPS disorders, MPSIIIA patients present primarily with a CNS phenotype, where the majority of the effects of the disease are brain-related, including severe behavioral disturbances, loss of speech by the age of 7 years, impaired walking leading to wheelchair existence by the age of 12, and death at a mean age of 18 years.2 Following the initial cloning and expression of human recombinant SGSH nearly 20 years ago,3 it was believed that MPSIIIA could be treated with weekly intravenous enzyme replacement therapy (ERT). However, patients with MPSIIIA presently have no therapy, as conventional ERT with recombinant SGSH would not be effective because the SGSH enzyme does not cross the blood–brain barrier (BBB).4 In an attempt to bypass the BBB, the enzyme has been administered to animals by intracerebroventricular (ICV) injections.5 However, SGSH only distributes to the surface of the brain following ICV injection,5 owing to the rapid rate of drug exit from the cerebrospinal fluid (CSF) compartment, as CSF courses by bulk flow from brain to blood.6
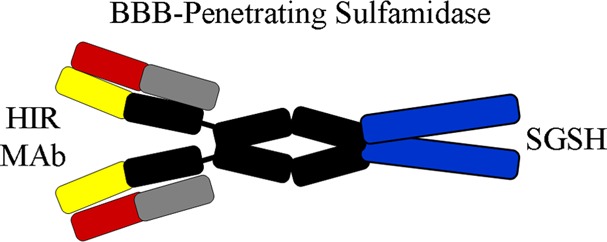
Large molecule pharmaceuticals, such as SGSH, can be re-engineered for BBB penetration, as IgG–enzyme fusion proteins, where the IgG domain is a molecular Trojan horse that crosses the BBB.7 The IgG domain is a monoclonal antibody (mAb) against an endogenous BBB receptor-transporter such as the insulin receptor or the transferrin receptor (TfR). The receptor-specific mAb binds the BBB receptor, which leads to receptor-mediated transport across the BBB of the mAb as well as the fused enzyme pharmaceutical.6 IgG–SGSH fusion proteins have not been previously engineered, and it is not known if an enzymatically active SGSH enzyme can be re-engineered to enable BBB transport. In the present study, human SGSH was fused to the carboxyl terminus of the heavy chain of a genetically engineered mAb against the human insulin receptor (HIR), and this fusion protein is designated the HIRMAb–SGSH fusion protein (Figure 1). This investigation describes the genetic engineering and expression of the HIRMAb–SGSH fusion protein, the purity, identity, and potency of the purified fusion protein, the reduction of GAGs in MPSIIIA fibroblasts, and the rapid penetration of the BBB in the Rhesus monkey following intravenous (IV) injection of the fusion protein.
Figure 1.
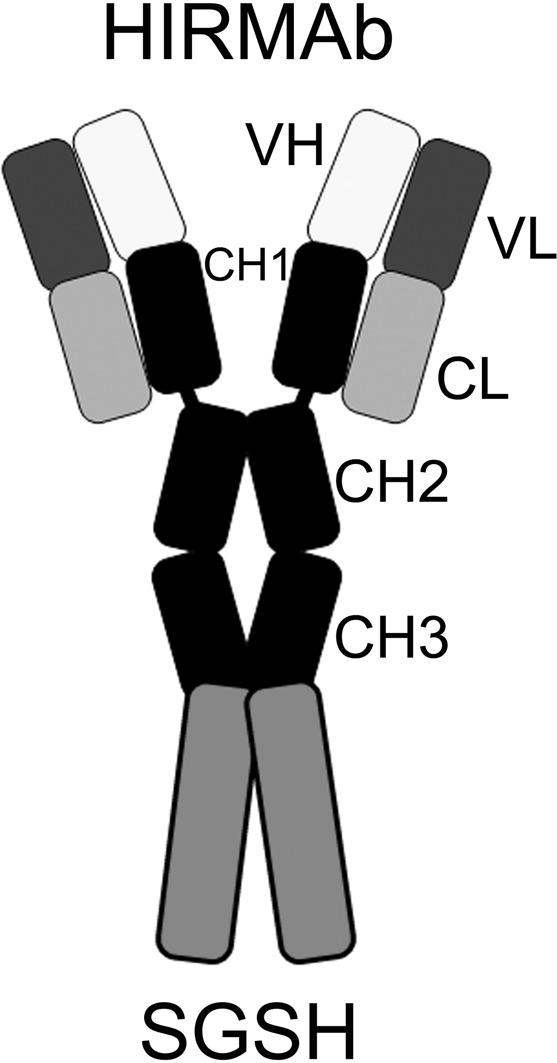
Structure of the HIRMAb–SGSH fusion protein, which is formed by two light chains and two heavy chains, where the SGSH monomer is fused to the carboxy terminus of each heavy chain. The domains of the light chain include the variable region of the light chain (VL) of the HIRMAb and the human κ constant region of the light chain (CL). The domains of the heavy chain include the variable region of the heavy chain (VH) of the HIRMAb, the the subdomains of the human IgG1 constant region (CH1, CH2, CH3), and the human SGSH enzyme (sulfamidase).
Experimental Section
Engineering of Tandem Vector and Production of Stably Expressing CHO Line
A 1.5 kb cDNA encoding the human SGSH cDNA was produced by reverse transcriptase polymerase chain reaction using the following forward and reverse oligodeoxynucleotide primers: 5′-phosphate- CACGTCCCCGGAACGCACTGCTG-3′ and 5′-phosphate-TCACAGCTCATTGTGGAGGGGCTG-3′, and polyA+RNA from human liver. The cDNA encoding the SGSH sequence, absent the enzyme signal peptide, was fused to the 3′-flank of the cDNA encoding the HIRMAb heavy chain so as to produce a heavy chain (HC) fusion protein wherein the amino terminus of the mature SGSH was fused, via a short linker, to the carboxyl terminus of the CH3 region of the HC of the HIRMAb. A tandem vector was engineered as previously described.8,9 Bidirectional DNA sequencing was performed, which showed the three expression cassettes spanned 10000 nucleotides. The light chain was described previously.9 The predicted molecular weight of the light chain is 23398 Da with a predicted isoelectric point (pI) of 5.45. The fusion protein of the HIRMAb heavy chain and SGSH was comprised of 946 AA, which included a 19 AA signal peptide, the 443 AA HIRMAb heavy chain, a 2 AA linker (Ser-Ser), and the 482 AA mature SGSH enzyme. The predicted pI of the heavy chain is 7.44, and the predicted molecular weight of the heavy chain, without glycosylation, is 103 412 Da, which includes 54674 Da within the SGSH domain. The AA sequence of the SGSH domain is 100% identical with the sequence of mature human SGSH (Genbank NP_000190), with the exception of a histidine at position 456 of the SGSH domain. The R456H substitution is a common polymorphism, which has no effect on enzyme activity.10 The predicted molecular weight of the heterotetramer, without glycosylation, is 253620 Da.
The tandem vector plasmid DNA was linearized and stably transfected Chinese hamster ovary (CHO) cells in serum free medium (SFM) were engineered and isolated by limited dilutional cloning as previously described.9
Purification and Analysis of Fusion Protein
The HIRMAb–SGSH fusion protein was affinity purified by protein A chromatography from SFM conditioned by the CHO cells as previously described.9
The identity of the HIRMAb–SGSH fusion protein was verified by human IgG and human SGSH Western blotting. For the human IgG Western blot, the primary antibody was a goat antihuman IgG (H+L) (Vector Laboratories, Burlingame, CA). For the human SGSH Western blot, the primary antibody was a rabbit antihuman SGSH antibody (Abcam, Cambridge, MA). The secondary antibody was a biotinylated horse antigoat IgG or biotinylated goat antirabbit IgG antibody (Vector Laboratories). The purity of the HIRMAb–SGSH fusion protein was verified by reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) as previously described.9 The molecular weight (MW) standards were obtained from Thermo Fisher Scientific, Inc. (Rockford, IL), and Biorad Laboratories, Inc. (Hercules, CA). Samples tested in the Western blotting include the protein A purified HIRMAb–SGSH fusion protein, the protein A purified HIRMAb, and a recombinant fusion protein of amino terminal glutathione S-transferase and human SGSH (Novus Biologicals, Littleton, CO). The potency of HIRMAb–SGSH fusion protein binding to the HIR was evaluated by measurement of the affinity of binding of the fusion protein to the HIR extracellular domain (ECD) using an ELISA, as described previously.8,9
SGSH Enzyme Activity Assay
The SGSH enzyme specific activity of the HIRMAb–SGSH fusion protein was measured with a two-step fluorometric enzymatic assay using 4-methylumbelliferyl-α-N-sulfo-d-glucosaminide (MU-αGlcNS) as the assay substrate.11 This substrate was custom synthesized and HPLC purified by Sigma-Aldrich (St Louis, MO), and the structure was confirmed by mass spectrometry. The MU-αGlcNS substrate was hydrolyzed by SGSH to 4-methylumbelliferyl-α-d-glucosaminide (MU-αGlcNH2), which was then hydrolyzed to the fluorescent product, 4-methylumbelliferone (4-MU), by the α-glucosaminidase side activity in commercial yeast α-glucosidiase.11 The assay was performed by incubation of the HIRMAb–SGSH fusion protein (30, 100, or 300 ng) and 3.4 mM MU-αGlcNS substrate in 0.03 M sodium barbital/0.03 M sodium acetate/0.13 M NaCl/pH = 5.5/0.02% sodium azide for 37 °C for 17 h. McIlvaine’s buffer and 0.1 unit of yeast α-glucosidase was added, followed by incubation at 37C for an additional 24 h. The reaction was stopped by the addition of 0.5 M sodium carbonate/pH = 10.7. Fluorescence was measured with a Farrand filter fluorometer with a 365 nm excitation filter and a 450 nm emission filter. A standard curve was generated with 0.03–3.0 nmol/tube of the 4-MU product (Sigma), which allowed for conversion of fluorescent units to nmol/tube. The enzyme activity was measured as units/mg protein of the HIRMAb–SGSH fusion protein, where 1 unit = nmol of 4-MU product formed during the 17 h primary incubation.11
GAG Reduction in Sanfilippo Type A Fibroblasts
The SGSH enzyme hydrolyzes the sulfate group from heparan sulfate GAGs,1 which allows for subsequent degradation of the GAG by other enzymes in the cell. The effect of treatment with the HIRMAb–SGSH fusion protein on the release of sulfate from GAGs was examined in Sanfilippo Type A fibroblasts (MPS-IIIA fibroblasts), which were obtained from the Coriell Institute for Medical Research (Camden, NJ). The cells were pulsed with 35S-sulfate, which is incorporated into both lysosomal GAGs, as well as cell surface GAGs. The lysosomal enzyme only reduces GAG accumulation in the lysosomal compartment, not the cell surface. Therefore, a pulse-chase experiment was designed to lower the cell surface 35S-labeled GAGs, as described previously.12 Sanfilippo A fibroblasts were grown in 6-well cluster dishes to confluency, and then a pulse-chase experiment is performed with [35S]-sodium sulfate (PerkinElmer, Waltham, MA). For the pulse phase, the confluent cells were washed with phosphate buffered saline (PBS), and incubated with 1 mL of low sulfate Ham’s F12 medium with 10% dialyzed fetal bovine serum (FBS) and 47 μCi/mL of [35S]sodium sulfate for 48 h at 37 °C in a humidified incubator. The medium was aspirated, and the wells were washed with PBS, and the cells were incubated with 1 mL/well of radioactivity-free DMEM/F12 medium with 10% regular FBS and different concentrations of the HIRMAb–SGSH fusion protein for 48 h at 37 °C in a humidified incubator. The medium was aspirated and the cells washed with PBS. The cells were removed from the well with 0.4 mL/well of 0.05% trypsin/EDTA at 37 °C for 4 min, as this step removes radioactivity adhered to the cell surface and not incorporated in lysosomal GAGs. The cells are suspended in serum-free medium to stop the trypsin reaction and centrifuged at 1000g. The supernatant was removed and discarded, and the cell pellet was solubilized in 0.4 mL of 1N NaOH at 60C for 1 h. The protein content was measured with the bicinchoninic acid (BCA) protein assay. The 35S radioactivity was measured with a PerkinElmer liquid scintillation counter in Ultima-gold (PerkinElmer). The CPM radioactivity was divided by the mg protein well and the data reported as CPM/mg protein.
Confocal Microscopy in Sanfilippo Type A Fibroblasts
Sanfilippo A fibroblasts were grown overnight in DMEM with 10% fetal bovine serum to >80% confluency on autoclaved 22 mm × 22 mm Fisherfinest glass coverslips (number 1 thickness) and placed on Costar 3516 6-well cluster dishes (Thermo Scientific, Rochester, NY). The cells were treated with 10 μg/mL of the HIRMAb–SGSH fusion protein, as previously described.9 Following a 6 h incubation at 37 °C, the medium was aspirated, the wells were washed extensively with cold PBS, and the cells were fixed with 4% paraformaldehyde for 40 min at 4 °C and permeabilized with 0.5% Triton X-100 for 15 min at room temperature. Following a PBS wash, the plates were blocked with 10% donkey serum and then colabeled with 10 μg/mL of a rabbit anti-SGSH antibody (Abcam) and 10 μg/mL of a mouse mAb to human lysosomal associated membrane protein (LAMP)-1 (Developmental Studies Hybridoma Bank, Iowa City, IA). Negative control antibodies were previously described.9 The secondary antibodies (Life Technologies, Carlsbad, CA) were 5 μg/mL each of Alexa Fluor-488 conjugated donkey antimouse IgG (green channel) and Alexa Fluor-555 conjugated donkey antirabbit IgG (red channel). The washed slides were air-dried and mounted in Vectashield Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Confocal microscopy was performed as described previously.9
Rhesus Monkey Brain Uptake and Pharmacokinetics
The HIRMAb–SGSH fusion protein was radiolabeled with [125I]-monoiodinated-Bolton–Hunter reagent (PerkinElmer). The specific activity of the Bolton–Hunter reagent was 2200 μCi/nmol, and the final specific activity of the [125I]-HIRMAb–SGSH fusion protein was 3.7 μCi/μg. The trichloroacetic acid (TCA) precipitability of the [125I]-HIRMAb–SGSH fusion protein was >97% for least 8 days after labeling during storage at −70 °C. Prior to labeling, the fusion protein was buffer exchanged with 0.01 M sodium acetate/140 mM NaCl/pH = 5.5/0.001% Tween-80, as described previously.9 The labeled HIRMAb–SGSH fusion protein was purified by gel filtration, as described previously.9 An adult male Rhesus monkey, 17.3 kg, was investigated at MPI Research (Mattawan, MI). The animal was injected intravenously (IV) with 1200 μCi of [125I]-HIRMAb–SGSH fusion protein by bolus intravenous injection, as described previously.9 The injection dose (ID) of the HIRMAb–SGSH fusion protein was 19 μg/kg. Anesthesia was induced with intramuscular ketamine, as described previously.9 All procedures were carried out in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the U.S. National Institutes of Health. Following intravenous drug administration, femoral venous plasma was obtained between 2 and 140 min for determination of total plasma [125I] radioactivity (DPM/mL) and plasma radioactivity that is precipitated by 10% cold TCA, as described previously.9 The animal was euthanized, and samples of brain and major organs (heart, liver, spleen, lung, skeletal muscle, and omental fat) were processed for determination of radioactivity, as described previously.9 Samples (∼2 g) of frontal cortex were removed for capillary depletion analysis to confirm transport of the HIRMAb–SGSH fusion protein across the BBB, as described previously.9,13
The TCA precipitable [125I] radioactivity in plasma, DPM/mL, was converted to ng/mL, based on the specific activity of the injected fusion protein, and a biexponential equation, %ID/mL = A1e–k1t + A2e–k2t, was fit to the plasma fusion protein concentration. The intercepts (A1, A2) and the slopes (k1, k2) were used to compute the median residence time (MRT), the central volume of distribution (Vc), the steady state volume of distribution (Vss), the area under the plasma concentration curve (AUC), and the systemic clearance (CL), as described previously.14 Nonlinear regression analysis used the AR subroutine of the BMDP Statistical Software (Statistical Solutions Ltd., Cork, Ireland). Data were weighted by 1/(ng/mL)2.
The BBB permeability-surface area (PS) product was computed from the terminal brain uptake, %ID/g, divided by the terminal (140 min) plasma AUC, %ID·min/mL, and reported as μL/min/g.
Statistical Analysis
Statistical differences were determined by Analysis of Variance (ANOVA) with Bonferroni correction.
Results
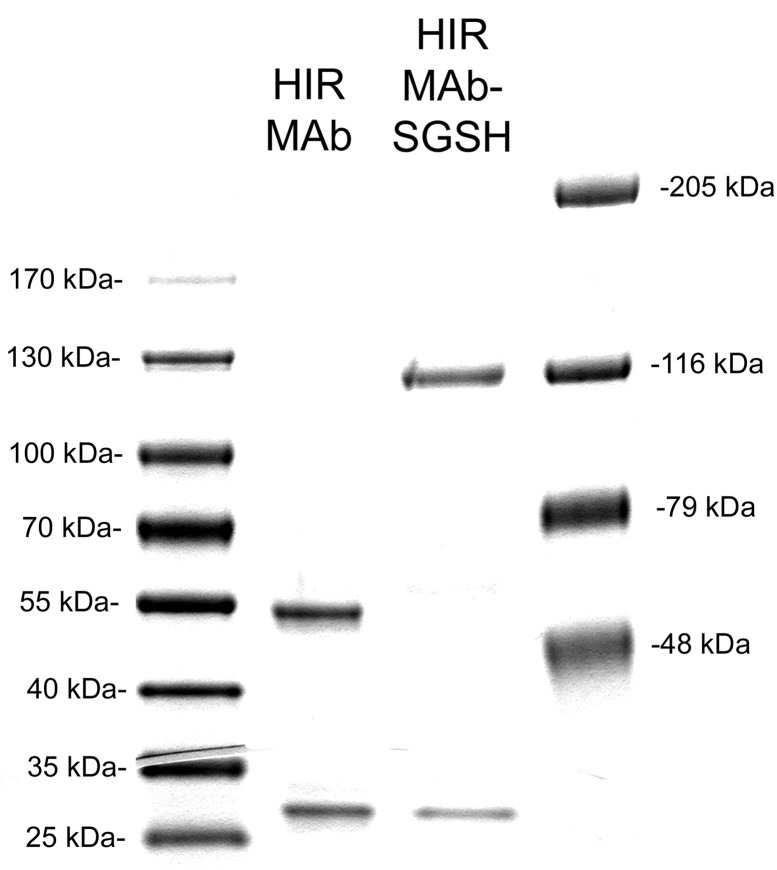
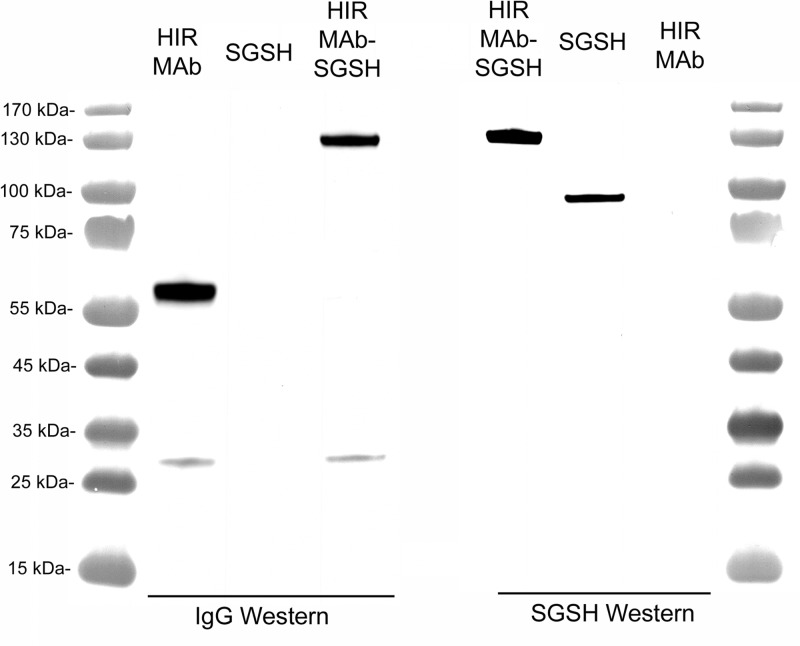
The CHO-derived HIRMAb–SGSH fusion protein was purified by protein A affinity chromatography to homogeneity on SDS-PAGE (Figure 2). The HIRMAb–SGSH fusion protein and the HIRMAb use the same LC, which migrates with the same mobility on SDS-PAGE (Figure 2). The size of the HC of the HIRMAb–SGSH fusion protein is larger than the size of the HC of the HIRMAb, owing to fusion of the SGSH to the antibody HC (Figure 2). The antihuman IgG antibody reacts with the LC and HC of both the HIRMAb and the HIRMAb–SGSH fusion protein but not with recombinant SGSH (Figure 3, left). The antihuman SGSH antibody reacts with recombinant SGSH and the HC of the HIRMAb–SGSH fusion protein but not with the HC of the HIRMAb (Figure 3, right). On the basis of the migration of molecular weight (MW) standards on the Western blot (lane 1, Figure 3), the MW of the LC and the HC of the HIRMAb–SGSH fusion protein is 27 and 133 kDa, respectively. The MW of the heterotetrameric IgG–enzyme fusion protein shown in Figure 1 is estimated to be 320 kDa based on mobility in the Western blotting.
Figure 2.
Reducing SDS-PAGE and Coomassie blue staining of the chimeric HIRMAb and the HIRMAb–SGSH fusion protein.
Figure 3.
Western blot with a primary antibody against human IgG (left) or human SGSH (right). The proteins tested are the HIRMAb alone, recombinant SGSH alone, and the HIRMAb–SGSH fusion protein.
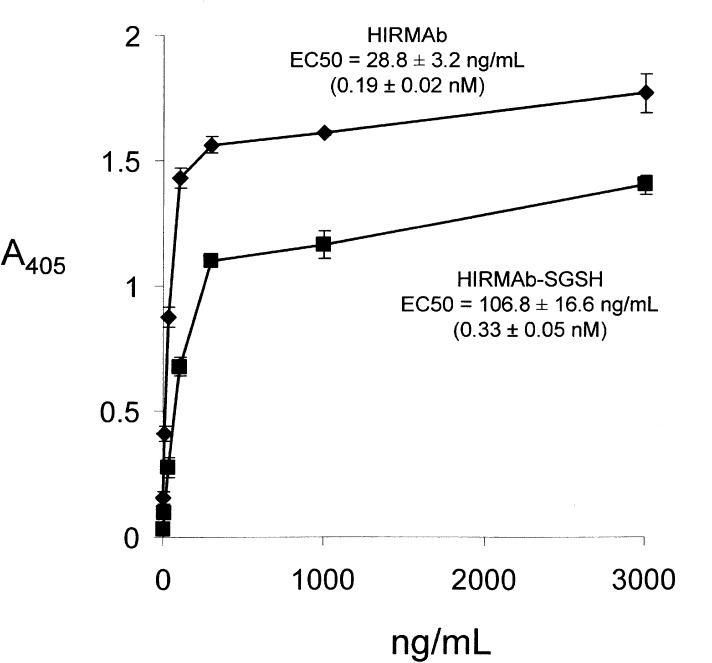
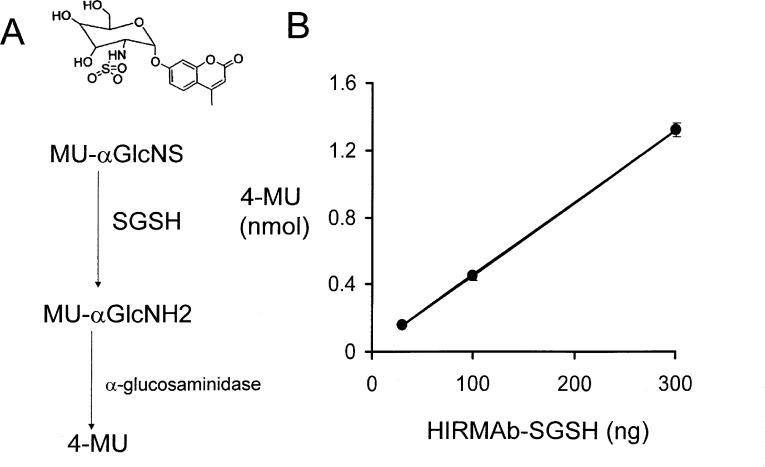
The binding of the HIRMAb–SGSH fusion protein, and the HIRMAb alone, to the HIR was measured with an ELISA (Experimental Section). The EC50 of the HIRMAb–SGSH fusion protein binding to the HIR, 0.33 ± 0.05 nM, is comparable to the EC50, 0.19 ± 0.02 nM, of the HIRMAb binding to the HIR (Figure 4). The SGSH enzyme activity of the HIRMAb–SGSH fusion protein was measured with a two-step fluorometric assay (Figure 5A), using the custom synthesized substrate shown in Figure 5A. The assay was linear with respect to mass of fusion protein (Figure 5B), and the average enzyme activity was 4712 ± 388 units/mg protein.
Figure 4.
Binding to the HIR is saturable for the HIRMAb and the HIRMAb–SGSH fusion protein. The EC50 was determined by nonlinear regression analysis and the value in ng/mL was converted to nM, based on a molecular weight of 150 kDa for the HIRMAb and a molecular weight of 320 kDa for the HIRMAb–SGSH fusion protein.
Figure 5.
(A) The SGSH fluorometric enzyme assay is a two-step assay. The substrate, 4-methylumbelliferyl-α-d-N-sulphoglucosaminide (MU-α-GlcNS), is converted by SGSH to methylumbelliferyl-α-d-glucosaminide (MU-α-GlcNH2), which is then converted to the fluorescent product, 4-methyl umbelliferone (4-MU), by the second step enzyme, α-glucosaminidase. (B) Linear formation of the 4-MU product by the addition of the 30–300 ng of the HIRMAb–SGSH fusion protein. Data are mean ± SD of three replicates; error bars are shown.
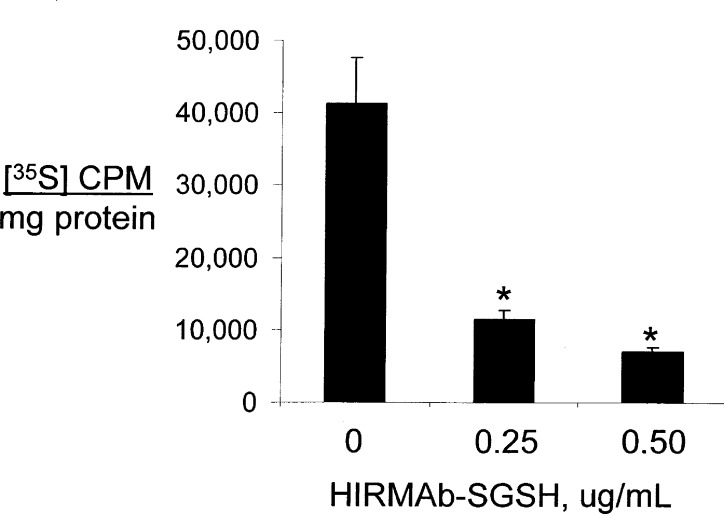
MPSIIIA fibroblasts were incubated with 10 μg/mL HIRMAb–SGSH fusion protein for 6 h followed by fixation and immune labeling with a mouse monoclonal antibody to LAMP1, a lysosomal marker, and a rabbit polyclonal antibody to human SGSH. The SGSH immunoreactivity is detected in the red channel (Figure 6A), and the LAMP1 immunoreactivity within the cell is detected in the green channel (Figure 6B). The overlap of the SGSH and LAMP1 immunoreactivity is shown in Figure 6C and indicates the HIRMAb–SGSH fusion protein is triaged to the lysosomal compartment following uptake into MPSIIIA fibroblasts. Nuclear labeling with DAPI is shown in Figure 6D. There was no immunoreactivity in the cells labeled with isotype control antibodies. The HIRMAb–SGSH fusion protein was pharmacologically active in MPSIIIA fibroblasts, as low concentrations of the fusion protein, 0.25 and 0.5 μg/mL, cause a 72% and 83% reduction in lysosomal GAGs labeled with sulfate in MPS-IIIA fibroblasts (Figure 7).
Figure 6.
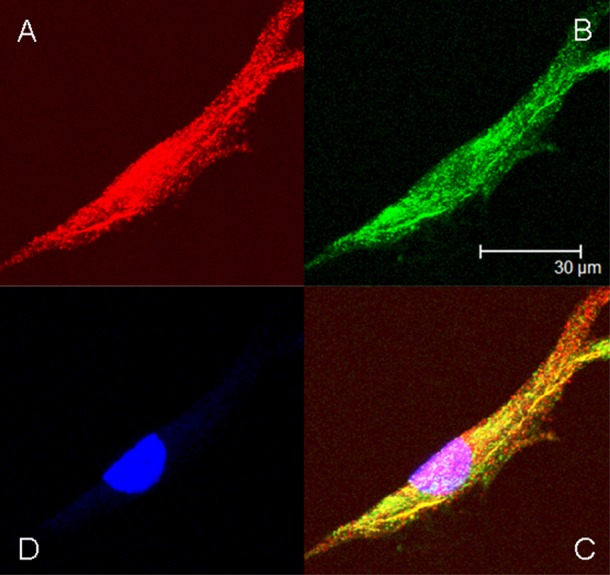
Confocal microscopy of MPSIIIA fibroblasts treated with the HIRMAb–SGSH fusion protein and immuno-stained with a primary antibody against either SGSH (red channel (A)) or LAMP1 (green channel (B)). Overlap of red and green images of the immune staining yields a yellow image as shown in (C). (D) nuclear labeling with DAPI.
Figure 7.
Incorporation of 35S-sulfate into lysosomal GAGs in MPSIIIA fibroblasts is reduced by 48 h incubations with 0.25–0.5 μg/mL HIRMAb–SGSH fusion protein. Mean ± SD (n = 4). *P < 0.01 difference from control.
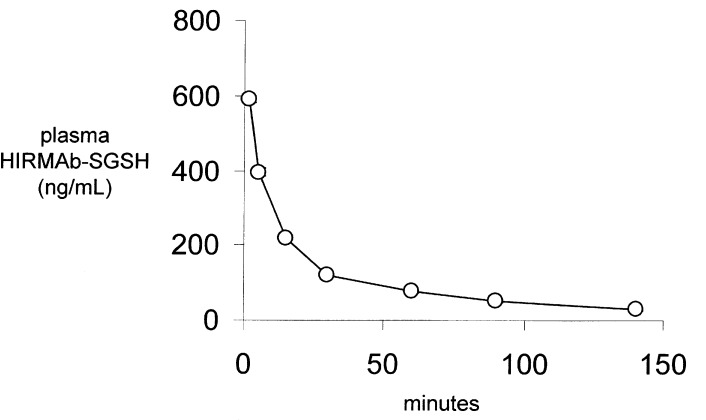
The HIRMAb–SGSH fusion protein was radiolabeled with the [125I]-Bolton–Hunter reagent to a specific activity of 3.7 μCi/μg and a TCA precipitation of 97%. The [125I]-HIRMAb–SGSH fusion protein (1200 μCi, 324 μg) was injected IV in a male Rhesus monkey. The time course of TCA precipitable [125I]-HIRMAb–SGSH fusion protein is shown in Figure 8. The percent of total plasma radioactivity that was precipitable by TCA was 96 ± 1%, 95 ± 1%, 94 ± 1%, 89 ± 1%, 84 ± 2%, 79 ± 1%, and 72 ± 2%, respectively, at 2, 5, 15, 30, 60, 90, and 140 min after IV injection. A 2-exponential equation was fit to the plasma profile of TCA-precipitable fusion protein (Experimental Section) to yield the pharmacokinetic (PK) parameters shown in Table 1. The [125I]-HIRMAb–SGSH fusion protein is rapidly cleared from plasma with a mean residence time of 62 ± 4 min, a systemic volume of distribution (Vss) that is 2.5-fold greater the central compartment volume (Vc), and a high rate of systemic clearance, 1.11 ± 0.03 mL/min/kg (Table 1).
Figure 8.
Plasma TCA-precipitable [125I]-HIRMAb–SGSH fusion protein concentration, ng/mL, in the adult Rhesus monkey, is plotted vs time over a 140 min period after a single IV injection of 19 μg/kg the fusion protein.
Table 1. Pharmacokinetic Parameters of the HIRMAb–SGSH Fusion Proteina.
| parameter | units | value |
|---|---|---|
| T1/21 | min | 5.5 ± 0.8 |
| T1/22 | min | 55 ± 4 |
| MRT | min | 62 ± 4 |
| Vc | mL/kg | 28 ± 2 |
| Vss | mL/kg | 69 ± 4 |
| AUCss | μg·min/mL | 16.8 ± 0.5 |
| CL | mL/min/kg | 1.11 ± 0.03 |
Parameters computed from the plasma profile in Figure 8. T1/21 and T1/22 are the half-times of plasma clearance for the first phase (α) and second phase (β) phases.
The volume of distribution (VD) of the HIRMAb–SGSH fusion protein in total brain homogenate at 140 min after injection is high, 782 ± 36 μL/g, compared to the brain VD of a nonspecific human IgG1 isotype control antibody, 20 ± 6 μL/g (Table 2). The brain VD of the IgG1 isotype control antibody represents the brain uptake of a molecule that is sequestered within the blood volume of brain, and which does not cross the BBB, as described previously.9 The VD of the HIRMAb–SGSH fusion protein in the postvascular supernatant, 666 ± 71 μL/g, is greater than the VD of the HIRMAb–SGSH fusion protein in the vascular pellet of brain, 24 ± 17 μL/g (Table 2), which indicates that the majority of the HIRMAb–SGSH fusion protein has traversed the BBB and penetrated the brain parenchyma. The radioactivity in the postvascular supernatant represents intact HIRMAb–SGSH fusion protein, and not labeled metabolites, as the TCA precipitation of the postvascular supernatant radioactivity is 95.9 ± 0.7% (Table 2).
Table 2. Capillary Depletion Analysis of the Brain Uptake of the HIRMAb–SGSH Fusion Proteina.
| molecule | brain fraction | VD (μL/g) |
|---|---|---|
| HIRMAb–SGSH fusion protein | brain homogenate | 782 ± 36 |
| postvascular supernatant | 666 ± 71 | |
| vascular pellet | 24 ± 17 | |
| human IgG1 isotype control | brain homogenate | 20 ± 6 |
Mean ± SD. The fusion protein was administered by IV injection, and brain measurements made 140 min following injection. The radioactivity in the postvascular supernatant was 95.9 ± 0.7% precipitable by cold 10% trichloroacetic acid. The homogenate VD for the human IgG1 isotype control antibody was reported previously.23
The organ uptake of the HIRMAb–SGSH fusion protein is expressed as % of injected dose (ID) per 100 g wet organ weight (Table 3) because the brain of the adult Rhesus monkey weighs 100 g.14 The major organs accounting for the removal of the HIRMAb–SGSH fusion protein from plasma are liver and spleen (Table 3). The brain cortical uptake of the HIRMAb–SGSH fusion protein is 0.81 ± 0.07% ID/100 g brain (Table 3). The BBB PS product, a measure of brain clearance (Experimental Section), for the HIRMAb–SGSH fusion protein is 1.8 ± 0.2 μL/min/g.
Table 3. Organ Uptake of the HIRMAb–SGSH Fusion Protein in the Rhesus Monkeya.
| organ | organ uptake (%ID/100 g) |
|---|---|
| frontal cortex | 0.81 ± 0.07 |
| cerebellar cortex | 0.60 ± 0.04 |
| choroid plexus | 1.13 ± 0.06 |
| liver | 16.2 ± 0.4 |
| spleen | 14.7 ± 0.5 |
| lung | 1.1 ± 0.2 |
| heart | 0.93 ± 0.11 |
| fat | 0.044 ± 0.004 |
| skeletal muscle | 0.31 ± 0.21 |
Data are mean ± SD of triplicate samples.
Discussion
The results of these studies are consistent with the following conclusions. First, fusion of the SGSH enzyme to the carboxyl terminus of the heavy chain of the HIRMAb (Figure 1) results in a bifunctional HIRMAb–SGSH fusion protein that retains both high affinity binding to the HIR (Figure 4) and high SGSH enzyme activity (Figure 5). Second, the HIRMAb–SGSH fusion protein is taken up by MPSIIIA fibroblasts (Figure 6C) and is pharmacologically active in these cells with 72–83% reduction in lysosomal GAGs (Figure 7). Third, the HIRMAb–SGSH fusion protein is rapidly cleared from plasma following IV injection (Table 1, Figure 8), owing to rapid uptake by peripheral tissues (Table 3). Fourth, the HIRMAb–SGSH fusion protein is taken up by brain at a rate of ∼1% ID/brain (Table 3), and capillary depletion results show rapid movement of the fusion protein into the postvascular parenchyma of brain (Table 2).
The SGSH lysosomal enzyme could be fused to either the amino terminus or the carboxyl terminus of the HIRMAb. Fusion of the enzyme to the amino terminus would enable folding of the enzyme cotranslationally prior to folding of the antibody heavy chain, which could promote retention of enzyme activity in the form of the fusion protein. However, the HIR binding site of the HIRMAb is near the amino terminus of the IgG chains, and fusion of a lysosomal enzyme to the heavy chain amino terminus of the HIRMAb results in a >95% reduction in HIRMAb binding to the HIR.15 High affinity binding to the HIR is retained following lysosomal enzyme fusion to the carboxyl terminus of the HIRMAb heavy chain, but this then resulted in a >95% loss of enzyme activity of a model lysosomal enzyme, β-glucuronidase.15 In the present study, SGSH was fused to the carboxyl terminus of the IgG, and the SGSH enzyme activity of the fusion protein is 4712 ± 388 units/mg protein (Results). The SGSH enzyme specific activity of the 60 kDa recombinant human SGSH, using the same two-step fluorometric enzyme assay, is 15000 units/mg protein.4 However, following re-engineering of the SGSH as a heterotetrameric IgG–SGSH fusion protein with a MW of 320 kDa (Results), the effective MW of the SGSH domain of the fusion protein is 160 kDa, whereas the MW of SGSH is 60 kDa. After normalization for MW differences, the effective SGSH specific activity of the fusion protein is equivalent to 12800 units/mg recombinant SGSH, which is 85% of the enzyme activity of recombinant SGSH. Therefore, fusion of the SGSH to the carboxyl terminus of the HC of the HIRMAb had minimal effect on the enzyme activity of the SGSH enzyme. The sulfatase activity of SGSH requires a post-translational conversion of a cysteine residue in the near amino terminal region of the enzyme to a N-formyl glycine residue via the enzymatic action of sulfatase modifying factor type 1, or SUMF1.16 Expression of SGSH in transfected host cells is dependent on cotransfection of the host cell with genes for both SGSH and SUMF1.16 However, when the SGSH is stably expressed in CHO cells as an IgG–SGSH fusion protein, without cotransfection with SUMF1, the CHO cells secrete an enzymatically active IgG–SGSH fusion protein. The retention of SGSH enzyme activity following fusion to the HIRMAb is also demonstrated by the 83% reduction in lysosomal GAGs following treatment of MPSIIIA fibroblasts with the HIRMAb–SGSH fusion protein (Figure 7). Triage of the HIRMAb–SGSH fusion protein into the lysosomal compartment of MPSIIIA fibroblasts is shown by confocal microscopy (Figure 6C).
The HIRMAb–SGSH fusion protein is rapidly cleared from plasma following IV injection, owing to uptake by peripheral tissues, similar to the rapid plasma clearance of recombinant SGSH.17 However, recombinant SGSH does not enter the brain from blood,17 which is problematic in the treatment of MPSIIIA, because the primary clinical symptoms of MPSIIIA involves the CNS.2 The present work shows the brain uptake of the HIRMAb–SGSH fusion protein is 0.8% of injected dose (ID)/100 g brain in the Rhesus monkey. Therefore, an injection dose (ID) of 3 mg/kg HIRMAb–SGSH fusion protein in a 17 kg Rhesus monkey is equal to 50 mg, or an ID of about 250000 units of enzyme activity. With a brain uptake of 1% ID/100 g brain (Table 3), the brain SGSH enzyme activity is 2500 units/100 g brain, or 25 units/g brain. There are 100 mg protein per g brain.18 Therefore, the SGSH enzyme activity is predicted to be 0.25 units/mg protein in brain following administration of a 3 mg/kg dose, which is comparable to the endogenous SGSH enzyme activity in brain, 0.12 units/mg protein, as determined by the same two-step fluorometric enzyme assay.19 These calculations suggest it is possible to achieve therapeutic SGSH enzyme activity in the brain following administration of the enzyme in the form of the BBB-penetrating HIRMAb–SGSH fusion protein. The HIRMAb resides in primate brain with a half-time of 16 h.20 However, the tissue half-time of HIRMAb–sulfatase enzyme activity is longer, 3 days,21 due to sequestration within the lysosome.
Penetration of the BBB followed by distribution of the HIRMAb–SGSH fusion protein into the postvascular parenchyma of brain is demonstrated by the capillary depletion method (Table 2). If the fusion protein was retained within the endothelial compartment of the brain microvasculature, then the VD in the vascular pellet would be high and the VD in the postvascular supernatant would be low. However, the opposite is observed, as the VD in the postvascular supernatant is 28-fold higher than the VD in the vascular compartment (Table 2). The radioactivity in the postvascular supernatant is not an artifact caused by brain uptake of low MW metabolites generated by rapid systemic degradation of the fusion protein because low MW metabolites labeled with the [125I]-Bolton–Hunter reagent do not cross the BBB.9,22 This is also demonstrated in the present study, where the TCA precipitability of the radioactivity in the postvascular supernatant of brain is high, 96 ± 1% (Table 2), compared to the TCA precipitability of the radioactivity in the terminal plasma, 72 ± 2% (Results). The results obtained with the capillary depletion method have been corroborated with emulsion autoradiography of Rhesus monkey brain, which shows broad distribution of a HIRMAb–sulfatase fusion protein in brain parenchyma following IV administration.9
In summary, the present study describes the re-engineering of the SGSH lysosomal enzyme as an IgG–SGSH fusion protein, wherein the IgG domain is the HIRMAb that mediates transport across the BBB, as well as uptake into target cells, which is followed by triage to the lysosomal compartment of target cells. The HIRMAb–SGSH fusion protein is bifunctional and retains both high affinity binding for the HIR and high SGSH enzyme activity. The HIRMAb–SGSH fusion protein is a new form of receptor-mediated enzyme replacement therapy of the brain in MPSIIIA.
Acknowledgments
This research was supported by NIH grant R43-NS-088868-01. The authors are indebted to Winnie Tai, Phuong Tram, and Stephanie Lee for technical support and to MPI Research, Inc. (Mattawan, MI) for collaboration with the primate study. Drs. Boado, Lu, and Hui are employees, and Dr. Pardridge is a consultant, of ArmaGen Technologies. Confocal laser scanning microscopy was performed at the CNSI Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA.
The authors declare the following competing financial interest(s): Drs. Boado, Lu, and Hui are employees, and Dr. Pardridge is a consultant, of ArmaGen Technologies.
Funding Statement
National Institutes of Health, United States
References
- Valstar M. J.; Ruijter G. J. G.; Van Diggelen O. P.; Poorthuis B. J.; Wijburg F. A. Sanfilippo syndrome: a mini-review. J. Inherited Metab. Dis. 2008, 31, 240–252. [DOI] [PubMed] [Google Scholar]
- Valstar M. J.; Neijs S.; Bruggenwirth H. T.; Olmer R.; Ruijter G. J. G.; Wevers R. A.; Van Diggelen O. P.; Poorthuis B. J.; Halley D. J.; Wijburg F. A. Mucopolysaccharidosis type IIIA: clinical spectrum and genotype–phenotype correlations. Ann. Neurol. 2010, 68, 876–887. [DOI] [PubMed] [Google Scholar]
- Scott H. S.; Blanch L.; Guo X.-H.; Freeman C.; Orsborn A.; Baker E.; Sutherland G. R.; Morris C. P.; Hopwood J. J. Cloning of the sulphamidase gene and identification of mutations in Sanfilippo A syndrome. Nature Genet. 1995, 11, 465–467. [DOI] [PubMed] [Google Scholar]
- Urayama A.; Grubb J. H.; Sly W. S.; Banks W. A. Mannose 6-phosphate receptor-mediated transport of sulfamidase across the blood–brain barrier in the newborn mouse. Mol. Ther. 2008, 16, 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly R. D.; Marshall N. R.; Perrott M. R.; Dittmer K. E.; Hemsley K. M.; Beard H. Intracisternal enzyme replacement therapy in lysosomal storage diseases: routes of absorption into brain. Neuropathol. Appl. Neurobiol. 2011, 37, 414–422. [DOI] [PubMed] [Google Scholar]
- Pardridge W. M. Drug transport across the blood–brain barrier. J. Cereb. Blood Flow Metabol. 2012, 32, 1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardridge W. M.; Boado R. J. Reengineering biopharmaceuticals for targeted delivery across the blood–brain barrier. Methods Enzymol. 2012, 503, 269–292. [DOI] [PubMed] [Google Scholar]
- Boado R. J.; Zhang Y.-F.; Zhang Y.; Pardridge W. M. Humanization of anti-human insulin receptor antibody for drug targeting across the human blood–brain barrier. Biotechnol. Bioeng. 2007, 96, 381–391. [DOI] [PubMed] [Google Scholar]
- Boado R. J.; Lu J. Z.; Hui W.K.-W.; Sumbria R. K.; Pardridge W. M. Pharmacokinetics and brain uptake in the Rhesus monkey of a fusion protein of arylsulfatase A and a monoclonal antibody against the human insulin receptor. Biotechnol. Bioeng. 2013, 110, 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montfort M.; Garrido E.; Hopwood J. J.; Grinberg D.; Chabas A.; Vilageliu L. Expression and functional characterization of human mutant sulfamidase in insect cells. Mol. Genet. Metab. 2004, 83, 246–251. [DOI] [PubMed] [Google Scholar]
- Karpova E. A.; Voznyi Y. V.; Keulemans J. L. M.; Hoogeveen A. T.; Winchester B.; Tsvetkova I. V.; Van Diggelen O. P. A fluorimetric enzyme assay for the diagnosis of Sanfilippo disease type A (MPS IIIA). J. Inherited Metab. Dis. 1996, 19, 278–285. [DOI] [PubMed] [Google Scholar]
- LaManna W. C.; Lawrence R.; Sarrazin S.; Esko J. D. Secondary storage of dermatan sulfate in Sanfilippo disease. J. Biol. Chem. 2011, 286, 6955–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triguero D.; Buciak J. B.; Pardridge W. M. Capillary depletion method for quantifying blood–brain barrier transcytosis of circulating peptides and plasma proteins. J. Neurochem. 1990, 54, 1882–1888. [DOI] [PubMed] [Google Scholar]
- Pardridge W. M.; Kang Y. S.; Buciak J. L.; Yang J. Human insulin receptor monoclonal antibody undergoes high affinity binding to human brain capillaries in vitro and rapid transcytosis through the blood–brain barrier in vivo in the primate. Pharm. Res. 1995, 12, 807–816. [DOI] [PubMed] [Google Scholar]
- Boado R. J.; Pardridge W. M. Genetic engineering of IgG–glucuronidase fusion proteins. J. Drug Targeting 2010, 18, 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraldi A.; Biffi A.; Lomardi A.; Visigalli I.; Pepe S.; Settembre C.; Nusco E.; Auricchio A.; Naldini L.; Ballabio A.; Cosma M. P. SUMF1 enhances sulfatase activities in vivo in five sulfatase deficiencies. Biochem. J. 2007, 403, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozaklis T.; Beard H.; Hassiotis S.; Garcia A. R.; Tonini M.; Luck A.; Pan J.; Lamsa J. C.; Hopwood J. J.; Hemsley K. M. Impact of high-dose, chemically modified sulfamidase on pathology in a murine model of MPS IIIA. Exp. Neurol. 2011, 230, 123–130. [DOI] [PubMed] [Google Scholar]
- Dunlop D. S.; Yang W.-R.; Lajtha A. The effect of elevated plasma phenylalanine levels on protein synthesis rates in adult rat brain. Biochem. J. 1994, 302, 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatsu S.; Vogler C.; Montano A. M.; Gutierrez M.; Oikawa H.; Dung V. C.; Orii T.; Noguchi A.; Sly W. S. Murine model (Galnstm(C76S)slu) of MPS IVA with missense mutation at the active site cysteine conserved among sulfatase proteins. Mol. Genet. Metab. 2007, 91, 251–258. [DOI] [PubMed] [Google Scholar]
- Wu D.; Yang J.; Pardridge W. M. Drug targeting of a peptide radiopharmaceutical through the primate blood–brain barrier in vivo with a monoclonal antibody to the human insulin receptor. J. Clin. Invest. 1997, 100, 1804–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. Z.; Boado R. J.; Hui E.K.-W.; Zhou Q.-H.; Pardridge W. M. Expression in CHO cells and pharmacokinetics and brain uptake in the Rhesus monkey of an IgG–iduronate-2-sulfatase fusion protein. Biotechnol. Bioeng. 2011, 108, 1954–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boado R. J.; Hui E. K.-W.; Lu J. Z.; Sumbria R. K.; Pardridge W. M. Blood–brain barrier molecular Trojan horse enables brain imaging of radioiodinated recombinant protein in the Rhesus monkey. Bioconjugate Chem. 2013, 24, 1741–1749. [DOI] [PubMed] [Google Scholar]
- Boado R. J.; Pardridge W. M. Comparison of blood–brain barrier transport of GDNF and an IgG–GDNF fusion protein in the Rhesus monkey. Drug Metab. Dispos. 2009, 37, 2299–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]