SUMMARY
Tissue-resident macrophages are highly heterogeneous in terms of their functions and phenotypes as a consequence of adaptation to different tissue environments. Local tissue-derived signals are thought to control functional polarization of resident macrophages; however, the identity of these signals remains largely unknown. It is also unknown whether functional heterogeneity is a result of irreversible lineage-specific differentiation or a consequence of continuous but reversible induction of diverse functional programs. Here, we identified retinoic acid as a signal that induces tissue-specific localization and functional polarization of peritoneal macrophages through the reversible induction of transcription factor GATA6. We further found that GATA6 in macrophages regulates gut IgA production through peritoneal B-1 cells. These results provide insight into the regulation of tissue-resident macrophage functional specialization by tissue-derived signals.
INTRODUCTION
Macrophages are among the most multifunctional and heterogeneous cell types, present in virtually every mammalian tissue, where they monitor local environment and maintain homeostasis (Davies et al., 2013; Hume et al., 1983; Wynn et al., 2013). They express a broad array of sensing molecules, including scavenger receptors, pattern recognition receptors, nuclear hormone receptors, and cytokine receptors, which allows macrophages to monitor tissue microenvironments and act as sentinel cells for infection and tissue damage. In addition, macrophages perform many tissue-specific functions, which is reflected in their phenotypic diversity. Thus, alveolar macrophages, Kupffer cells, microglia, and osteoclasts all have specialized functions and phenotypes, suggesting that local tissue-derived signals may control the development of tissue-specific phenotypes (Gordon and Taylor, 2005; Murray and Wynn, 2011). However, with some exceptions (Boyle et al., 2003), these signals remain largely unknown.
It is also increasingly appreciated that distinct transcriptional master regulators control the development of tissue-specific macrophage phenotypes (Gautier et al., 2012). Several examples of transcription factors that dictate tissue-specific transcription programs in macrophages have been reported, and the deletion of these transcription factors resulted in the ablation of particular tissue macrophage subsets (Kohyama et al., 2009; A-Gonzalez et al., 2013; Takayanagi et al., 2002), suggesting their involvement in the differentiation of the corresponding macrophage populations. In addition, mature macrophages can undergo functional polarization in response to environmental signals (Stout et al., 2005). Two well-appreciated macrophage polarization programs are classically activated (M1) and alternative activated (M2) macrophages that are induced by different stimuli such as LPS+IFNγ and IL-4, respectively (Biswas and Mantovani, 2010; Gordon and Martinez, 2010). Transcription factors, including STAT1, STAT6, C/EBPβ, IRF-4, IRF5, and PPARγ, have been shown to regulate transcription programs that control M1/M2 macrophage polarizations (Lawrence and Natoli, 2011). It is also increasingly appreciated that many other functional polarization programs of macrophages likely exist, which may be expressed in either an inducible or constitutive and tissue-specific manner. However, the signals and transcription factors that control most of these programs remain to be defined.
In principle, tissue-specific phenotypes of macrophages (or any other cell type) can be generated by hard-wired, irreversible differentiation programs that are controlled by lineage-specific master regulators. Alternatively, they can be based on functional polarization programs, which are reversible and inducible on demand, analogous to M1 and M2 polarizations. In the latter scenario, one can expect that multiple transcriptional regulators may be induced to control specific functional programs at times and places specified by diverse functional requirements in different tissues.
Macrophages of the mouse peritoneal cavity are among the best-studied tissue macrophage in terms of cell biology and inflammatory responses (Cain et al., 2013). However, the tissue-specific function of macrophages in this site remains poorly defined. Peritoneal cavity is a unique body compartment for B-1 cell distribution. B-1 cells are a subtype of B cells that account for 35%–70% of B cells in peritoneal cavity, whereas they are almost absent in lymphoid tissues (0.1%–2%) (Baumgarth, 2011). Peritoneal B-1 cells generate the majority of the natural IgM antibodies, including antibody specific for phosphorylcholine (PC). B-1 cells thus constitute a key component of early immune responses to pathogens. Additionally, B-1 cells in peritoneal cavity continuously migrate to intestinal lamina propria, where they give rise to IgA-secreting cells (Baumgarth, 2011; Fagarasan et al., 2010). The tissue-specific role of macrophages in body cavity immunity is not clear in terms of B-1 cell regulation. However, CXCL13, a chemokine that is essential for B-1 cell migration to peritoneal cavity, is abundantly expressed by peritoneal macrophages (Ansel et al., 2002), suggesting that peritoneal macrophages may have a pivotal role in B-1 cell regulation.
Here, we used peritoneal macrophage as an experimental model to investigate the tissue-specific functions and external cues that control their specific gene expression program. Based on the whole-genome gene expression analysis comparing six tissue-resident macrophages, we identified zinc finger transcription factor GATA6 as a regulator of a tissue-specific gene expression program in peritoneal macrophages. GATA6 controls anatomical localization of peritoneal macrophages, but not their development. In addition, we found that GATA6 expression and other peritoneal macrophage-specific gene expression programs are induced by local tissue-derived retinoic acid. Lastly, we show that GATA6 in peritoneal macrophages regulates gut IgA response mediated by peritoneal B-1 cells. Together, our study provides new insight into the mechanism of generation of tissue macrophage diversity.
RESULTS
Identification of GATA6 in Peritoneal Macrophages
The aim of the study was to characterize tissue-derived signals that control diversity of macrophage phenotypes. To address this, we first examined gene expression profiles of tissue-resident macrophages. We purified macrophages from peritoneal cavity, lung, liver, spleen, intestine, and adipose tissue from C57BL/6 mice (Figure S1A available online), and whole-genome gene expression was determined by DNA microarray (Figure 1A). Microarray analysis revealed diversity of tissue macrophages in terms of gene expression (Figure S1B).
Figure 1. Identification of GATA6 in Peritoneal Macrophages.
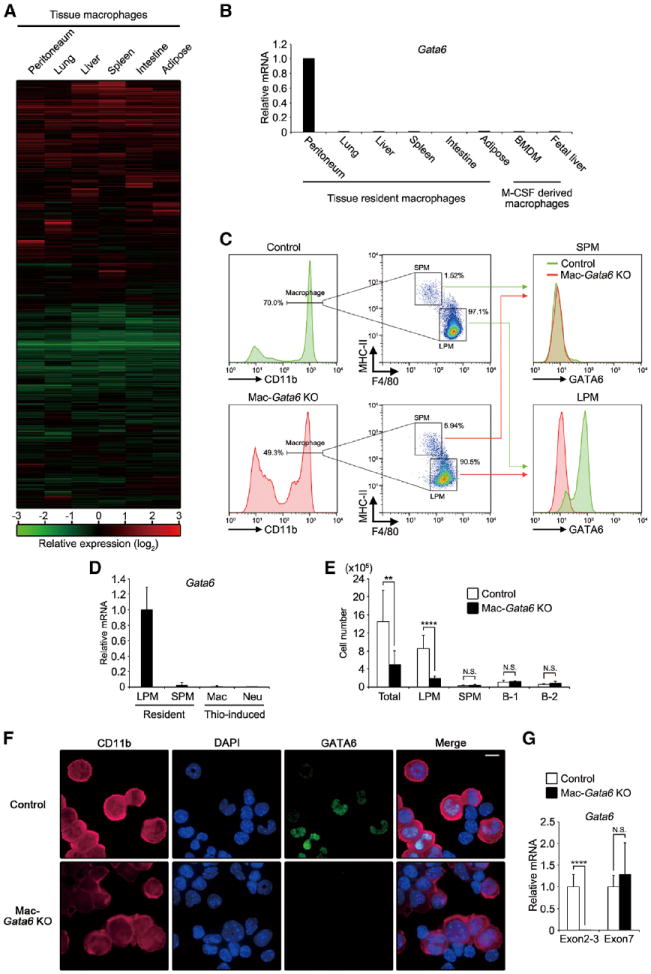
(A) Heatmap displaying hierarchical clustering results from microarray expression data derived from tissue macrophages. Expression levels were normalized by that of BMDM and expressed by relative values (log-2). Genes whose signal was under detection limit were excluded, and 17,513 genes were shown.
(B) Tissue macrophages and in-vitro-cultured macrophages were determined for Gata6 mRNA by quantitative PCR and were expressed as relative values normalized by Gapdh mRNA (n = 1). Graph is representative of two independent experiments.
(C) (Left and middle) Flow cytometry analysis for macrophage subsets in peritoneal exudate cells of controls and Mac-Gata6 KO mice. (Right) Staining of GATA6 protein in SPMs and LPMs.
(D) Quantitative PCR analysis for Gata6 mRNA in LPMs, SPMs, Thio-pMacs, and neutrophils (n = 3–12).
(E) Numbers of total cells, LPM, SPM, B-1 cell, and B-2 cell in peritoneal exudate cells from controls and Mac-Gata6 KO mice (n = 6–12). Data were pooled from three independent experiments with similar results.
(F) Immunofluorescence microscopy of peritoneal exudate cells from control and Mac-Gata6 KO mice stained for CD11b, DAPI, and GATA6. Scale bar, 10 μm.
(G) Quantitative PCR analysis of LPMs for Gata6 mRNA targeting exons 2–3 and exon 7 (n = 6–12).
Errors bars represent SD. **p < 0.01, ****p < 0.0001. N.S., not significant. See also Figure S1.
Consistent with the previous study (Gautier et al., 2012), we found that zinc finger transcription factor GATA6 is uniquely expressed at a high level in peritoneal macrophages compared to tissue-derived, bone-marrow-derived (BMDM) and fetal-liver-derived macrophages (Figure 1B). Previous studies showed that transcription factors NFATc1 and Spic, which regulate tissue-specific transcription programs in osteoclast and splenic red pulp macrophages, respectively, had restricted expression among tissue macrophages (Kohyama et al., 2009; Takayanagi et al., 2002), suggesting that GATA6 may control peritoneal macrophage-specific gene expression. Mouse peritoneal macrophages are made up of two subsets (Ghosn et al., 2010): large peritoneal macrophages (LPMs) and small peritoneal macrophages (SPMs). LPMs make up the majority of peritoneal macrophages and express high levels of F4/80 but low MHC class II (MHC-II); SPMs express lower F4/80 but high levels of MHC-II (Figure 1C, left and middle). The expression of GATA6 in LPMs had bimodal distribution (Figure 1C, lower-right), but it was negligible in SPMs, thioglycollate-induced peritoneal macrophages (Thio-pMacs), and neutrophils (Figures 1C, lower-right, and 1D). In contrast, mRNA of chemokine receptor Ccr2, which plays a critical role in monocyte recruitment during inflammation (Kurihara et al., 1997), was highly expressed in SPMs (Figure S1C). The cell number of SPM, but not of LPM, was significantly reduced in peritoneal exudate from Ccr2 KO mice (Figures S1D and S1E), suggesting that the majority of SPMs are originated from inflammatory monocyte population.
To examine the role of GATA6 in peritoneal macrophages, we crossed Gata6-floxed mice with LysM-cre mice (with macrophage and neutrophil specific Cre expression) to establish mice specifically deficient for Gata6 gene in macrophage lineage (Mac-Gata6 KO), as neutrophils do not express GATA6 (Clausen et al., 1999; Sodhi et al., 2006). Mac-Gata6 KO mice developed LPMs in peritoneal cavity with reduced F4/80 expression (Figure 1C). Furthermore, the number of LPMs, but not SPMs, harvested from peritoneal exudate was greatly reduced in Mac-Gata6 KO mice (Figure 1E). This was consistent with ex vivo analysis of mice genetically labeled with Yfp reporter for macrophage lineage (LysM-Cre;R26-stop-Yfp), which revealed reduction in the numbers of macrophages on parietal peritoneal membrane of Mac-Gata6 KO mice (Figure S1F). Despite the dramatic reduction in LPM numbers, their proliferative status was not affected by GATA6 deficiency (Figure S1G). In addition, blood leukocyte counts were normal in Mac-Gata6 KO mice (Figure S1H).
Immunofluorescence analysis of peritoneal exudate cells from control mice demonstrated restricted expression of GATA6 protein in macrophages and its absence in Mac-Gata6 KO mice (Figure 1F). The elimination of GATA6 protein in LPMs of Mac-Gata6 KO mice was also confirmed by flow cytometry (Figures 1C, lower-right), whereas truncated Gata6 mRNA, which lacks the targeted region, exon 2, was comparably detected in LPMs of Mac-Gata6 KO mice (Figure 1G). These results further confirm that GATA6 is not essential for LPM development.
GATA6-Dependent Tissue-Specific Gene Expression Program
We next determined the role of GATA6 in gene regulation of peritoneal macrophages. DNA microarray identified genes suppressed in Mac-Gata6 KO peritoneal macrophages, many of these genes being specific to peritoneal macrophages in WT mice (Figure S2A). To determine the role of GATA6 in the peritoneal macrophage-specific gene expression program, we selected 44 genes that had expression of at least 5-fold higher in peritoneal macrophages compared to all five other tissue macrophages studied here (Figure 2A). We provisionally termed these genes peritoneal macrophage-specific genes (PMSGs). Microarray analysis revealed that Mac-Gata6 KO peritoneal macrophages strongly downregulated the expression of 39% (17 out of 44 genes) of PMSGs (Figure 2A), which was further confirmed by quantitative PCR (Figures 2B and 2C). In contrast, the rest of PMSGs had comparable expression in Mac-Gata6 KO macrophages (Figures 2B, 2C, S2B, and S2C). Consistently, the reduction of protein expression of CD62P, CD49f, and CD73 was detected in Mac-Gata6 KO LPMs, whereas that of CD102 was intact (Figure 2D). These findings indicate that GATA6 is essential for the induction of a subset of PMSGs. Furthermore, retroviral transduction of Gata6 into fetal-liver-derived macrophages induced the expression of GATA6-dependent PMSGs such as Serpinb2, Cd62p, Thbs1, Tgfb2, and Ltbp1 (Figure 2E), indicating that GATA6 regulates these genes in a cell-autonomous manner.
Figure 2. GATA6-Dependent PMSG Induction.

(A) Heatmap of mRNA expressed at least five times over in peritoneal macrophages relative to their expression in all other tissue macrophages. Expression levels were shown as relative values normalized by that of BMDM. Note that apparent expression of Gata6 mRNA is due to the hybridization region (exon7) of microarray probe (refer to Figure 1G).
(B and C) The mRNA expression of the indicated genes in tissue macrophages (B, representative of two independent experiments) and LPMs of littermate controls and Mac-Gata6 KO mice (C, n = 6–12) was determined by quantitative PCR and is expressed as a relative value to Gapdh mRNA.
(D) Expression of indicated proteins in LPMs was analyzed by flow cytometry. Green, control; red, Mac-Gata6 KO; dotted line, unstained control.
(E) mRNA expression of the indicated genes was determined in fetal liver-derived macrophages after retrovirus-mediated transduction of Gata6 (n = 3).
Error bars represent SD. *p < 0.05, ***p < 0.001, ****p < 0.0001. N.S., not significant. See also Figure S2.
Retinoic Acid Regulates PMSG Program
We next addressed the extracellular signal(s) inducing the expression of PMSGs including GATA6. We found that peritoneal macrophages had an abundant expression of retinoic acid nuclear receptor RARβ (Figures 2A, S2B, and S2C). We also found the presence of retinoic acid response elements (RAREs) in the putative regulatory region of Gata6 gene (within 1 kb 5′ to the transcription start site) (Figure 3A). Rarb and two other GATA6-independent PMSGs, Rai14 and Arg1, were shown to be induced by retinoic acid in other cell types (Chang et al., 2013; de Thé et al., 1990; Kutty et al., 2001). Furthermore, the link between retinoic acid and GATA6 was previously described, though the exact molecular mechanism was unclear (Capo-Chichi et al., 2005; Mauney et al., 2010). Collectively, this suggested a possibility that retinoic acid in peritoneal macrophages may control expression of Gata6- and GATA6-dependent genes, as well as other PMSGs.
Figure 3. Activation of Gata6 Gene and Other PMSGs by Retinoic Acid.
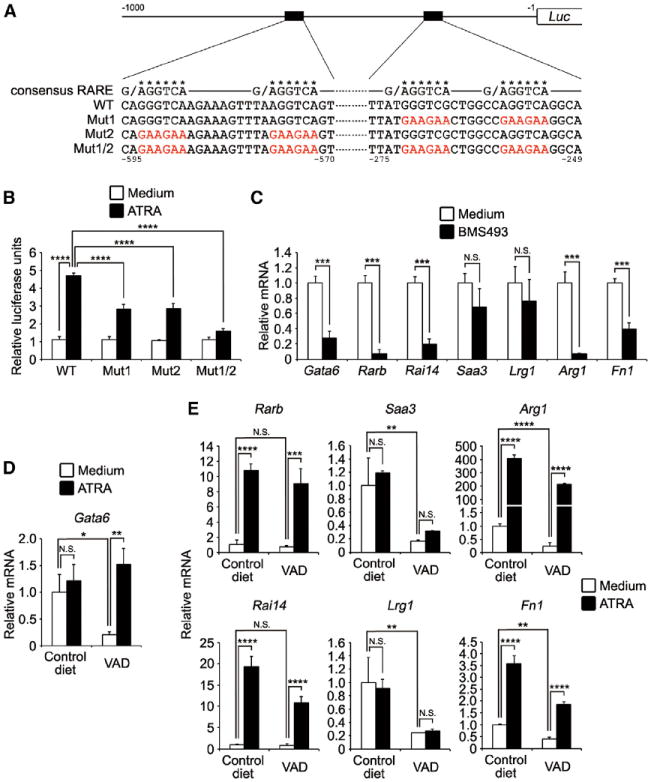
(A) Schematic diagram of Gata6 promoter constructs. Sequences of consensus retinoic acid response elements (RAREs) and putative RAR-binding regions in WT and mutants promoters were shown. Asterisks indicate positions of putative RAREs, and mutated nucleotides were shown by red. Numbers indicate position from Gata6 transcription start site.
(B) 3T3 cells were transfected with GATA6 reporter plasmids and expression plasmids for RARβ and were then stimulated with 1 μM ATRA for 6 hr. The luciferase activities are shown as relative values (n = 3).
(C) Peritoneal macrophages were cultured in the presence or absence of 1 μM BMS493 for 6 hr. The expression of PMSGs was determined by quantitative PCR (n = 3).
(D and E) Peritoneal macrophages from 6-week-old mice bred with control diet or VAD were stimulated with 1 μM ATRA for 6 hr (D) or 24 hr (E), and then the expression of indicated genes was quantified (n = 3).
Error bars represent SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. N.S., not significant.
To test whether retinoic acid could activate the Gata6 gene, genomic DNA covering 1 kbp upstream from Gata6 transcription start site was cloned into luciferase reporter plasmid (Figure 3A). The reporter constructs were transfected into 3T3 cells together with RARβ expression plasmid, and the cells were stimulated with all trans-retinoic acid (ATRA), which is the most abundant form of retinoic acid in vivo. Reporter activity of WT promoter was increased about 4-fold post-ATRA stimulation, whereas mutations in either or both RAREs reduced or eliminated the reporter activity, respectively (Figure 3B). We next treated freshly isolated peritoneal macrophages with pan-retinoic acid receptor inverse agonist BMS493 to determine whether blocking retinoic acid signaling affected PMSG induction. Interestingly, the expression of Gata6 and several GATA6-independent PMSGs (Rarb, Rai14, Arg1, and Fn1) was significantly suppressed by BMS493 (Figure 3C). To further explore the role of retinoic acid signal in PMSG induction, we carried out in vivo depletion of vitamin A, which is the precursor of retinoic acid, from mice. In the previous study, mouse breeding on vitamin-A-depleted diet was shown to result in a steady decline of serum vitamin A. At 6 weeks of age, it is <50% of control value; at 8 weeks, < 20%; and at 11 weeks, < 10% (Smith et al., 1987). Six-week-old vitamin-A-depleted (VAD) mice had a comparable or slightly reduced number of peritoneal macrophages to control diet fed mice. The expression of Gata6 mRNA was significantly downregulated in peritoneal macrophages from 6-week-old VAD mice, whereas ATRA treatment recovered the expression (Figure 3D). In addition, the mRNA expression of Saa3, Lrg1, Arg1, and Fn1 was significantly suppressed in peritoneal macrophages from 6-week-old VAD mice (Figure 3E). ATRA stimulation also induced the expression of PMSGs (Rarb, Rai14, Arg1, and Fn1). These results indicate that retinoic acid inducibly and reversibly regulates gene expression of GATA6 and other PMSGs in peritoneal macrophages.
Longer periods of vitamin A deprivation (9 and 12 week old ages) showed further reduction of GATA6 expression in LPMs (Figure 4A). In a delayed fashion, downregulation of CD102 and CD11b, which are GATA6-independent PMSGs (Figures 2A-2C), was also observed (Figures 4B and 4C). In addition, peritoneal exudate cells from VAD mice revealed age-dependent reduction in the frequencies and numbers of LPMs (Figures 4C and 4D). In contrast, CD11b-F4/80 intermediate population started appearing at 9 weeks of age (Figures 4C and 4E). Flow cytometry and morphological analyses revealed that this population consists of at least three cell types: eosinophils (P1), macrophages (P2), and monocytes (P3) (Figure 4E). P2 macrophages displayed similar protein expression profiles with SPMs in terms of GATA6, CCR2, CD102, and MHC-II (Figure 4F), suggesting that continuous vitamin A deprivation results in recruitment of inflammatory macrophages. Indeed, further vitamin A deprivation (18 weeks old) significantly increased peripheral neutrophils (Figure S3), consistent with the previous study (Kuwata et al., 2000). Because it has been reported that peritoneal inflammation induces the disappearance of macrophages from peritoneal cavity, a phenomenon known as macrophage disappearance reaction (MDR) (Barth et al., 1995), these results suggest that long-term vitamin A deprivation, in addition to control of GATA6 expression, can lead to LPM disappearance through inflammation.
Figure 4. Essential Role of Vitamin A in PMSG Induction.
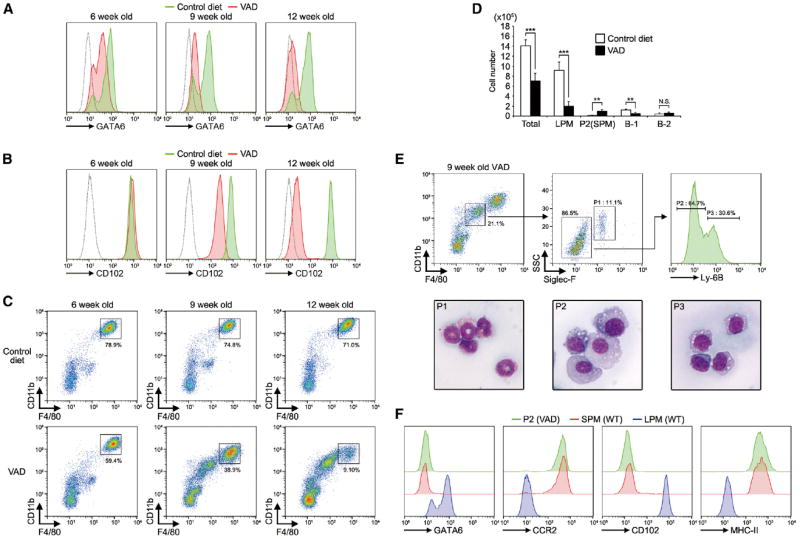
(A and B) LPMs from mice fed with indicated diets were analyzed for GATA6 (A) and CD102 (B). Green, control diet; red, vitamin-A-deficient diet (VAD); dotted line, unstained control. The data are representative of at least three different mice in each group.
(C) Flow cytometry profiles of peritoneal exudate cells from 6-, 9-, and 12-week-old mice bred with control diet or VAD. The data are representative of 3–8 different mice in each group.
(D) Numbers of total cells, LPM, P2 (SPM), B-1 cells, and B-2 cells in peritoneal exudate cells from 9-week-old control diet and VAD mice (n = 5). Error bars represent SD. **p < 0.01, ***p < 0.001. N.S., not significant.
(E) (Top) Flow cytometric gating strategy for characterization of F4/80-CD11b intermediate population. (Bottom) Wright and Giemsa staining of each of sorted subsets.
(F) P2 population in (E) and SPMs and LPMs from WT C57BL/6 mice were analyzed for GATA6, CCR2, CD102, and MHC-II by flow cytometry. The data are representative of at least two different mice in each group.
See also Figure S3.
Although some tissues had severely reduced their size (e.g., lung and adipose tissue) at the late stage of vitamin A deprivation, frequency of tissue macrophages in the spleen and small intestine was comparable to that of mice bred on control diet (Figure S3), indicating that early macrophage differentiation was not affected by vitamin A deprivation.
Macrophage Accumulation in Omenta of Mac-Gata6 and VAD Mice
Most of the vitamin A in the body is stored in liver as transcriptionally inactive metabolites (e.g., retinyl esters) and is continuously deployed into circulation (Hall et al., 2011). Vitamin-A-mediated transcriptional activation requires local conversion of these inactive vitamin A metabolites into biologically active retinoic acid (Duester, 2008; Gudas, 2012). The expression of neither Gata6 nor retinoic-acid-inducible genes (Rarb and Rai14) was detected in circulating leukocytes, including monocytes (Figures S4A and S4B), suggesting that LPMs or their precursor cells (if they originate from circulating progenitors) receive retinoic acid signal after recruitment into peritoneal cavity or its associated tissues. Retinoic-acid-converting enzymes are abundantly expressed in peritoneum-associated adipose tissue, omentum, which is formed by a double layer of mesothelial cells that connects the stomach, pancreas, spleen, and colon (Maruya et al., 2011 and Figure 5A, left). Indeed, Raldh2, which is the rate-limiting enzyme for the last step of retinoic acid synthesis (retinaldehyde to retinoic acid) (Gudas, 2012), is highly expressed in omentum compared to other tissues (Figure 5B), suggesting a high local concentration of retinoic acid at this anatomical location.
Figure 5. Accumulation of Macrophages in Omenta of Mac-Gata6 and VAD Mice.

(A) (Left) Omentum was illustrated by intraperitoneal injection of black carbon particles. (Right) Paraffin section of omentum from WT mouse was stained with hematoxylin and eosin (H&E). Clusters of leukocytes (milky spots) were indicated by arrows. Scale bar, 100 μm.
(B) Indicated tissues were determined for Raldh2 mRNA by quantitative PCR and were expressed as relative values normalized by Gapdh mRNA (n = 3).
(C) Milky spots of the omenta from control, Mac-Gata6 KO, 9-week-old VAD, and LPS-injected mice were stained as indicated color-coded lettering. Scale bars, 100 μm.
(D) (Top) Omental cells from indicated mice were analyzed by flow cytometry. (Bottom) Macrophage population in top panels (gated) was analyzed for GATA6. Dotted line showed unstained control. Plots are representative of at least five different mice in each group.
(E) Macrophages from omenta of VAD- or ATRA-treated VAD mice were analyzed for GATA6. Histogram is a representative of two different mice in each group.
(F) Absolute cell numbers of LPMs present in the peritoneal exudate cells of indicated mice (n = 3) were counted 3 hr post-IP injection of saline or 10 mg of LPS.
(G) Peritoneal macrophages from indicated mice were cultured for 2 hr on monolayers of mesothelial cells. Percentage of cells adherent to mesothelial cells was determined (n = 5).
(H) (Upper-left) Schematic of mixed bone marrow transfer. Bone marrow cells from CD45.2 WT and that from Mac-Gata6 KO mice in macrophage-specific YFP-expressing strain (Mac-Yfp/Mac-Gata6 KO) were mixed at a ratio of 1:1 and were then injected into lethally irradiated CD45.1 WT recipients. (Upper-right) Percent chimerism of YFP+ macrophages in indicated tissues was shown (n = 4). (Bottom) Representative flow cytometry profiles for YFP were shown. YFP-positive population was gated. Error bars represent SD. **p < 0.01, ****p < 0.0001.
See also Figure S4.
Omentum contains at regular intervals opaque structures called milky spots, which are clusters of leukocytes such as B-1 cells (Figure 5A, right) (Rangel-Moreno et al., 2009). Mac-Gata6 KO and 9-week-old VAD mice accumulated CD11b+ macrophages around milky spots, as illustrated by clusters of B cells marked by B220 signal, whereas relatively few macrophages were detected in the omentum of control mice (Figure 5C). Consistent with this observation, flow cytometry analysis of omentum cells revealed increased frequency of macrophages in Mac-Gata6 KO and VAD mice (Figure 5D, top). Omentum macrophages expressed GATA6 protein in control mice, but not in VAD or Mac-Gata6 KO mice (Figure 5D, bottom). Together with the reduction in LPM numbers in peritoneal lavage of Mac-Gata6 KO (Figure 1E) and VAD mice (Figure 4D), these results indicate that GATA6 and retinoic acid maintain macrophages in peritoneal cavity and that loss of these factors results in the accumulation of macrophages in the omentum. Moreover, omentum macrophages in VAD mice had reduced expression of CD102 (Figure 5D), similar to LPMs in peritoneal cavity (Figure 4B). Interestingly, administration of ATRA to VAD mice restored the expression of GATA6 in omentum (Figure 5E), confirming inducibility of GATA6 by retinoic acid in macrophages.
Intraperitoneal (IP) challenge with lipopolysaccharide (LPS) induces MDR—macrophage disappearance from peritoneal cavity (Figures 5F and S4C; Barth et al., 1995). We found that, following LPS injection, GATA6+ macrophages rapidly accumulated around milky spots in omentum (Figures 5C and 5D). Macrophage interaction with mesothelial cells was proposed to be a key step in MDR (Jonjić et al., 1992). Consistent with that, peritoneal macrophages from Mac-Gata6 KO mice had enhanced interaction with mesothelial cells in vitro (Figure 5G), as well as with tissue culture plastic (Figure S4D), suggesting that alteration of adhesion property might be involved in macrophage redistribution in Mac-Gata6 KO mice.
There were two possible explanations for the accumulation of macrophages in the omenta in Mac-Gata6 KO and VAD mice. One is that cell-extrinsic signals, such as constitutive peritoneal inflammation, induce macrophage migration from peritoneal cavity to omentum in these mice. This possibility is suggested by the appearance of inflammatory macrophages (P2 or SPM) in VAD mice (Figures 4E and 4F), but not in Mac-Gata6 KO mice (Figure 1E). Another possibility is that GATA6 controls macrophage localization in a cell-autonomous manner. As bone marrow (BM) transfer could establish GATA6+ macrophage population in peritoneal cavity (Figure S4E), we examined mixed BM chimeric mice to distinguish between these possibilities. We prepared BM cells isolated from WT and Mac-Yfp/Mac-Gata6 KO mice (LysM-Cre;R26-stop-Yfp; Gata6-floxed) and cotransferred into lethally irradiated mice at the 1:1 ratio (Figure 5H). Alveolar macrophages in lungs were observed to maintain initial chimerism (50%) at 5 weeks post-BM transfer (47.5% ± 3.2% for Mac-Gata6 KO). In contrast, Mac-Gata6 KO macrophages greatly reduced frequency in peritoneal cavity (17.6% ± 0.2%) but increased in omentum (62.0% ± 2.7%). This indicates that the accumulation of macrophages in omentum of Mac-Gata6 KO mice is caused by cell-autonomous defect of macrophage phenotype. In addition, these results suggest that macrophages receive retinoic acid provided by omentum and migrate to peritoneal cavity in a GATA6-dependent manner.
Coordinated Induction of PMSGs by Retinoic Acid and Omentum-Derived Factor(s)
We next examined whether ATRA alone or together with other local tissue-derived signals controls functional polarization of peritoneal macrophages. We treated BMDMs with ATRA and/or omentum culture supernatant (OMsup) as a source of additional tissue-derived signals. OMsup was prepared with serum-free medium, which did not contain any vitamin A metabolites. Genes induced by these stimuli were analyzed by microarray (Figure 6A). Interestingly, several GATA6-independent PMSGs (Saa3, Lrg1, Arg1, and Prtn3) were strongly induced in BMDMs stimulated with OMsup in the presence or absence of ATRA. Quantitative PCR analysis revealed that some PMSGs (Rarb, Rai14, and Apoc2) were induced by ATRA, whereas other PMSGs (Saa3, Lrg1, and Hp) were induced by OMsup in a retinoic-acid-independent manner (Figures 6B, S5A, and S5B). Furthermore, the induction of Arg1, Fn1, and Prtn3 genes was only detected in the presence of both ATRA and OMsup. This result suggests that the retinoic acid and omentum-derived factor(s) play a role, alone and in combination, to control distinct subsets of PMSGs. In contrast to these GATA6-independent PMSGs, Gata6 was not induced in BMDMs by either or both ATRA and OMsup. In addition, it was also unclear why GATA6 was only expressed in LPMs, but not in SPMs and Thio-pMacs that were present in peritoneal cavity, even though RAR mRNA was expressed in these cells (Figure S5C). Histone 3 lysine 4 trimethylation (H3K4me3) is associated with transcriptionally active or poised loci, whereas H3K27me3 is associated with gene silencing (Kouzarides, 2007). The presence of H3K4me3 modification was detected in Gata6 locus of LPMs, whereas BMDMs and Thio-pMacs had H3K27me3 modification (Figures 6C and S5D). This indicates that Gata6 locus of BMDMs and Thio-pMacs is epigenetically silenced, which presumably explains why Gata6 and its target genes were not expressed in these cell types.
Figure 6. Induction of PMSGs by Omentum Factor.
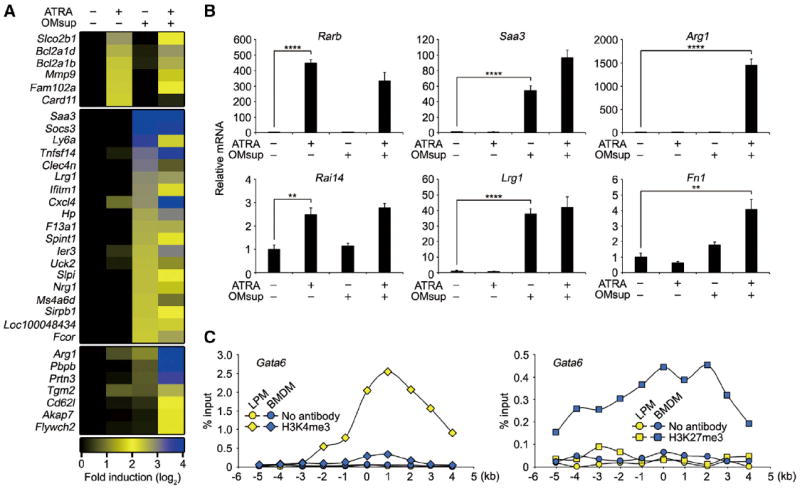
(A) Heatmap of microarray signals upregulated at least five times by 1 μM ATRA or OMsup stimulation compared to unstimulated sample and upregulated at least three times by a combination of ATRA and OMsup compared to individual stimulations after 24 hr. Expression levels were shown as fold induction to unstimulated BMDMs and were expressed by relative values (log-2).
(B) BMDMs were cultured with 1 μM ATRA and/or OMsup for 24 hr. The expression of indicated genes was analyzed by quantitative PCR and expressed as relative values normalized by Gapdh mRNA (n = 3).
(C) LPMs (yellow) and BMDM (blue) were analyzed by ChIP for Gata6 loci without antibody (circle) or with antibodies against H3K4me3 (diamond) or H3K27me3 (square). The data are represented as %input. The x axis depicts probe location on each loci relative to the transcription start site.
Error bars represent SD. **p < 0.01, ****p < 0.0001. See also Figure S5.
The expression of Arg1 was remarkably induced by the combination of retinoic acid and omentum factor(s). Although Arg1 is one of the signature genes for IL-4/IL-13-induced M2 macrophage polarization, which is mediated by IL-4 receptor α subunit (IL4Rα) and STAT6 (Chawla et al., 2011), the expression of Arg1 in peritoneal macrophages was intact in Il4ra KO and Stat6 KO mice (Figure S5E). Consistent with this, the M1 (Nos2) and M2 (Cd206, Retnla, Chi3l3, and Chi3l4) marker genes were differently expressed across different tissue macrophages (Figure S5F).
GATA6 in Macrophages Controls Gut IgA Response
We next asked whether the retinoic acid-GATA6 pathway controls peritoneal macrophage-specific functions and what these functions are. Among the genes that are highly and specifically expressed in peritoneal macrophages in a GATA6-dependent manner are Tgfb2; Ltbp1, which regulates extracellular matrix deposition of TGF-β; and Thbs1, which promotes activation of latent form of TGF-β (Fortunel et al., 2000) (Figures 2A–2C and 2E). TGF-β and retinoic acid are the most prominent factors inducing IgA class switching as well as gut-homing receptor expression on the B cell (Hall et al., 2011; Roy et al., 2013). In addition, peritoneal B-1 cells can give rise to IgA-secreting plasma cells in the gut (Baumgarth, 2011; Mora and von Andrian, 2009). Therefore, we asked whether GATA6 in peritoneal macrophages plays a role in gut IgA production through peritoneal B-1 cell regulation. B-1 cells of peritoneal cavity are thought to directly migrate into intestinal lamina propria and give rise to IgA-secreting cells in a manner that is independent of gut-associate lymphoid tissue (GALT) (Fagarasan et al., 2010; Uematsu et al., 2008). Because B-2 cell IgA production can mask the contribution of B-1 cells, we crossed Mac-Gata6 KO mice to Rorc-deficient (Rorcgfp/gfp) mice, which lack secondary lymphoid organs (Eberl et al., 2004). Deficiency of the Rorc gene resulted in a reduced but detectable amount of fecal IgA (Figure 7A). Mac-Gata6 KO/Rorc KO mice had significantly reduced fecal IgA compared to Rorc KO mice (Figure 7A), whereas serum natural IgM-, IgA-, and PC-specific IgM were comparably detected (Figure S6A). Consistent with this, the number of IgA+ cells in the lamina propria was much fewer in Mac-Gata6 KO/Rorc KO mice (Figures 7B and S6B). GATA6 deficiency did not affect peritoneal B-1 cell population (Figure S6C), and the expression of LysM and Gata6 was not detected in peritoneal B-1 cells (Figures S6D and S6E), indicating that a reduction in IgA production was not due to B-1 cell-intrinsic alteration. Additionally, GATA6 protein in small intestine was only detected in intestinal epithelial cells, but not in lamina propria cells, and the epithelial expression was not affected in Mac-Gata6 KO mice (Figure S6F). This excludes the possibility of the contribution of lamina propria cells (e.g., macrophages and dendritic cells) to the IgA phenotype.
Figure 7. GATA6 in Macrophage-Dependent Regulation of Gut IgA.
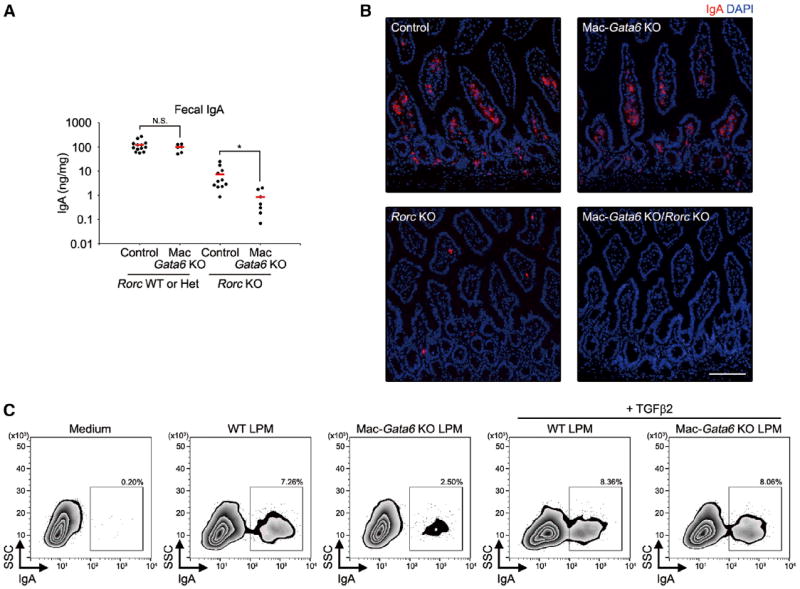
(A) Fecal supernatant from unimmunized indicated mice (8 weeks old) were analyzed for IgA. Each point represents one mouse. Error bars represent mean values. *p < 0.05. N.S., not significant.
(B) Immunohistochemistry analysis for small intestines of indicated mice. Red, IgA; blue, DAPI. Scale bars, 100 μm.
(C) Flow cytometry analysis of IgA-positive peritoneal B-1 cells cultured with or without LPMs from WT or Mac-Gata6 KO mice and recombinant TGF-β2 in the presence of BAFF/LPS/ATRA for 4 days.
See also Figure S6.
Lastly, to determine the role of GATA6-dependent expression of TGF-β in the generation of IgA, we examined coculture of peritoneal B-1 cells with LPMs. Peritoneal B-1 cells underwent IgA class switching by coculture with WT LPMs in the presence of ATRA (Figure 7C). Mac-Gata6 KO LPMs were deficient in the generation of IgA+ B-1 cells, and this defect was restored by the addition of recombinant TGF-β2. In contrast, GATA6 deficiency in LPMs did not affect the expression of gut-homing receptors (CCR9 and integrin-α4β7) on B-1 cells (Figure S6G). These results indicate that GATA6 in peritoneal macrophages is critical for GALT-independent IgA production by peritoneal B-1 cells.
DISCUSSION
Although accumulating evidence highlights the diversity of tissue-specific macrophage phenotypes, the extracellular signals that regulate specialized macrophage functions are largely unknown. Here, we show how local tissue-derived signals, including retinoic acid, control peritoneal macrophage-specific transcriptional program. A transcription factor GATA6, which is uniquely expressed in peritoneal, but not other macrophage subsets, is induced by retinoic acid and controls a subset of peritoneal macrophage functions, including their compartmentalization and control of IgA production by B-1 cells.
Previous studies showed the presence of macrophage precursor cells in omentum (Daems and de Bakker, 1982), local proliferation of omentum macrophages (Wijffels et al., 1992), and the production of macrophage colony-stimulating factor (M-CSF) in milky spot stromal cells (Ratajczak et al., 1987), suggesting that omentum may serve as a site for peritoneal macrophage development. Together with these earlier observations, a high level of Raldh2 expression in omentum (Figure 5B) suggests that, during macrophage development, omentum provides retinoic acid, which is required for the efficient migration of macrophages to peritoneal cavity through the induction of GATA6. In addition, our data suggest that omentum provides not only retinoic acid, but also additional factor(s) for the induction of a subset of PMSGs (Figure 6B). Thus, multiple signals that are present in the local tissue environment may control different gene expression programs in macrophages.
Although the expression of several PMSGs was induced by ATRA, OMsup, or a combination of both factors in BMDMs (Figures 6B and S5A), the induction of the Gata6 gene by any of these signals was not detected in BMDMs, even though hematopoietic progenitors that give rise to BMDMs can generate GATA6-positive LPMs in vivo, as demonstrated by BM chimera experiments (Figure S4E). Interestingly, we found the Gata6 locus to have silencing chromatin modifications (H3K27me3) in BMDMs and inflammatory macrophages, whereas Gata6 locus in LPMs has an H3K4me3 mark associated with active chromatin (Figures 6C and S5D). In multipotent progenitor stage, developmental genes are bivalently marked by H3K4me3 and H3K27me3 and are thus primed for activation prior to differentiation (Kraushaar and Zhao, 2013). As cells differentiate into different lineages, these bivalent modifications resolve into monovalent H3K27me3 or H3K4me3 modifications. Thus, our findings suggest that induction of Gata6 gene in peritoneal macrophages requires at least two steps. First, Gata6 locus is epigenetically modified to remove the silencing histone modification H3K27me3, making it competent for induction at the second step, when omentum-derived ATRA induces Gata6 expression through RAR. The signal involved in epigenetic modification of Gata6 locus and the anatomical location where this signal is provided will need to be determined in future studies because this mechanism may be applicable to other compartment-specific cell differentiation pathways.
GATA6 has a bimodal expression in LPMs (Figure 1C). Interestingly, the GATA6-high LPM population but not GATA6-low population is positive for proliferation marker Ki67 (Figure S1G), suggesting that GATA6-high LPMs might reflect newly migrated population from omentum after proliferation. GATA6 target gene(s) responsible for macrophage localization in peritoneal cavity and chemoattractive signal that might regulate macrophage egress from omentum (if such signals exist) remain to be identified.
Previous studies identified transcription factors that control tissue specific transcription programs in macrophages. However, as the deficiency of these transcription factors resulted in the disappearance of corresponding macrophage subsets from tissues, it was not possible to determine the role of these transcription factors in the tissue-specific gene regulation. In this study, we found that PMSGs fall into GATA6-dependent and GATA6-independent subsets. Thus, tissue-specific macrophage phenotypes can be defined by a combination of multiple transcription factors, each controlling different functional programs. For example, high expression of transcription factors RARβ and NFE2 was detected in peritoneal macrophages (Figures 2B and S2B), suggesting that these transcription factors may have a role in GATA6-independent gene regulation. Importantly, we found that GATA6 is not essential for the development of peritoneal macrophages but is required for their maintenance in the proper tissue compartment. Thus, some macrophage subset-specific transcription factors and associated gene expression programs can control macrophage localization and maintenance in a particular tissue compartment, rather than macrophage development.
Tissue macrophages are derived from two sources. Traditionally, all tissue macrophages have been considered to derive from circulating monocytes originated from hematopoietic stem cells (Gordon and Taylor, 2005). Recent studies uncovered that a substantial portion of tissue macrophages arises from yolk sac during embryogenesis, and these cells are maintained by local proliferation (Ginhoux et al., 2010; Schulz et al., 2012). Together with the previous report (Schulz et al., 2012), our data would suggest that LPMs are most likely yolk sac derived. However, our BM transfer experiment showed that hematopoietic-stem-cell-derived macrophages are also able to express GATA6 in peritoneal cavity. Although the details of the origin of LPMs will need to be established further, the data so far suggests that a GATA6-driven program can generate LPMs from either yolk sac or hematopoietic progenitors.
Vitamin A deficiency is an important public health problem in humans, particularly in developing countries, where it is associated with increased susceptibility to gastrointestinal and lung infections, poor response to vaccination, increased HIV pathogenesis, and overall increased mortality, especially in children (Cassani et al., 2012). Vitamin A can modulate the function and development of many immune cell types, including T cells (Iwata et al., 2004; Mucida et al., 2007), B cells (Mora et al., 2006), dendritic cells (Coombes et al., 2007), innate lymphoid cells (Spencer et al., 2014), and B-1 cells (Maruya et al., 2011). Our study provides additional insights into retinoic acid function in the immune system. Specifically, retinoic-acid-dependent GATA6-TGF-β induction in peritoneal macrophages regulates B-1 cell-mediated gut IgA production. Previous studies showed that TGF-β can be provided by dendritic cells and stromal cells in intestinal lamina propria for IgA class switching in peritoneal B-1 cells (Fagarasan et al., 2010). It will be interesting to determine the contribution or distinct roles of LPM-derived and lamina propria cell-derived TGF-β in the generation of intestinal IgA. It is possible that the preferential IgA class-switching property of peritoneal B-1 cells (Kaminski and Stavnezer, 2006; Roy et al., 2013) is mediated by the priming effect of TGF-β2 provided from LPMs. B-1 cell-derived IgA plays a dominant role in the recognition of commensal bacteria compared to B-2 cell-derived gut IgA (Macpherson et al., 2000). It will be interesting to determine the role of GATA6-dependent gut IgA production in the maintenance of intestinal microbial homeostasis.
The expression level of PMSGs, including GATA6, was gradually affected by the dietary vitamin A depletion (Figures 4A-4C) and was restored by exogenous ATRA (Figures 3D and 3E) and suppressed by RAR inhibition (Figure 3C), suggesting that the availability of the instructive signals can affect the degree of tissue-specific gene expression in macrophages. Macrophages may thus constantly survey local tissue status and dynamically change their phenotype to deal with continuously changing tissue environment. Thus, the diversity of tissue macrophage phenotypes may, at least in part, be due to a reversible transcriptional program activated on demand by tissue-derived signals.
EXPERIMENTAL PROCEDURES
Extended Experimental Procedures are included in the Supplemental Information.
Mice
All mice were bred in the Yale University School of Medicine animal facility in specific pathogen-free conditions, and experiments were performed in accordance with the institutional animal care and use guidelines. Unless specifically indicated, 8- to 16-week-old same-gender littermates were used and data were pooled where indicated. Ccr2–/–, Gata6-floxed, LysM-cre, Rosa26-floxed-Yfp, Rorcgfp/gfp, Il4ra–/–, and Stat6–/– mice were obtained from Jackson Laboratories. Gata6-floxed mice were backcrossed to C57BL/6 for 6–11 generations in our facility. Hematology analysis was performed by the animal healthcare service Antech.
Statistical Analysis
All experiments were performed at least twice. Results were statistically analyzed using an analysis of variance (ANOVA) test or Student’s t test. A p value of < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank members of the Medzhitov laboratory and the W.M. Keck facility for microarray analysis. Y.O. was supported by the Japan Society for the Promotion of Science (JSPS), Uehara Memorial Foundation, and Kanae Foundation for the Promotion of Medical Science. R.M. lab is supported by The Blavatnik Family Foundation, grants from NIH (AI046688, AI089771, CA157461) and the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
The GEO accession number for the microarray data reported in this paper is GSE56711.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.04.016.
References
- A-Gonzalez N, Guillen JA, Gallardo G, Diaz M, de la Rosa JV, Hernandez IH, Casanova-Acebes M, Lopez F, Tabraue C, Beceiro S, et al. The nuclear receptor LXRα controls the functional specialization of splenic macrophages. Nat Immunol. 2013;14:831–839. doi: 10.1038/ni.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel KM, Harris RB, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16:67–76. doi: 10.1016/s1074-7613(01)00257-6. [DOI] [PubMed] [Google Scholar]
- Barth MW, Hendrzak JA, Melnicoff MJ, Morahan PS. Review of the macrophage disappearance reaction. J Leukoc Biol. 1995;57:361–367. doi: 10.1002/jlb.57.3.361. [DOI] [PubMed] [Google Scholar]
- Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011;11:34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Cain DW, O’Koren EG, Kan MJ, Womble M, Sempowski GD, Hopper K, Gunn MD, Kelsoe G. Identification of a tissue-specific, C/ EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J Immunol. 2013;191:4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi CD, Rula ME, Smedberg JL, Vanderveer L, Parmacek MS, Morrisey EE, Godwin AK, Xu XX. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev Biol. 2005;286:574–586. doi: 10.1016/j.ydbio.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Cassani B, Villablanca EJ, De Calisto J, Wang S, Mora JR. Vitamin A and immune regulation: role of retinoic acid in gut-associated den-dritic cell education, immune protection and tolerance. Mol Aspects Med. 2012;33:63–76. doi: 10.1016/j.mam.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Thangamani S, Kim MH, Ulrich B, Morris SM, Jr, Kim CH. Retinoic acid promotes the development of Arg1-expressing dendritic cells for the regulation of T-cell differentiation. Eur J Immunol. 2013;43:967–978. doi: 10.1002/eji.201242772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Cárcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daems WT, de Bakker JM. Do resident macrophages proliferate? Immunobiology. 1982;161:204–211. doi: 10.1016/S0171-2985(82)80075-2. [DOI] [PubMed] [Google Scholar]
- Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013;14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Thé H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. 2010;28:243–273. doi: 10.1146/annurev-immunol-030409-101314. [DOI] [PubMed] [Google Scholar]
- Fortunel NO, Hatzfeld A, Hatzfeld JA. Transforming growth factor-beta: pleiotropic role in the regulation of hematopoiesis. Blood. 2000;96:2022–2036. [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Immunological Genome Consortium (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn EE, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, Bortoluci KR, Almeida SR, Herzenberg LA, Herzenberg LA. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci USA. 2010;107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Gudas LJ. Emerging roles for retinoids in regeneration and differentiation in normal and disease states. Biochim Biophys Acta. 2012;1821:213–221. doi: 10.1016/j.bbalip.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Grainger JR, Spencer SP, Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity. 2011;35:13–22. doi: 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA, Robinson AP, MacPherson GG, Gordon S. The mononuclear phagocyte system of the mouse defined by immunohisto-chemical localization of antigen F4/80. Relationship between macrophages, Langerhans cells, reticular cells, and dendritic cells in lymphoid and hemato-poietic organs. J Exp Med. 1983;158:1522–1536. doi: 10.1084/jem.158.5.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Jonjić N, Peri G, Bernasconi S, Sciacca FL, Colotta F, Pelicci P, Lanfrancone L, Mantovani A. Expression of adhesion molecules and chemotactic cytokines in cultured human mesothelial cells. J Exp Med. 1992;176:1165–1174. doi: 10.1084/jem.176.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski DA, Stavnezer J. Enhanced IgA class switching in marginal zone and B1 B cells relative to follicular/B2 B cells. J Immunol. 2006;177:6025–6029. doi: 10.4049/jimmunol.177.9.6025. [DOI] [PubMed] [Google Scholar]
- Kohyama M, Ise W, Edelson BT, Wilker PR, Hildner K, Mejia C, Frazier WA, Murphy TL, Murphy KM. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kraushaar DC, Zhao K. The epigenomics of embryonic stem cell differentiation. Int. J Biol Sci. 2013;9:1134–1144. doi: 10.7150/ijbs.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T, Warr G, Loy J, Bravo R. Defects in macrophage recruitment and host defense in mice lacking the CCR2 chemokine receptor. J Exp Med. 1997;186:1757–1762. doi: 10.1084/jem.186.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutty RK, Kutty G, Samuel W, Duncan T, Bridges CC, El-Sherbeeny A, Nagineni CN, Smith SB, Wiggert B. Molecular characterization and developmental expression of NORPEG, a novel gene induced by retinoic acid. J Biol Chem. 2001;276:2831–2840. doi: 10.1074/jbc.M007421200. [DOI] [PubMed] [Google Scholar]
- Kuwata T, Wang IM, Tamura T, Ponnamperuma RM, Levine R, Holmes KL, Morse HC, De Luca LM, Ozato K. Vitamin A deficiency in mice causes a systemic expansion of myeloid cells. Blood. 2000;95:3349–3356. [PubMed] [Google Scholar]
- Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- Maruya M, Suzuki K, Fujimoto H, Miyajima M, Kanagawa O, Wakayama T, Fagarasan S. Vitamin A-dependent transcriptional activation of the nuclear factor of activated T cells c1 (NFATc1) is critical for the development and survival of B1 cells. Proc Natl Acad Sci USA. 2011;108:722–727. doi: 10.1073/pnas.1014697108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauney JR, Ramachandran A, Yu RN, Daley GQ, Adam RM, Estrada CR. All-trans retinoic acid directs urothelial specification of murine embryonic stem cells via GATA4/6 signaling mechanisms. PLoS ONE. 2010;5:e11513. doi: 10.1371/journal.pone.0011513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora JR, von Andrian UH. Role of retinoic acid in the imprinting of gut-homing IgA-secreting cells. Semin Immunol. 2009;21:28–35. doi: 10.1016/j.smim.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora JR, Iwata M, Eksteen B, Song SY, Junt T, Senman B, Otipoby KL, Yokota A, Takeuchi H, Ricciardi-Castagnoli P, et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314:1157–1160. doi: 10.1126/science.1132742. [DOI] [PubMed] [Google Scholar]
- Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel-Moreno J, Moyron-Quiroz JE, Carragher DM, Kusser K, Hartson L, Moquin A, Randall TD. Omental milky spots develop in the absence of lymphoid tissue-inducer cells and support B and T cell responses to peritoneal antigens. Immunity. 2009;30:731–743. doi: 10.1016/j.immuni.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak MZ, Jaskulski D, Pojda Z, Wiktor-Jedrzejczak W. Omental lymphoid organ as a source of macrophage colony stimulating activity in peritoneal cavity. Clin Exp Immunol. 1987;69:198–203. [PMC free article] [PubMed] [Google Scholar]
- Roy B, Brennecke AM, Agarwal S, Krey M, D¨ber S, Weiss S. An intrinsic propensity of murine peritoneal B1b cells to switch to IgA in presence of TGF-β and retinoic acid. PLoS ONE. 2013;8:e82121. doi: 10.1371/journal.pone.0082121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Smith SM, Levy NS, Hayes CE. Impaired immunity in vitamin A-deficient mice. J Nutr. 1987;117:857–865. doi: 10.1093/jn/117.5.857. [DOI] [PubMed] [Google Scholar]
- Sodhi CP, Li J, Duncan SA. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev. Biol. 2006;6:19. doi: 10.1186/1471-213X-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SP, Wilhelm C, Yang Q, Hall JA, Bouladoux N, Boyd A, Nutman TB, Urban JF, Jr, Wang J, Ramalingam TR, et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–349. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, Nishiyama M, Sato S, Tsujimura T, Yamamoto M, Yokota Y, et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol. 2008;9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- Wijffels JF, Hendrickx RJ, Steenbergen JJ, Eestermans IL, Beelen RH. Milky spots in the mouse omentum may play an important role in the origin of peritoneal macrophages. Res Immunol. 1992;143:401–409. doi: 10.1016/s0923-2494(05)80072-0. [DOI] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


