Abstract
Cancer treatment is often hindered by inadequate methods for diagnosing the disease or insufficient predictive capacity regarding therapeutic efficacy. Targeted cancer treatments, including Bcr-Abl and EGFR kinase inhibitors, have increased survival for some cancer patients but are ineffective in other patients. In addition, many patients who initially respond to targeted inhibitor therapy develop resistance during the course of treatment. Molecular analysis of cancer cells has emerged as a means to tailor treatment to particular patients. While DNA analysis can provide important diagnostic information, protein analysis is particularly valuable because proteins are more direct mediators of normal and diseased cellular processes. In this review article, we discuss current and emerging protein assays for improving cancer treatment, including trends toward assay miniaturization and measurement of protein activity.
Keywords: cancer, diagnostic, protein assay, personalized cancer treatment, molecular diagnostic
1. INTRODUCTION
A generally accepted paradigm in cancer diagnosis and treatment is that early detection improves odds of survival [1,2]. However, inadequate diagnostics prohibit detection of certain types of cancer until late stages. For example, endoscopy is the typical screening method for esophageal cancer and is generally only performed once symptoms appear [3]. Other screens may be subjective or provide high levels of false positives or negatives. Hepatocellular carcinoma is typically detected by ultrasound, but this technique is subject to operator error and often cannot distinguish between malignant and benign nodules [4]. Although mammography is the standard screen for breast cancer, 20% of cases go undetected with this screen and specificity is 25%, leading to a large number of false positives [5].
A variety of biomarkers have been identified for molecular diagnosis and prognosis of cancer. For example, the MammaPrint and OncotypeDX multigene tests predict breast cancer subtypes and treatment response [6]. Other tests commonly measure human epidermal growth factor receptor type 2 (Her2) and estrogen receptor levels in breast cancer patients [6,7]. Elevated Her2 levels predict resistance to endocrine and chemotherapy treatment but susceptibility to Her2-targeted treatment, while elevated estrogen receptor levels correlate with increased endocrine response but chemotherapy resistance [7]. While many cancer screens utilize DNA analysis to detect and characterize disease, some applications require protein examination for greater accuracy because transcription at the gene level does not necessarily correspond to expression at the protein level [8]. For example, in cervical cancer screening, DNA testing can detect the presence of HPV, but protein methods are needed to determine whether HPV is active or latent [9].
Protein analysis offers opportunities for diagnosing, stratifying, and monitoring disease [10]. In order to be clinically useful, an assay must meet certain requirements. First, it must be sensitive enough to detect the protein or proteins of interest. Protein concentrations in blood vary by more than 12 orders of magnitude, and cancer biomarkers may be present at trace (pg/ml) levels [11]. In addition to sensitivity, an assay must be specific to the protein that it aims to detect. For example, in ovarian cancer, which occurs in 40 out of 100,000 individuals, an assay needs 99.6% specificity for the benefits of early detection to outweigh the harm of unnecessary surgery [12]. While the specificity required for the benefits of early detection to outweigh the harm of false positives is lower for more common types of cancer, false positives and false negatives have significant physical and emotional effects for patients [12]. These assays must also function with a sample size that is readily available from cancer patients. Minimally invasive assays, such as those using blood, urine, or saliva, are advantageous because they cause less discomfort to the patient, have higher compliance rates, and may be taken repeatedly during treatment to monitor results [13,14]. However, concentrations of biomarkers may be lower in blood than in other tissues and saliva has particularly low biomarker concentrations [11]. Additionally, the equipment to perform the assay must be clinically available or easily attainable and personnel must have the necessary training to perform these procedures [15,16]. Assays must also be cost and labor effective and robust enough to result in comparable results between different laboratories and different personnel [16,17].
The objective of this review is to provide an overview of current and emerging analytical cancer diagnostic and prognostic assays based on protein detection. We begin by discussing currently clinically implemented protein analytical techniques such as immunohistochemistry, enzyme linked-immunosorbent assay, and flow cytometry. Next we discuss emerging analytical techniques not yet applied in the clinic such as mass spectrometry and proximity assays followed by discussions of miniaturized detection assays and protein activity assays for kinases, proteases, and telomerase.
2. CURRENTLY IMPLEMENTED PROTEIN ASSAYS
A variety of protein assays for cancer diagnosis, prognosis, and treatment decisions, including those based on immunohistochemistry (IHC), enzyme linked-immunosorbent assay (ELISA), and flow cytometry, are currently available in the clinic [16,18,19]. Over 100 unique proteins, although not all cancer related, can be detected by these methods [20]. Even within these methods, however, work continues to be done to improve sensitivity and specificity as well as to improve the multiplexing and analysis capabilities for these methods. Improvements to these methods are particularly advantageous since the capabilities and knowledge to run these methods clinically already exists.
2.1 Immunohistochemistry
Immunohistochemistry is a well established method for clinical measurement of diagnostic and prognostic proteins and allows for the analysis of biomarker expression and localization [21]. It has been used to detect proteins such as the estrogen receptor and Her2 marker in breast cancer patients [7,16]. Tissue microarrays (TMA) allow for the simultaneous immunohistochemical analysis of hundreds of patient samples. In these arrays, a tissue core as small as 600 μm is embedded in paraffin and as little as 4–5 μm can be used for immunohistochemical analysis [22]. Several groups have developed protein signatures using TMAs consisting of 5–8 proteins for diagnostic and prognostic predictions. A five antibody TMA divided breast cancer patients into high, moderate, and low risk groups with 95% confidence for decisions regarding cytotoxic chemotherapy [23]. Another five antibody TMA was able to correctly identify lung cancer as adenocarcinoma, identifying patients for Bevacizumab treatment, with 88.6% sensitivity and a negative predictive value of 84.8% [24]. An additional five marker test has been developed for distinguishing between malignant melanomas and benign nevi with 95% specificity and 91% sensitivity [25]. A TMA measuring 8 biomarkers in breast cancer patients with axillary lymph node metastasis was able to significantly group patients into high, medium, and low 10 year survival rates [26].
While each of these TMAs offers advantages over current methods of prognosis or diagnosis, standardization of IHC assays is needed, including sample handling, assay performance, and data interpretation [16]. IHC results are often manually ranked on a scale of 0 to 3 based on staining intensity. This semi-quantitative method introduces subjectivity and inter-user variability into the analysis [25]. Quantitative automated image analysis tools are available, but are not widely used or standardized [22]. Additionally several groups including pathologists, technologists, and scientists are working to standardize protocols to reduce variability between facilities [7,16,22]. Despite the challenges of quantification and standardization, IHC is a time and cost effective method and is widely clinically available [22], making it a viable method for clinical protein analysis.
2.2 Flow Cytometry
Another method currently available to physicians for assessing protein expression in cancer cells is flow cytometry. Many hospitals have access to flow cytometers with as many as 8 colors to simultaneously assess multiple biomarkers in samples [15,17]. Flow cytometry is less subjective [27] and requires fewer cells than immunohistochemistry [14]. It can also detect as few as 1/10,000 cancer cells, and can be performed in a few hours and more cheaply than immunohistochemistry [15,27,28]. It is commonly used to diagnose chronic lymphocytic leukemia [19] and a variety of studies have been performed on its use in other leukemias and lymphomas [15,17,27–29]. However, flow cytometry is capital equipment intensive, and requires cell singularization, limiting its usefulness for solid tumors [30].
Several modifications and improvements have been suggested to improve flow cytometry as a diagnostic and prognostic tool. The use of the Quantibrite system, which measures the absolute number of antibodies bound per cell, has been investigated as an alternative to measuring the percentage positive cells in a sample [17,29]. In one study, a subgroup of patients who qualified as negative for CD38 expression based on percentage of cells expressing CD38, which indicates a poor prognosis in chronic lymphocytic leukemia, was moved to the percentage positive group based on the average antibodies bound per cell [17]. Patients with a low CD38+ population but high CD38 expression per cell had a similar poor prognosis to patients with a high CD38+ population. Another group used image flow cytometry, in which a digital image is taken of the cells in addition to the standard cell counting, to detect promyelocytic leukemia protein and diagnose acute promyelocytic leukemia [27]. Previously promyelocytic leukemia protein bodies could not be assessed using flow cytometry because of the inability to view cell staining patterns [27]. Another advance in flow cytometric cancer detection has been the identification of sets of biomarkers that predict cancers more efficiently than a single biomarker. One group developed a 9-color assay to detect Hodgkin and Reed Sternberg cells in classical Hodgkins Lymphoma with 88.7% sensitivity and 100% specificity [15]. Although this assay exceeds the capabilities of some current clinical flow cytometers, efforts to reduce the assay to 5 or 6 colors as well as advances in flow cytometry technology may improve the viability of this approach [15].
Less invasive and less costly assays are more accessible to patients. Fine needle aspirates reduce the burden on the patient and cost of the procedure when compared to surgical biopsies, but provide fewer cells for analysis. Several assays have used fine needle aspirate samples for flow cytometric analysis, including one assay to diagnose and classify Non-Hodgkin lymphoma that achieved 97% sensitivity and 94% specificity [15,28].
2.3 Enzyme Linked Immunosorbent Assays
Enzyme linked immunosorbent assays (ELISAs) are another established method for clinical assessment of cancer biomarkers [31]. In an ELISA, proteins are immobilized to a surface and then labeled with an enzyme that catalyzes a reaction resulting in a color change [32]. These assays are quantitative and antibodies that recognize both linear and conformational epitopes can be used [9,31]. Like flow cytometry, ELISAs can simultaneously analyze multiple markers with high sensitivity and specificity. For example, a six marker ELISA for ovarian cancer reported 95.3% sensitivity and 99.4% specificity [12].
Less invasive sampling techniques and rapid and simple analysis can increase patient compliance with diagnostic tests [33]. This is particularly true in developing countries where clinics may lack access to expensive equipment. As shown in Table 1, several modifications have been made to ELISAs to create point of care diagnostics while maintaining high sensitivity and specificity. One such example is a dot ELISA, in which all samples are absorbed onto a nitrocellulose membrane. A dot ELISA on urine samples to detect nuclear matrix protein-52 in bladder cancer could be performed in 5 minutes with 97% sensitivity and 94% specificity [34]. A paper-based ELISA using chitosan and wax screen printing was developed to detect α-fetoprotein (AFP), cancer antigen-125 (CA-125), and carcinoembryonic antigen (CEA) using chemiluminescence. This assay, which can be performed in minutes, offers a point of care testing option when quantitative signal is needed [35]. One group developed a microchip ELISA assay coupled to a portable cell phone/lensless charge-coupled device for point of care detection of ovarian cancer [33].
Table 1.
Advances in ELISA assays.
| Unique points of assay | Biomarkers | Assay test solution | Cancer types | Sensitivity/Specificity |
|---|---|---|---|---|
| 5 minute point of care detection with dot ELISA [34] | Nuclear matrix protein | Urine | Bladder | 97% positive predictive value |
| 6 biomarker panel [12] | Leptin, prolactin, osteopontin, insulin-like growth factor II, macrophage inhibitory factor, CA-125 | Blood | Ovarian | 95.3% sensitivity, 99.4% specificity |
| Modified surface, eliminate enzyme step, confocal optics [32] | IL-8 | Buffer, saliva | Oral | 4.0 fM in buffer |
| Aptamer-antibody sandwich ELISA [36] | MUC1 | Buffer | Epithelial | 1 μg/ml |
| Microfluidic chip, differential pulse voltammetry [37] | AFP | BSA solution, human serum | Hepatocellular carcinoma | 1 pg/ml in BSA solution |
| Electrokinetic concentrator, bead based, automated chip [38] | PSA, CA19-9 | Spiked donkey or human serum | Prostate, pancreatic, gastrointestinal | 1.85 pg/ml PSA, 0.002 U/ml CA19-9 |
| Microchip point of care, cell phone/charge coupled device [33] | HE4 | urine | Ovarian | 89.5% sensitivity, 90% specificity |
| Gold nanoparticle layer [39] | CEA | Purified CEA solution | Many | 2ng |
| Microfluidic paper based analytical device, point of care [35] | AFP, CA-125. CEA | Purified biomarker solutions | Many | 0.06 ng/ml AFP 0.33 U/ml CA-125 0.05 ng/ml CEA |
| Point of care multiplex detection [40] | Thioredoxin, IL-8 | Saliva | Oral | 50 pg/ml |
| Immuno-gold silver staining, microbeads, microbiochips [41] | AFP | Purified AFP solution | Hepatocellular carcinoma | 1 ng/ml |
In addition to improving access to diagnostics, increased detection sensitivity offers promise for improving the early detection of disease. One such example is an aptamer-based ELISA for detecting mucin1 (MUC1), a marker of epithelial malignancies. This assay had a sensitivity of 1 μg/ml, and additional optimization using higher affinity aptamers and by reduction of non-specific binding to coating and blocking reagents may further improve sensitivity [36]. To simplify the ELISA assay and improve sensitivity, another group removed the enzyme activity step by directly labeling immobilized proteins with fluorophores and decreasing the assay volume to increase surface to volume ratio. The sensitivity for interleukin-8 (IL-8) in oral cancer was further lowered to 4.0 fM using confocal detection optics [32].
3. EMERGING PROTEIN ANALYSIS TECHNIQUES
Other analysis techniques are currently only used in laboratory settings but are being investigated for clinical applications. While used extensively for research purposes, mass spectrometry has received increasing attention as a potential clinical diagnostic [42]. Proximity assays detect proteins using multiple epitopes and are also being investigated for clinical applications.
3.1 Matrix Assisted Laser Desorption/Ionization
Mass spectrometry can detect a wide range of proteins in a single measurement without labeling and can be performed from a simple blood test [5,42]. To establish biomarker sets for clinical diagnosis, a set of peaks is often identified in an evaluation set and further narrowed through a test set. This approach allows for analysis of multiple markers in determining a cancer diagnosis [3,4,43,44].
Matrix assisted laser desorption/ionization (MALDI) is performed by mixing a sample with a matrix solution and allowing the solution to crystallize (Figure 1) [45]. The mass detection range for this technique is 1 kDa to 50 kDa. While it cannot directly detect larger proteins, it can detect peptides that correspond to these proteins [3,42]. Protein profile peaks for esophageal and oral cancer have been analyzed using MALDI time of flight mass spectrometry with sensitivity of at least 89% and specificity of at least 84% [3,46]. An adaptation of MALDI, MALDI imaging mass spectrometry can analyze tissues with spatial resolution. In this method, samples are taken from a thin tissue slice along a 2-dimensional grid to generate an array of ion concentrations at each point on the grid [42,47]. This assay detected Her2 positive status, as determined using IHC as a comparison, with 83% sensitivity and 92% specificity [42].
Figure 1.
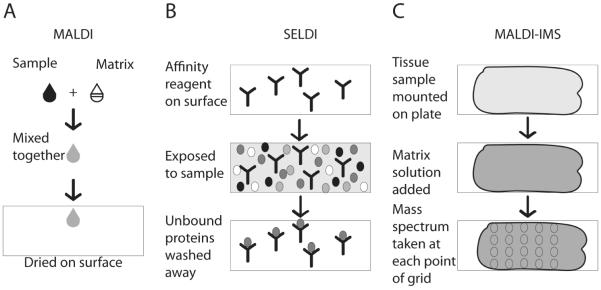
Mass spectrometry preparations for potential cancer diagnostics. A) In matrix assisted laser desorption/ionization (MALDI), a sample is mixed with a matrix solution and then spotted onto a surface to dry. B) In surface enhanced laser desorption/ionization (SELDI), an affinity reagent is bound to the surface. The sample is exposed to the surface and then unbound portions of the sample are washed away. C) In MALDI-imaging mass spectrometry (MALDI-IMS), a tissue sample is mounted on a surface. A matrix solution is added to the sample and mass spectra are taken from defined points of the tissue to provide spatial ion concentration data.
3.2 Surface Enhanced Laser Desorption/Ionization Mass Spectroscopy
In surface enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF), samples are spotted onto a surface containing a binding reagent. This binding reagent allows for the separation of less abundant biomarkers from common proteins in the cell lysate as well as the use of unpurified samples (Figure 1) [45]. The monolayer of proteins and peptides formed in this method also allows for clearer signal detection [48]. SELDI-TOF has identified protein peaks for the diagnosis of a variety of cancers including breast cancer, hepatocellular carcinoma, and renal cell carcinoma, often with greater accuracy than current diagnostic methods [4,5,43,48].
Challenges in mass spectrometry include data analysis and procedural consistencies between laboratories. Mass spectrometry generates large data sets for each sample, creating difficulty in assessing the significant features of each data set [49]. Improved or automated data analysis methods may make mass spectrometry more feasible for clinical applications [4,49]. One approach to increase high throughput analysis of SELDI-TOF data has been to decrease the dimensionality of the data through four statistical moments: mean, variance, skewness, and kurtosis [49]. Inconsistencies in peak presence between laboratories and even between tests sets in a single laboratory also complicate the clinical application of mass spectrometry [50]. Further peak validation, standardized methods for sample handling, and larger test sets may ameliorate this challenge [50,51] and further division of cancer patients into subgroups based on disease characteristics may decrease heterogeneity within patient test populations [44]. Studies have optimized lysis procedures to minimize residual peaks and maximize signal. Laser microdissection is one option to obtain a more homogeneous sample [51]. While heterogeneity exists in the peaks identified by different experiments, the correlation between experiments is statistically significant. Further validation and standardization are needed before mass spectrometry becomes a clinically useful diagnostic technique [50].
3.3 Proximity Assays
Proximity assays allow for the detection of multiple epitopes within a certain distance. This multipronged detection increases specificity by decreasing background from non-specific antibody binding since it is less likely that multiple antibodies will non-specifically bind nearby [52]. Additionally, the detection of proximal epitopes creates a functional assay to assess protein-protein interactions [52]. In proximity ligation, antibodies that recognize two or more epitopes are attached to oligonucleotides. These oligonucleotides hybridize and ligate at close distances and can then be amplified with real time PCR or rolling circle amplification (Figure 2) [52]. Amplification of hybridized oligonucleotides increases sensitivity and allows the reaction to detect trace levels of protein [52]. A proximity ligation strategy called 3 PLA uses 3 oligonucleotides attached to antibodies that bind to three different epitopes. The three antibodies bind to epitopes in proximity to each other and the first two oligonucleotides are connected to each other using the third oligonucleotide as the template and detected with RT-PCR. This method detected as few as 100 molecules of vascular endothelial growth factor (VEGF) [53]. Proximity ligation has also been performed in solid phase by immobilizing antibodies to PCR tubes or magnetic beads. The oligonucleotides attached to these antibodies were then amplified using loop-mediated isothermal amplification, which isothermally amplifies the DNA in place of PCR. This method detected as little as 0.001 pM PSA [54].
Figure 2.
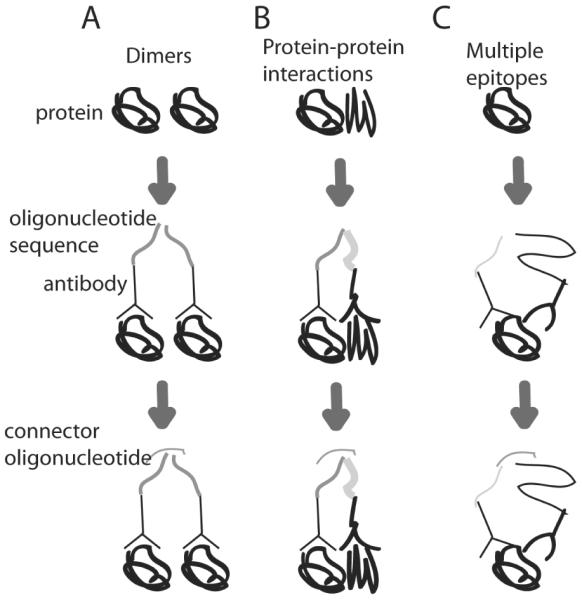
Proximity ligation detects binding of nearby epitopes. Protein is incubated with antibodies conjugated to oligonucleotides sequences. These antibodies bind to the protein(s), bringing the oligonucleotides sequences in proximity with each other. A connector oligonucleotide then facilitates ligation and replication through methods such as RT-PCR. A) Protein homodimerization results in nearby epitopes, causing oligonucleotide amplification. B) Protein heterodimerization results in nearby epitopes, causing oligonucleotides amplification. C) Detection of multiple epitopes on the same protein allows for increased sensitivity. Nearby binding to multiple epitopes causes oligonucleotide amplification.
In the VeraTag proximity assay, two antibodies bind to nearby epitopes, such as the two molecules in Her2 dimers [55,56]. One of these antibodies contains a fluorescent tag while the other antibody is biotin conjugated. A streptavidin-bound photosensitizer binds to the biotin conjugated antibody, and, in the presence of light, cleaves thioester bonds within 30 to 100 nm, including the fluorescent tag from the proximal antibody. Capillary electrophoresis then detects the released fluorescent tags [55,57]. This method uses formalin fixed paraffin embedded tumor specimens, the most common way of preserving solid tumor samples for analysis such as IHC [45,46,48].
4. MINIATURIZATION
One emphasis in cancer diagnostics is the move to methods that use increasingly smaller sample sizes and more portable equipment. Miniature devices for assaying cancer cells reduce costs by requiring less sample volume and use smaller samples that can be obtained less invasively from patients. Often, these devices are easier to transport for point of care application.
4.1 Microarrays
Protein microarrays have been used extensively to identify protein biomarkers for cancer, but are yet to be used in a clinical setting. These arrays can analyze 5000 or more proteins in the space of a single microscope slide and require only a few microliters of sample, which allows large numbers of proteins to be measured even with limited sample size [10,58]. Because of the specificity of these arrays, even low abundance proteins can be profiled [10,58]. Protein arrays include both forward phase arrays, in which a capture molecule is bound to the array, and reverse phase arrays, in which the analyte itself is bound to the array. The most common type of protein microarrays are forward phase antibody arrays [59].
A variety of antibody microarrays have been developed to identify protein signatures of cancer and as potential diagnostics for cancer. A 378 spot antibody microarray detected the upregulation of 8 proteins associated with breast cancer [8]. Protein signatures using scFv antibody arrays have been developed for diagnosis, monitoring and prognosis of breast and pancreatic cancer [10,60]. Other microarrays have been used for improved analysis of other techniques. An IHC-based microarray used an automatic image analysis application in place of a traditional microscope and a microarray format to increase throughput, reduce sample size, and decrease bias during analysis [61]. A microarray ELISA was developed to quantify expression of five proteins, hepatocyte growth factor (HGF), VEGF, CA15-3, Fas ligand, and PSA, using tyramide signal amplification on a glass microscope slide and was able to detect as little as 0.5 pg/ml HGF. All components for this array are commercially available, making it more feasible to implement in the clinic [58]. As with other techniques, standardization is important in antibody microarrays due to varying antibody affinity. To better standardize results, a dual color microarray was developed to assess 741 cancer related proteins. This assay used one color for a control sample and another color for the test sample. The use of dual colors allowed the assay to more reliably detect protein profile differences between pancreatic cancer patients and healthy patients [62]. These arrays have potential for multiplexed, reliable analysis of patient protein profiles. However, antibody specificity and variability in antibody affinity present challenges for this technology [59]. Improved antibodies or capture reagents as well as improvements in sensitivity offer promise for the clinical use of protein microarrays [59].
4.2 Microfluidics
Microfluidic devices minimize sample volume and reagent consumption by several orders of magnitude as compared to conventional techniques such as 96 well plates, and allow for high throughput analysis and precise spatial and temporal control over reagent and sample delivery [30,63]. They also provide closed and often automated systems to minimize human labor, variability, and sample loss and contamination [64]. Microfluidic systems have been used to capture low abundance cancer cells. One microfluidic system captured intact cervical cancer cells using an antibody to immobilized α6 integrin, a cervical cancer cell surface marker. More than 30% of cancer cells were isolated while less than 5% of non-cancerous cells were captured [65]. Another system captured 97% of breast cancer circulating tumor cells (CTCs) from as much as 1 ml of blood using antibodies against epithelial cell adhesion molecule. These cells were released from the microchannels and counted using conductivity measurements, removing the need for labeling or complex instruments [64].
The combination of microfluidics with traditional immunoassays, including IHC, flow cytometry, and ELISA, reduces antibody consumption by several orders of magnitude and offers the potential for assay automation. These features make microfluidic assays cheaper, more consistent, and less labor intensive than traditional forms of the assay [21,30,37,41]. One example is a microfluidic multiplexed IHC assay that measures 10 biomarkers from a smaller sample size than a traditional single biomarker IHC assay. Automatic staining and analysis provide more consistent and objective results as compared to the semi-quantitative traditional IHC. A drawback of this technique is that the small sample size may be less representative than the larger sample used for traditional IHC [21,66]. Microfluidics has also been combined with imaging cytometry to analyze the expression of EGFR, PTEN, phospho-Akt, and phospho-S6 from as few as 1000 cells from solid tumor biopsies including single cell measurements, whereas traditional flow cytometry is generally limited to hematological cancers [30]. Another microfluidic device detected IL-8 from saliva using an automated ELISA sandwich assay on a self assembled monolayer surface [40].
ELISAs have been adapted to microfluidic formats to create point of care devices that are self-contained and eliminate the need for extraneous detection equipment [37,41]. In one configuration, AFP was detected in a sandwich assay using a gold-conjugated secondary antibody. A silver enhancer solution then bound to these gold-conjugated antibodies, and platinum electrodes detected the electrical current from this silver bridge. This assay detected as little as 1 ng/ml AFP but used purified AFP rather than patient samples or cell lysate [41]. Another microfluidic sandwich ELISA point of care device used a horseradish peroxidase (HRP) conjugated antibody and differential pulse voltammetry detection to detect as little as 1 pg/ml of AFP. The assay successfully detected 16 pg/ml of AFP in healthy human serum, giving it a clinically significant sensitivity [37]. The use of commonly available fabrication procedures enhances the adaptability of a diagnostic into the clinic. Another microfluidic immunoassay used standard production procedures to create electrochemical sensor arrays integrated into antibody functionalized microfluidic devices. These arrays detected purified cancer biomarkers including CEA, PSA, estrogen receptor-α, plasminogen activator urokinase receptor, EGFR, and erythroblastic leukemia viral oncogene homolog-2, but were not implemented for more complex samples such as cell lysates [67].
5. MICRO AND NANO PARTICLE BASED APPROACHES
Microparticles and nanoparticles have found a variety of applications in cancer diagnostics. Use of these particles in suspension can simplify and standardize assays. Unique properties of certain nanoparticles provide an expanded toolbox of protein analysis techniques.
5.1 Microparticles
While capture probes in microarrays must be synthesized for each microarray, microparticles are often in suspension and may be synthesized in large batches, which reduces labor and allows for higher standardization between assays [68–70]. Suspension assays also allow for easier mixing, leading to faster reaction times on microspheres. For multiplexed analyses, microspheres can be added or removed to individually choose each protein for the assay. Fluorescence microbeads can be analyzed by flow cytometry, laser scanning cytometer or confocal laser scanning microspectrometer [68,70]. Table 2 lists some of the features of the common types of microparticles and nanoparticles.
Table 2.
Types and features of microparticles and nanoparticles.
| Types of particles | Detection Methods | Advantages | Disadvantages |
|---|---|---|---|
| Polystyrene beads | Immunodetection methods | Batch synthesis [68–70], cheaper [69], concentrating analytes [71,72] | Photobleaching, wide emission spectrum [68] |
| Quantum Dots | Fluorescence | Excited over wide range, emit over narrow range, brighter than fluorescent dyes [68] | Cellular toxicity, emit in same range as natural cellular fluorescence [73], lack of established protocols [74] |
| Gold particles | SPR, SERS, two photon scattering, dynamic light scattering | Non-toxic [75], emit in infrared away from natural fluorescence, unique electronic properties [73] | High fabrication costs, availability of analysis equipment [39], size and shape inconsistencies [74] |
| Carbon nanotubes | electrochemical | Increase binding surface area, maintain conductivity [76], amenable to composite material [77] | Metallic impurities, insolubility, lack of fabrication techniques [74] |
Luminex polystyrene beads have been applied to a variety of immunoassays. Because each bead serves as a separate replicate and many beads can be fabricated simultaneously in batch, bead-based immunoassays may be more reliable, more reproducible, and cheaper than traditional ELISA [69]. Similarly to traditional immunoassays, however, these assays still suffer from non-specific binding, in this case to the polystyrene beads, and with cross reactions between antibodies during multiplexing [69]. These beads have been used to measure cytokine profiles in breast cancer cells [68], to detect breast cancer from 35 analytes with 91.8% accuracy [69], to detect cystatin B in hepatocellular carcinoma serum [78], and to simultaneously measure free and total PSA in prostate cancer [69]. These beads have also been used to detect autoantibodies, or antibodies to cancer antigens present in a patient's blood. To eliminate the need to purify recombinant proteins, peptide epitopes were conjugated to Luminex beads to detect PSA and 6 autoantibodies associated with prostate cancer in the same assay [79]. Other polystyrene beads, functional fluorescence encoded polystyrene beads, were created by the incorporation of rhodamine 6G and carboxyl functional groups around a core bead. Varying the rhodamine 6G dye concentration allowed for multiplex detection of AFP, CEA, and PSA at concentrations below 1 ng/ml, and serum sample measurements corresponded well with a reference electrochemiluminescence immunoassay [70]
Another advantage of microparticles is the ability to trap and concentrate analyte molecules for improved sensitivity [11]. This improved sensitivity is particularly important in microfluidic and point of care devices where detector sensitivity may be compromised because of size and cost limitations [71]. Additionally, the short optical paths in microfluidic devices further decrease fluorescence detection sensitivity [71]. A microfluidic packed bed consisting of microparticles in a point of care microchip concentrated C-reactive protein for 20-fold improvement in fluorescence sensitivity detection to 1.4 nM limit of detection [71]. Improvement in microfluidic bead handling techniques may further enhance bead based microfluidic devices [63]. Another packed bed using a flow injection system captured antibody-bound AFP to detect as little as 5 ng/ml AFP. A challenge of packed beds is inconsistent packing of beads, leading to variability between assays [72]. Molecules can also be concentrated with electrokinetic accumulation. In this case, an ion selective membrane trapped enzyme product from a bead based microchannel ELISA to detect 0.002 U/ml of CA19-9 corresponding to 65-fold sensitivity enhancement over non-concentrated ELISA [38].
Because they are unbound to any surface, microparticles conjugated to different detection molecules can be used to detect the same analyte. This approach increases assay specificity since two antibodies must recognize the same molecule [80]. A microparticle proximity ligation assay requiring recognition of the target molecule by 3 antibodies detected VEGF, IL-8 and IL-6 in 5 μl samples of serum and whole blood [11]. Leukemia often results from the fusion of two normal proteins in the cell. Binding of antibody-conjugated beads to each of the two proteins in a leukemia-linked fusion protein and detection by flow cytometry enabled the detection of leukemic cells [80].
5.2 Quantum Dots
Quantum dots are semiconductor nanocrystals most commonly consisting of a CdSe core and a ZnS coat [68,81]. These nanocrystals are more resistant to photobleaching and 20–40 times brighter than beads containing organic dyes. While organic dyes must be individually excited and often emit at overlapping wavelengths, quantum dots emit in a narrow spectrum determined by their size, and one excitement wavelength excites quantum dots of many sizes, allowing for the simultaneous measurement of many analytes [68]. One drawback to quantum dots, however, is their toxicity to cells [73]. Quantum dots also generally lack the ability to emit in the near infrared range where cellular natural autofluorescence is low [73].
The charge level and size properties of quantum dots present a challenge for highly specific binding and accessibility. Charge neutral quantum dots may aggregate together while highly charged dots non-specifically bind to cells [82]. Addition of a hydrophilic polyethylene glycol (PEG) spacer to these dots has been shown to decrease background binding to cells [83]. PEG-capped quantum dots were streptavidin labeled and used to detect biotin-labeled anti CD-146 antibody on the surface of human melanoma cells [84]. Lipid-coated quantum dots measured absolute quantitative levels of prostate stem cell antigen, CLDN4, and mesothelin in pancreatic cells [82]. To address size constraints and improve accessibility to in vivo tumor samples, scFv antibodies, rather than complete antibodies, to Her2 and prostate stem cell antigen were conjugated to quantum dots. Addition of cysteine residues to the c-terminus of the scFv allowed thiol conjugation chemistry to occur specifically at the c-terminus and avoided interference with the antigen recognition sites of these small fragments [83].
Quantum dots often deposit on surfaces, but microfluidic fluid flow inhibits deposition and accelerates reaction with the quantum dots [85]. An immunoassay using quantum dots in a microfluidic nanobiochip detected 0.02 ng/ml CEA antigen, a 2 order of magnitude reduction in limit of detection relative to ELISA, and 50 ng/ml CEA in patient saliva samples [81]. Aqueous quantum dots conjugated to a secondary antibody in a microfluidic chip detected 2.5 pM CEA in serum and simultaneously detected both CEA and AFP [85].
5.3 Gold Nanoparticles
The unique properties of gold nanoparticles provide opportunities for additional types of protein analysis in cancer cells. Gold nanoparticles are not inherently toxic to cells [73] and can be conjugated to antibodies [86]. They emit in the near infrared range, where cellular fluorescence is minimal, and do not suffer from photobleaching [73]. However, high fabrication costs and expensive equipment required for assays that take advantage of the unique properties of gold nanoparticles hinder their use [39]. Size distribution of gold nanoparticles can be measured with dynamic light scattering. Particles were coupled to either capture or detection PSA antibodies and used to quantify free PSA. Depending on the PSA concentrations, these nanoparticles remained as monomers or formed dimers or oligomers, which could be measured in a serum matrix at 0.5 ng/ml with dynamic light scattering [87]. Two photon scattering (TPS) measures symmetry and size changes caused by nanoparticle aggregates. Gold nanoparticle ovals coated with both Her2 antibodies and Her2 aptamers detected Her2 on the cell surface of only 100 breast cancer cells using TPS. The combination of both Her2 antibodies and Her2 aptamers improved sensitivity over single molecule detection [88].
Gold nanoclusters can enhance more traditional analysis techniques. CD33 conjugated gold nanoclusters detected acute myeloid leukemia with flow cytometry [73]. By functionalizing an ELISA plate with a gold nanoparticle layer, the micro and nanostructure of the particles increased surface area to volume ratio and increased binding to the plate. Using this method, as little as 2 ng purified CEA could be detected compared to greater than 10 ng in a traditional ELISA [39].
5.4 Localized Surface Plasmon Resonance
Localized surface plasmon resonance (LSPR) is a label-free, real-time detection method [89–92] based on electromagnetic resonance of gold particles. In this method, an electromagnetic wave induces oscillations at the surface of gold nanoparticles [92]. When a change occurs in the environment of these particles, it results in a change in the refractive index [90].
Several approaches have been taken to multiplex LSPR assays. One assay used antibody functionalized gold nanorods with different aspect ratios for the multiplexed detection of 3 cancer stem cell surface biomarkers, with a possibility of detecting up to 15 biomarkers [93]. For even higher levels of multiplexing, an antibody microarray consisting of 2000 spots/cm2 measured concentrations of human chorionic gonadotropin and activated leukocyte cell adhesion molecule in a background of human serum using LSPR [94]. One challenge with LSPR is the size variation of gold nanoparticle aggregates. While these aggregates increase resonance signal, they also result in assay variability. To control aggregation, antibody conjugated particles were labeled with single stranded DNA. Once these particles were captured on substrates, additional particles with complementary DNA were added resulting in hybridization and controlled particle aggregation. Sequences with different hybridization rates and real-time LSPR monitoring allowed for dual detection of both CD44 and CD24 [91].
LSPR signal could be increased by using both a gold surface and gold nanoparticles to amplify the signal. EGFR antibody-conjugated gold nanoparticles on a gold nanoparticle surface identified oral cancer from saliva samples [75]. A sandwich immunoassay captured TNF-α on a gold film and then detected the captured molecules using antibody conjugated gold nanorods. Coupling gold film and gold nanorods increased sensitivity to 0.03 pM, a 40 fold increase over traditional SPR methods [92].
In addition to detecting cancer antigens, autoantibodies and antigen modifications can also be detected. A sandwich LSPR sensor detected CEA autoantibody levels by immobilizing CEA on a gold-coated slide followed by capture of CEA autoantibodies [89]. In some cases, protein modification may be as predictive as or more predictive than the presence of the protein itself. Glycosylation status of haptoglobin was measured in prostate cancer blood samples using a haptoglobin capture antibody and a lectin to identify glycosylation status [95].
In addition to measuring the presence of particular proteins based on antibody antigen capture, LSPR assays also measure antigen localization, acoustic signals, and competitive adsorption of antigens. EGFR antibody-conjugated gold nanoparticles identified cancer cells in a sample by localizing to nasopharyngeal and lung cancer cells in tissue samples, increasing the LSPR of these cells [86]. Photoacoustic detection is advantageous because it can measure protein concentrations up to several centimeters in depth. For photoacoustic detection, cancer tissue samples mixed with EGFR antibody conjugated gold nanoparticles were implanted into a mouse. The samples were excited with light, and an ultrasound detector sensed the resulting acoustic waves [96]. Competitive adsorption of proteins onto a surface can avoid the need for antibodies since proteins vary in the strength of their adsorbance on the surface and in their LSPR. Thyroglobin, a biomarker for thyroid cancer was detected in a mixture of three proteins in a competitive adsorption microfluidic device. However, this device requires extensive sample preparation since only a small number of proteins can be simultaneously distinguished with this method [90].
5.5 Surface Enhanced Raman Spectroscopy
Surface enhanced Raman spectroscopy (SERS) is a vibrational technique [97] in which molecules bound to the surface of silver or gold nanoparticles are excited at one wavelength and emit at another wavelength [98], providing a unique signature for a sample [14]. Inclusion of rough surfaced metals such as gold or silver nanoparticles enhances SERS spectra up to 14 orders of magnitude over Raman spectra, possibly because of an enhanced electromagnetic field at the surface of the nanoparticles [97,99]. Compared to fluorescence, sharp narrow peaks in SERS spectra correspond to true signal whereas broad peaks correspond to background signal [98]. Background is further reduced in SERS because excitation generally occurs at red to infrared wavelengths, which minimizes cellular autofluorescence [100,101].
A key consideration in SERS is signal intensity, which can often be less than fluorescence [98]. A variety of approaches have been taken to maximize signal intensity. Since only molecules close to noble metal nanoparticles can be measured in SERS, colloidal nanoparticles can increase contact points with the sample and thus the number of molecules detected [101]. Composite organic-inorganic nanoprobes (COIN) are made by aggregating silver particles around an organic label molecule [98]. COIN-antibody conjugates detected PSA in an IHC format with sensitivity comparable to Alexa Fluor fluorescence sensitivity [98]. Coating gold nanoparticles with polyethylene glycol (PEG) molecules increased stability of the particles under extreme pH and high salt conditions and enhanced signal intensity. Neither the addition of PEG molecules nor scFv antibodies changed the SERS properties of these particles, and they were able to detect EGFR on human cancer cells and in mouse tumors [97]. New triphenylmethine and cyanine Raman reporter molecules and a lipoic acid linker also improved Raman sensitivity [100,102]. Conjugation of these nanotags to Her2 and EGFR antibodies resulted in detection of Her2 and EGFR positive cells [100,102]. Silver particles generate a stronger Raman signal than gold, but must form aggregates for this signal enhancement, which can create non-homogeneity in the sample. Hollow gold nanospheres overcome this challenge by increasing Raman signal with a single particle. Functionalization of these nanospheres with CEA or AFP antibodies simultaneously detected both molecules in patient blood samples [103]. A gold capture substrate used in conjunction with gold nanoparticles further amplifies SERS signal because of the oscillations between the substrate and nanoparticles [99]. A SERS immunoassay captured MUC4 on a gold substrate and then detected MUC4 via SERS on an antibody-labeled gold nanoparticle [99]. Specificity of detection can also increase the signal to background ratio. Magnetic bead EpCam tags were used to bind epithelial cells circulating in whole blood, and SERS-Her2 tags detected breast cancer cells from these epithelial cells. As few as 5 cells could be detected with this method [104].
In addition to antibody detection, other methods have also been used for biomarker isolation prior to SERS analysis. The biomarker p53 was linked to a gold nanoparticle and captured for SERS detection with the protein azurin, which specifically reacts with p53. Even in human serum, p53 could be linked to the nanoparticle and specifically captured with azurin [105]. In another example, whole blood samples were purified to isolate albumin and globulin of gastric cancer patients via membrane electrophoresis before SERS analysis [14].
While many methods isolate a particular protein or proteins, other methods have sought to evaluate the complete SERS spectrum. Although more complicated analysis is required and background may be higher, these spectra contain additional information that may be lost with the purification of samples [13,14]. SERS detection with colloidal gold particles identified colorectal cancer patient blood samples versus normal patient samples by statistical analysis of the whole SERS spectrum [13]. Healthy and tumor regions of brain tissue samples were also differentiated by analyzing the whole SERS spectrum [101].
5.6 Carbon Nanotubes
Carbon single walled nanotubes (SWNTs) can be either metals or semiconductors and increase the surface area of electrodes while maintaining conductivity and providing a surface for biomolecule conjugation. SWNT forests, or dense orthogonal arrays of SWNTs, were labeled with PSA antibodies. Prostate cancer serum was captured with these SWNTs and secondary carbon nanotube-linked HRP antibodies detected the PSA upon incubation in an electrochemical cell with H2O2. Carbon nanotube-linked HRP antibodies increased sensitivity over HRP antibodies because more antibodies could bind per PSA molecule [76]. To adapt detection to proteins at different expression levels, some assays can modify signal amplification based on expected expression levels. A SWNT sandwich array measured four different protein markers: PSA, prostate specific membrane antigen (PSMA), platelet factor-4 (PF-4) and IL-6. While HRP conjugated detection antibodies were sufficient for the more abundant proteins PSMA and PSA, biotinylated detection antibodies that bound to streptavidin HRP further amplified the signal 14–16 fold for the less abundant proteins PF-4 and IL-6 [106]. An IL-6 SWNT sandwich array also used two different secondary antibody detection systems. For the highest sensitivity, multi-walled carbon nanotubes bound 106 HRP labels per 100 nm while samples with higher expression levels could be detected using streptavidin with 14–16 HRP labels per molecule [107]. Improved material properties are important for enhanced sensitivity and consistency [77,108]. Gold nanoparticles and carbon nanotubes have been combined to create a material with high electron transfer rate and electrocatalytic stability. Chitosan has also been added as a matrix agent with functional sites for immobilization. These hybrid material immunosensors detected AFP and CEA from human serum [77,108]. Further nanodevices are discussed in a review on emerging nanoproteomics [74].
6. FUNCTIONAL ASSAYS OF PROTEIN ACTIVITY
Measurement of protein activity, rather than abundance, supplies additional information about the role of a particular protein in disease. Kinase activity, protease activity, and telomerase activity have each been linked to cancer diagnosis and metastasis. As shown in Figure 4, the activity of each of these enzymes may be measured by the enzyme's effect on its substrate.
Figure 4.
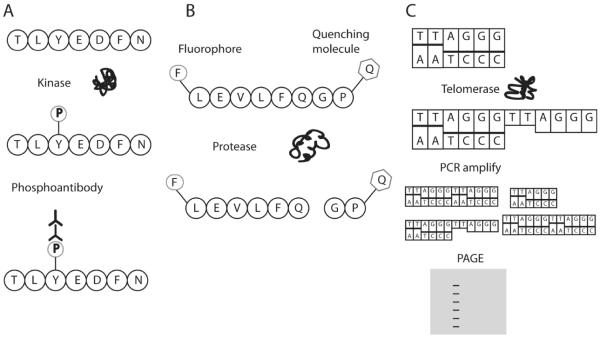
Detection of enzyme activity via substrate modification. A) Kinase phosphorylation of a substrate can be detected with phosphoantibodies. B) Protease cleavage of a substrate can be detected by the release of a fluorophore from a quenching molecule. C) In the telomere repeat amplification protocol (TRAP), telomerase extends oligonucleotides sequences by addition of the telomere sequence TTAGGG. This extension can then be detected by PCR amplification and polyacrylamide gel electrophoresis (PAGE).
6.1 Kinase Assays
Deregulated kinase activity and cellular signaling is involved in most forms of cancer [63]. Kinase inhibitors are used clinically to correct overactive kinases, including Bcr-Abl and EGFR, and are very effective for some patients [109,110]. However, overexpression of these kinases does not always correspond to effective treatment by these inhibitors [109,110]. The activity of a kinase and the effects of that activity may vary based on mutational status and cellular environment. Therefore, measurement of kinase activity, rather than expression, may provide a better gauge of the functional impact of a kinase in the cell and the potential therapeutic values of kinase inhibitors [111–114].
An assay to determine kinase activity in patient cells could improve patient specific treatment [111,112,115]. These assays typically measure either kinase activation, through phosphorylation, dimerization or ligand interactions, or substrate phosphorylation by the kinase. Measurement of kinase activation avoids the need for identifying a kinase-specific substrate. A proximity ligation assay detected PDGFRβ with one antibody and phosphorylated PDGFRβ with an additional antibody in tissue samples. Secondary, species specific antibodies were conjugated to DNA strands to make the method more generally applicable with commercial primary antibodies [113]. The VeraTag assay measured association of Met kinase with its ligand HGF, which has been shown to correspond to activating phosphorylation of Met kinase [57]. Another method identified phosphorylated Her1 kinase, a member of the EGFR kinase family, in a single cell with three different antibodies. Following immunocapture of Her1 from cell lysate, an additional Her1 antibody conjugated to glucose oxidase and a Her1 phospho-antibody conjugated to HRP completed the immune complex resulting in tyramide mediated signal amplification [116]. Both heterodimerization and phosphorylation of the four EGFR kinase family members leads to downstream signaling effects, but these effects and disease prognosis vary depending on the combination of overexpressed kinases. To detect these signaling variations, a microbead immunoassay detected expression, phosphorylation, and heterodimerization of the four EGFR kinase family members in breast cancer cell lines [117].
Other assays have measured substrate phosphorylation to quantify kinase activity. Additionally, many of these assays detected kinase inhibition by specific kinase inhibitors. This function could give the assays utility in predicting patient resistance to a current inhibitor or to the potential benefits of switching to a different kinase inhibitor [63,111,114,115]. Radioassays are commonly used to detect phosphorylation and are beneficial because radiolabeling is sensitive and quantitative and does not change molecular interaction properties. In one approach, an immunocapture bead based radioassay detected Bcr-Abl kinase activity from as few as 3000 cells. Immunocaptured Bcr-Abl was exposed to 32P-ATP and a kinase specific bead-bound substrate followed by real-time radioassay detection with a microfluidics embedded camera [63]. Changes in FRET signal upon substrate phosphorylation have also been detected. In this sensor, two fluorescent probes were attached to Crkl, a substrate for Bcr-Abl, such that the phosphorylation induced conformational change of Crkl brought the probes closer together to generate a FRET signal [112]. However, delivery of the labeled Crkl molecule into cells with this approach was only 20–30% efficient [112,118]. In another FRET assay, a his-tagged Her2 substrate was incubated with a Her2 containing enzyme mixture followed by binding to a quantum dot and incubation with a fluorescently tagged phosphoantibody. Phosphorylated substrate resulted in proximity of the antibody and quantum dot and a change in the emission spectrum. This assay detected as little as 7.5 nM Her2 and did not require the washing steps of traditional immunoassays [119]. To avoid antibodies, another substrate phosphorylation assay attached a PKCα peptide substrate to a cationic molecule containing a near infrared fluorophore. When the peptide was not phosphorylated, an anionic fluorescence quenching molecule complexed with the cationic molecule and silenced the fluorophore. Upon peptide phosphorylation, the complex dissociated, resulting in fluorescence [120].
Since a substrate must be recognizable by a kinase for phosphorylation to occur, it is important to maintain the substrate as close as possible to its native conformation. Hydrogels often improve surface-based assay sensitivity by providing a three dimensional hydrated environment for the protein and minimizing hydrophobic interactions between the substrate and assay components. An EGFR substrate was immobilized in a hydrogel on a microscope slide and incubated with cell lysate, followed by antibody detection of the phosphorylation of the EGFR substrate [114]. A similar array using a peptide substrate for EGFR measured the effects of acquired resistance in cell lines to the EGFR inhibitors erlotinib and gefitinib [111], showing the assay could be used for detecting resistance to inhibitors [111]. A microfluidic device used macroporous hydrogel pillars to immobilize Crkl to detect the kinase activity of Bcr-Abl. Diffusion of large molecules such as proteins, including both kinases and antibodies, is limited in typical hydrogels. By creating macroporous hydrogels, phosphorylation signal increased by an order of magnitude, allowing the detection of kinase activity from as few as 500 cells [115].
6.2 Protease Assays
Proteases have also been associated with cancer diagnosis and metastasis. They often play a role in degrading extracellular matrix (ECM) so that metastasis can occur or in cleaving other cancer-associated substrates [119,121,122]. As with kinases, the activity of these enzymes can be quantified through detection of enzyme activation or substrate modification. The membrane type serine protease 1 (MT-SP1) cleaves certain cancer related substrates and upregulation in its activity has been correlated to epithelial cancers. Antibodies to the active form of MT-SP1 were produced and fluorescently tagged for imaging of activated MT-SP1 [122]. Matrix metalloproteinases (MMP) are often overexpressed in cancer and degrade ECM, allowing for cancer metastasis [121,123]. The activity level of these proteins often corresponds to tumor grade and metastasis level [121,123]. To measure the protease activity of MMP-2, a fine needle aspirate sample was placed on gelatin covered film. Biebrich scarlet staining identified only intact areas of the gelatin that had not been degraded by MMP-2 in a process called film in situ zymography [121]. Another assay measured MMP-2 and MMP-13 by immobilizing a MMP-13 and MMP-2 specific peptide substrate onto a glycol polymer chitosan, giving the peptide a flexible, three-dimensional environment. The peptide was linked to a near infrared fluorophore and a quencher molecule. When MMP activity was present in synovial fluid samples, the peptide was cleaved and fluorescence could be observed [123]. The serine protease urokinase type plasminogen activator (uPA) and its substrate plasmin are known to degrade the ECM. A uPA specific peptide substrate was attached to a gold nanoparticle, which quenches quantum dot fluorescence. When uPA was present, the peptide substrate was cleaved, releasing the gold nanoparticle. Attachment of the peptide sequence to a quantum dot by biotin streptavidin labeling generated fluorescence in the presence of uPA [119]. Although a variety of assays detect the expression levels of PSA, PSA serine protease activity is a better tumor marker for prostate cancer [124]. A PSA-specific cleavable peptide was attached to a gold shelled nanocrescent and SERS detected the spectrum differences between cleaved and uncleaved peptide [124].
6.3 Telomerase Activity
Telomerase catalyzes the extension of telomeric repeats at the ends of chromosomes and is associated with cellular immortality and cancer [125]. While rarely expressed in most normal cells, telomerase is overactive in more than 85% of tumors, making it a possible general cancer biomarker [126]. Telomerase activity is most commonly measured by the telomere repeat amplification protocol (TRAP). In this procedure, a substrate primer is PCR extended by telomerase addition of repeat telomere sequences and analyzed by gel electrophoresis. The assay can detect telomerase activity from as few as 10 cells, and real time TRAP allows the assay to be quantitative [125]. To analyze CTCs in prostate cancer patients, epithelial cells from the blood were immunocaptured and subjected to TRAP. Telomerase activity was detected in 79% of advanced prostate cancer samples but no healthy samples [127]. Analysis of TRAP activity in live cells is beneficial for measuring the maximum level of telomerase activity present. TRAP assays often rely on the presence of epithelial markers for identifying cancer cells for the assay. To assess telomerase in live CTC and avoid reliance on epithelial marker expression to isolate cells, a Parylene-C slot microfilter was created. This filter captured 90% of circulating tumor cells based on the increased size of these cells with 90% viability and 200-fold enrichment over background. The TRAP assay could then be performed on these cells to measure telomerase activity [128].
While measuring telomerase activity is potentially useful for cancer diagnostics, several features of the TRAP assay are unsuitable for clinical use. The TRAP assay requires radioisotopes and relies on PCR, which can be contaminated by telomerase inhibitors, heme components from blood, or other molecules inhibiting the reaction and causing false negatives [126]. It is also time consuming and can generate false positives due to primer dimers [129]. Several strategies have improved these limitations of the TRAP assay. Magnetic bead capture offers a more automated and high throughput procedure and decreased non-specific signal using additional washes. In this method, telomerase recognition sequences were conjugated to magnetic beads. Exposure to telomerase containing cell lysate in the presence of biotin labeled dUTP resulted in extension of these sequences. Streptavidin-HRP labeling of biotin then detected the biotinylated sequences [126]. Another magnetic bead assay eliminated the need for PCR amplification by increasing sensitivity with an ECL probe containing 100 ECL signaling molecules per telomerase product. A biotinylated telomerase substrate was incubated with cell lysate followed by addition of the ECL probe to bind the telomerase reaction products. Finally, a streptavidin magnetic bead bound the biotinylated primers of the ECL probes. This method could detect telomerase activity from as few as 500 cancer cells without PCR [130].
Another assay uses a looped DNA sequence as the primer for telomerase activity. This stem loop contains a fluorescent molecule and a quencher molecule on each end. Upon hybridization, the stem loop opens up allowing fluorescent signal. Exonuclease III is present and cleaves 3' blunt ended double stranded DNA, such as the telomere primer pair, recycling the stem loop for further telomere addition. To avoid exonuclease digestion, digestion resistant locked nucleic acids were included in the 5' end of the stem loop DNA. The assay detected telomerase activity of as few as 30 cells and was simpler than TRAP [129].
7. CONCLUSIONS
Cancer diagnosis has moved from morphology based cellular analysis to incorporating molecular characterization. Cancer now has the possibility to be diagnosed and subtyped based on protein expression or even protein activity. Established clinical techniques such as IHC, flow cytometry, and ELISA offer trustworthy systems on which to build and assess other techniques. Miniaturization reduces cost and allows for easier multiplexing. Microfluidics reduces assay variability by controlling fluid flow and reduces labor through the possibility of automation. Micro and nanoparticles improve consistency between reactions and concentrate products. Optimized materials such as quantum dots and gold nanoparticles provide new and more efficient detection methods. Protein activity assays provide additional information about the functions occurring in the cells.
An interdisciplinary approach will be needed to advance cancer diagnostics. Improved biorecognition molecules such as antibodies and aptamers as well as strategies for using these molecules will continue to increase detection specificity and sensitivity. Integration of microfluidics with these themes will improve affordability by miniaturizing the system and repeatability by providing a more controlled environment. New materials and electronics may reduce background, decrease cost, and improve sensitivity. Commercially available fabrication components and similarity to current assays will assist in translating these assays into the clinic. Assays with each of these components are being developed, but now the challenge is to combine these techniques to create sensitive, simple, accurate, convenient, and cost effective assays that will increase patient quantity and quality of life.
Figure 3.
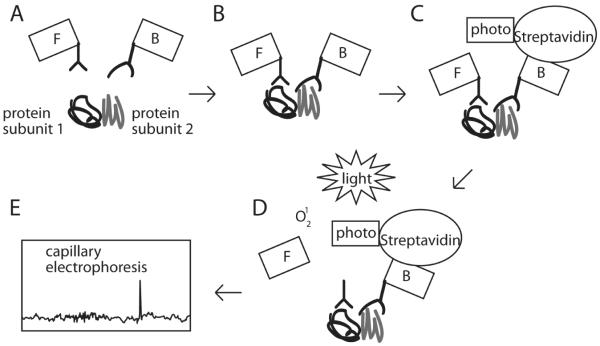
The VeraTag assay detects binding of nearby epitopes. A) Two proteins that bind to each other are incubated with a fluorescently tagged antibody (labeled F) and a biotin tagged antibody (labeled B). B) The tagged antibodies bind the proteins, bringing them into proximity with each other. C) A streptavidin probe (labeled S) linked to a photosensitizer binds to the biotin linked antibody. D) Exposure to light in the presence of a photosensitizer excites oxygen into the singlet state, causing localized cleavage of the fluorescent tag from the fluorescently tagged antibody. E) The released fluorescent tag can be detected by capillary electrophoresis.
Acknowledgements
Support was provided by a NIH grant R01GM074691 to SPP and an NSF Graduate Research Fellowship to ADP.
Footnotes
Conflict of Interest The authors declare no conflicts of interest.
References
- [1].Pollack LA, Li J, Berkowitz Z, Weir HK, Wu XC, Ajani UA, Ekwueme DU, Li C, P.B. Melanoma survival in the United States, 1992 to 2005. Journal of the American Academy of Dermatology. 2011:S78–86. doi: 10.1016/j.jaad.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bohndiek SE, Brindle KM. Imaging and “omic” methods for the molecular diagnosis of cancer. Expert review of molecular diagnostics. 2010;10:417–34. doi: 10.1586/erm.10.20. [DOI] [PubMed] [Google Scholar]
- [3].Guo R, Pan C, Shen J, Liu C. New serum biomarkers for detection of esophageal carcinoma using Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Journal of cancer research and clinical oncology. 2011;137:513–9. doi: 10.1007/s00432-010-0899-3. [DOI] [PubMed] [Google Scholar]
- [4].Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos, Dimitrios Kalmowitz B, Wells M, Guerrero M, Asara JM, Libermann T, Afdhal Nezam H. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clinical cancer research. 2008;14:470–7. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- [5].Vlahou A, Laronga C, Wilson L, Gregory B, Fournier K, McGaughey D, Perry RR, Wright GL, Semmes OJ. A novel approach toward development of a rapid blood test for breast cancer. Clinical breast cancer. 2003;4:203–9. doi: 10.3816/cbc.2003.n.026. [DOI] [PubMed] [Google Scholar]
- [6].Galanina N, Bossuyt V, Harris LN. Molecular predictors of response to therapy for breast cancer. Cancer journal (Sudbury, Mass.) 2011;17:96–103. doi: 10.1097/PPO.0b013e318212dee3. [DOI] [PubMed] [Google Scholar]
- [7].Wolff AC, Hammond MEH, Schwartz JN, Hagerty KL, Allred D, Craig Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez E, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes Daniel F. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Archives of pathology & laboratory medicine. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- [8].Hudelist G, Pacher-Zavisin M, Singer CF, Holper T, Kubista E, Schreiber M, Manavi M, Bilban M, Czerwenka Klaus. Use of high-throughput protein array for profiling of differentially expressed proteins in normal and malignant breast tissue. Breast cancer research and treatment. 2004;86:281–91. doi: 10.1023/b:brea.0000036901.16346.83. [DOI] [PubMed] [Google Scholar]
- [9].Gutiérrez-Xicoténcatl L, Plett-Torres T, Madrid-González CL, Madrid-Marina V. Molecular diagnosis of human papillomavirus in the development of cervical cancer. Salud pública de México. 2009;51(Suppl 3):S479–88. doi: 10.1590/s0036-36342009000900014. [DOI] [PubMed] [Google Scholar]
- [10].Ingvarsson J, Wingren C, Carlsson A, Ellmark P, Wahren B, Engström G, Harmenberg U, Krogh M, Peterson C, Borrebaeck CK. Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics. 2008;8:2211–9. doi: 10.1002/pmic.200701167. [DOI] [PubMed] [Google Scholar]
- [11].Darmanis S, Nong RY, Hammond M, Gu J, Alderborn A, Vänelid J, Siegbahn A, Gustafsdottir S, Ericsson O, Landegren U, Kamali-Moghaddam M. Sensitive plasma protein analysis by microparticle-based proximity ligation assays. Molecular & cellular proteomics. 2010;9:327–35. doi: 10.1074/mcp.M900248-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Visintin I, Feng Z, Longton G, Ward DC, Alvero AB, Lai Y, Tenthorey J, Leiser A, Flores-Saaib R, Yu H, Azori M, Rutherford T, Schwartz PE, Mor G. Diagnostic markers for early detection of ovarian cancer. Clinical cancer research. 2008;14:1065–72. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- [13].Lin D, Feng S, Pan J, Chen Y, Lin J, Chen G, Xie S, Zeng H, Chen R. Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis. Optics express. 2011;19:13565–77. doi: 10.1364/OE.19.013565. [DOI] [PubMed] [Google Scholar]
- [14].Lin J, Chen R, Feng S, Pan J, Li Y, Chen G, Cheng M, Huang Z, Yu Y, Zeng H. A novel blood plasma analysis technique combining membrane electrophoresis with silver nanoparticle-based SERS spectroscopy for potential applications in noninvasive cancer detection. Nanomedicine. 2011;7:655–63. doi: 10.1016/j.nano.2011.01.012. [DOI] [PubMed] [Google Scholar]
- [15].Fromm JR, Thomas A, Wood BL. Flow cytometry can diagnose classical hodgkin lymphoma in lymph nodes with high sensitivity and specificity. American journal of clinical pathology. 2009;131:322–32. doi: 10.1309/AJCPW3UN9DYLDSPB. [DOI] [PubMed] [Google Scholar]
- [16].Yaziji H, Taylor CR, Goldstein NS, Dabbs DJ, Hammond EH, Hewlett B, Floyd AD, Barry TS, Martin AW, Badve S, Baehner F, Cartun RW, Eisen RN, Swanson PE, Hewitt SM, Vyberg M, Hicks DG. Consensus recommendations on estrogen receptor testing in breast cancer by immunohistochemistry. Applied immunohistochemistry & molecular morphology. 2008;16:513–20. doi: 10.1097/PAI.0b013e31818a9d3a. [DOI] [PubMed] [Google Scholar]
- [17].Hsi ED, Kopecky KJ, Appelbaum FR, Boldt D, Frey T, Loftus M, Hussein M. Prognostic significance of CD38 and CD20 expression as assessed by quantitative flow cytometry in chronic lymphocytic leukaemia. British journal of haematology. 2003;120:1017–25. doi: 10.1046/j.1365-2141.2003.04213.x. [DOI] [PubMed] [Google Scholar]
- [18].Schmitt M, Sturmheit AS, Welk A, Schnelldorfer C, Harbeck N. Procedures for the quantitative protein determination of urokinase and its inhibitor, PAI-1, in human breast cancer tissue extracts by ELISA. Methods in molecular medicine. 2006;120:245–65. doi: 10.1385/1-59259-969-9:245. [DOI] [PubMed] [Google Scholar]
- [19].Uhrmacher S, Schmidt C, Erdfelder F, Poll-Wolbeck SJ, Gehrke I, Hallek M, Kreuzer K-A. Use of the receptor tyrosine kinase-like orphan receptor 1 (ROR1) as a diagnostic tool in chronic lymphocytic leukemia (CLL) Leukemia research. 2011;35:1360–6. doi: 10.1016/j.leukres.2011.04.006. [DOI] [PubMed] [Google Scholar]
- [20].Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clinical chemistry. 2010;56:177–85. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- [21].Kim MS, Kim T, Kong S-Y, Kwon S, Bae CY, Choi J, Kim Chul H., Lee ES, Park J-K. Breast cancer diagnosis using a microfluidic multiplexed immunohistochemistry platform. PloS one. 2010;5:e10441. doi: 10.1371/journal.pone.0010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Decaestecker C, Lopez XM, D'Haene N, Roland I, Guendouz S, Duponchelle C, Berton A, Debeir O, Salmon I. Requirements for the valid quantification of immunostains on tissue microarray materials using image analysis. Proteomics. 2009;9:4478–94. doi: 10.1002/pmic.200800936. [DOI] [PubMed] [Google Scholar]
- [23].Ross DT, Kim C-Y, Tang G, Bohn OL, Beck R, Ring BZ, Seitz RS, Paik S, Costantino JP, Wolmark N. Chemosensitivity and stratification by a five monoclonal antibody immunohistochemistry test in the NSABP B14 and B20 trials. Clinical cancer research. 2008;14:6602–9. doi: 10.1158/1078-0432.CCR-08-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ring BZ, Seitz RS, Beck R. a, Shasteen WJ, Soltermann A, Arbogast S, Robert F, Schreeder MT, Ross DT. A novel five-antibody immunohistochemical test for subclassification of lung carcinoma. Modern pathology. 2009;22:1032–43. doi: 10.1038/modpathol.2009.60. [DOI] [PubMed] [Google Scholar]
- [25].Kashani-Sabet M, Rangel J, Torabian S, Nosrati M, Simko J, Jablons DM, Moore DH, Haqq C, Miller JR, Sagebiel RW. A multi-marker assay to distinguish malignant melanomas from benign nevi. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6268–72. doi: 10.1073/pnas.0901185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Crabb SJ, Bajdik CD, Leung S, Speers CH, Kennecke H, Huntsman DG, Gelmon K. Can clinically relevant prognostic subsets of breast cancer patients with four or more involved axillary lymph nodes be identified through immunohistochemical biomarkers? A tissue microarray feasibility study. Breast cancer research. 2008;10:R6. doi: 10.1186/bcr1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Grimwade L, Gudgin E, Bloxham D, Scott M. a, Erber WN. PML protein analysis using imaging flow cytometry. Journal of clinical pathology. 2011;64:447–50. doi: 10.1136/jcp.2010.085662. [DOI] [PubMed] [Google Scholar]
- [28].Demurtas A, Accinelli G, Pacchioni D, Godio L, Novero D, Bussolati G, Palestro G, Papotti M, Stacchini A. Utility of flow cytometry immunophenotyping in fine-needle aspirate cytologic diagnosis of non-Hodgkin lymphoma: A series of 252 cases and review of the literature. Applied immunohistochemistry & molecular morphology : AIMM / official publication of the Society for Applied Immunohistochemistry. 2010;18:311–22. doi: 10.1097/PAI.0b013e3181827da8. [DOI] [PubMed] [Google Scholar]
- [29].D'Arena G, Musto P, Cascavilla N, Dell'Olio M, Di Renzo N, Carotenuto M. Quantitative flow cytometry for the differential diagnosis of leukemic B-cell chronic lymphoproliferative disorders. American journal of hematology. 2000;64:275–81. doi: 10.1002/1096-8652(200008)64:4<275::aid-ajh7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- [30].Sun J, Masterman-Smith MD, Graham N. a, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K.-i., Lee K-B, Wang H, Yu ZTF, Lu Y-T, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS, Lazareff J, Kornblum HI, Yong WH, Graeber TG, Tseng H-R. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer research. 2010;70:6128–38. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ferrier CM, de Witte HH, Straatman H, van Tienoven DH, van Geloof WL, Rietveld FJ, Sweep CG, Ruiter DJ, van Muijen GN. Comparison of immunohistochemistry with immunoassay (ELISA) for the detection of components of the plasminogen activation system in human tumour tissue. British journal of cancer. 1999;79:1534–41. doi: 10.1038/sj.bjc.6690245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tan W, Sabet L, Li Y, Yu T, Klokkevold PR, Wong DT, Ho C-M. Optical protein sensor for detecting cancer markers in saliva. Biosensors & bioelectronics. 2008;24:266–71. doi: 10.1016/j.bios.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang S, Zhao X, Khimji I, Akbas R, Qiu W, Edwards D, Cramer DW, Ye B, Demirci U. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab on a chip. 2011;11:3411–8. doi: 10.1039/c1lc20479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Attallah AM, Sakr H. a, Ismail H, Abdel-Hady E-SK, El-Dosoky I. An office-based immunodiagnostic assay for detecting urinary nuclear matrix protein 52 in patients with bladder cancer. BJU international. 2005;96:334–9. doi: 10.1111/j.1464-410X.2005.05627.x. [DOI] [PubMed] [Google Scholar]
- [35].Wang S, Ge L, Song X, Yu J, Ge S, Huang J, Zeng F. Paper-based chemiluminescence ELISA: lab-on-paper based on chitosan modified paper device and wax-screen-printing. Biosensors & bioelectronics. 2012;31:212–8. doi: 10.1016/j.bios.2011.10.019. [DOI] [PubMed] [Google Scholar]
- [36].Ferreira CSM, Papamichael K, Guilbault G, Schwarzacher T, Gariepy J, Missailidis S. DNA aptamers against the MUC1 tumour marker: design of aptamer-antibody sandwich ELISA for the early diagnosis of epithelial tumours. Analytical and bioanalytical chemistry. 2008;390:1039–50. doi: 10.1007/s00216-007-1470-1. [DOI] [PubMed] [Google Scholar]
- [37].Liu Y, Wang H, Huang J, Yang J, Liu B, Yang P. Microchip-based ELISA strategy for the detection of low-level disease biomarker in serum. Analytica chimica acta. 2009;650:77–82. doi: 10.1016/j.aca.2009.06.048. [DOI] [PubMed] [Google Scholar]
- [38].Cheow LF, Ko SH, Kim SJ, Kang KH, Han J. Increasing the sensitivity of enzyme-linked immunosorbent assay using multiplexed electrokinetic concentrator. Analytical chemistry. 2010;82:3383–8. doi: 10.1021/ac9024335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhou F, Yuan L, Wang H, Li D, Chen H. Gold Nanoparticle Layer: A Promising Platform for Ultra-Sensitive Cancer Detection. Langmuir : the ACS journal of surfaces and colloids. 2011:2155–2158. doi: 10.1021/la1049937. [DOI] [PubMed] [Google Scholar]
- [40].Gau V, Wong D. Oral fluid nanosensor test (OFNASET) with advanced electrochemical-based molecular analysis platform. Annals of the New York Academy of Sciences. 2007;1098:401–10. doi: 10.1196/annals.1384.005. [DOI] [PubMed] [Google Scholar]
- [41].Maeng J-H, Lee B-C, Ko Y-J, Cho W, Ahn Y, Cho N-G, Lee Seoung-Hwan H., Seung Y. A novel microfluidic biosensor based on an electrical detection system for alpha-fetoprotein. Biosensors & bioelectronics. 2008;23:1319–25. doi: 10.1016/j.bios.2007.11.019. [DOI] [PubMed] [Google Scholar]
- [42].Rauser S, Marquardt C, Balluff B, Deininger S-O, Albers C, Belau E, Hartmer R, Suckau D, Specht K, Ebert MP, Schmitt M, Aubele M, Höfler H, Walch A. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. Journal of proteome research. 2010;9:1854–63. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- [43].Hu Q, Huang Y, Wang Z, Tao H, Liu J, Yan L, Wang K. Application of Surface-Enhanced Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Coupled With an Artificial Neural Network Model for the Diagnosis of Hepatocellular Carcinoma. Hepato-gastroenterology. 2011:59. doi: 10.5754/hge11771. [DOI] [PubMed] [Google Scholar]
- [44].Gast M-CW, Gils CHVAN, Wessels LFA, Harris N, Bonfrer JMG, Rutgers EJTH, Schellens JHM, Beijnen JH. Serum protein profiling for diagnosis of breast cancer using SELDI-TOF MS. Oncology Reports. 2009;22:205–213. doi: 10.3892/or_00000426. [DOI] [PubMed] [Google Scholar]
- [45].Liu C. The application of SELDI-TOF-MS in clinical diagnosis of cancers. Journal of biomedicine & biotechnology. 2011;2011:245821. doi: 10.1155/2011/245821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Szanto I, Mark L, Bona A, Maasz G, Sandor B, Gelencser G, Turi Z, Gallyas F. High-throughput screening of saliva for early detection of oral cancer: a pilot study. Technology in cancer research & treatment. 2012;11:181–8. doi: 10.7785/tcrt.2012.500248. [DOI] [PubMed] [Google Scholar]
- [47].Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nature methods. 2007;4:828–33. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- [48].Walter M, Heinze C, Steiner T, Pilchowski R, von Eggeling F, Wunderlich H, Junker K. Immunochemotherapy-associated protein patterns in tumour tissue and serum of patients with metastatic renal cell carcinoma. Archives of physiology and biochemistry. 2010;116:197–207. doi: 10.3109/13813455.2010.513392. [DOI] [PubMed] [Google Scholar]
- [49].Tang K-L, Li T-H, Xiong W-W, Chen K. Ovarian cancer classification based on dimensionality reduction for SELDI-TOF data. BMC bioinformatics. 2010;11:109. doi: 10.1186/1471-2105-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Callesen AK, Vach W, Jørgensen PE, Cold S, Mogensen O, Kruse T, Jensen ON, Madsen JS. Reproducibility of mass spectrometry based protein profiles for diagnosis of breast cancer across clinical studies: a systematic review. Journal of proteome research. 2008;7:1395–402. doi: 10.1021/pr800115f. [DOI] [PubMed] [Google Scholar]
- [51].Cadron I, Van Gorp T, Amant F, Vergote I, Moerman P, Waelkens E, Daemen A, Van De Plas R, De Moor B, Zeillinger R. The use of laser microdissection and SELDI-TOF MS in ovarian cancer tissue to identify protein profiles. Anticancer research. 2009;29:1039–45. [PubMed] [Google Scholar]
- [52].Blokzijl a, Friedman M, Pontén F, Landegren U. Profiling protein expression and interactions: proximity ligation as a tool for personalized medicine. Journal of internal medicine. 2010;268:232–45. doi: 10.1111/j.1365-2796.2010.02256.x. [DOI] [PubMed] [Google Scholar]
- [53].Schallmeiner E, Oksanen E, Ericsson O, Spångberg L, Eriksson S, Stenman U-H, Pettersson K, Landegren U. Sensitive protein detection via triple-binder proximity ligation assays. Nature methods. 2007;4:135–7. doi: 10.1038/nmeth974. [DOI] [PubMed] [Google Scholar]
- [54].Jiang X, Cheng S, Chen W, Wang L, Shi F, Zhu C. Comparison of oligonucleotide-labeled antibody probe assays for prostate-specific antigen detection. Analytical biochemistry. 2012;424:1–7. doi: 10.1016/j.ab.2012.02.004. [DOI] [PubMed] [Google Scholar]
- [55].Shi Y, Huang W, Tan Y, Jin X, Dua R, Penuel E, Mukherjee A, Sperinde J, Pannu H, Chenna A, DeFazio-Eli L, Pidaparthi S, Badal Y, Wallweber G, Chen L, Williams S, Tahir H, Larson J, Goodman L, Whitcomb J, Petropoulos C, Winslow J. A novel proximity assay for the detection of proteins and protein complexes: quantitation of HER1 and HER2 total protein expression and homodimerization in formalin-fixed, paraffin-embedded cell lines and breast cancer tissue. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 2009;18:11–21. doi: 10.1097/PDM.0b013e31818cbdb2. [DOI] [PubMed] [Google Scholar]
- [56].Huang W, Reinholz M, Weidler J, Yolanda L, Paquet A, Whitcomb J, Lingle W, Jenkins RB, Chen B, Larson JS, Tan Y, Sherwood T, Bates M, Perez E. Comparison of central HER2 testing with quantitative total HER2 expression and HER2 homodimer measurements using a novel proximity-based assay. American journal of clinical pathology. 2010;134:303–11. doi: 10.1309/AJCP3BZY4YAFNTRG. [DOI] [PubMed] [Google Scholar]
- [57].Dua R, Zhang J, Parry G, Penuel E. Detection of hepatocyte growth factor (HGF) ligand-c-MET receptor activation in formalin-fixed paraffin embedded specimens by a novel proximity assay. PloS one. 2011;6:e15932. doi: 10.1371/journal.pone.0015932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Woodbury RL, Varnum SM, Zangar RC. Elevated HGF levels in sera from breast cancer patients detected using a protein microarray ELISA. Journal of proteome research. 2002;1:233–7. doi: 10.1021/pr025506q. [DOI] [PubMed] [Google Scholar]
- [59].Lin JC-H. Protein microarrays for cancer diagnostics and therapy. Medical principles and practice. 2010;19:247–54. doi: 10.1159/000312709. [DOI] [PubMed] [Google Scholar]
- [60].Carlsson A, Wingren C, Ingvarsson J, Ellmark P, Baldertorp B, Fernö M, Olsson H, Borrebaeck CAK. Serum proteome profiling of metastatic breast cancer using recombinant antibody microarrays. European journal of cancer. 2008;44:472–80. doi: 10.1016/j.ejca.2007.11.025. [DOI] [PubMed] [Google Scholar]
- [61].Strömberg S, Björklund MG, Asplund C, Sköllermo A, Persson A, Wester K, Kampf C, Nilsson P, Andersson A-C, Uhlen M, Kononen J, Ponten F, Asplund A. A high-throughput strategy for protein profiling in cell microarrays using automated image analysis. Proteomics. 2007;7:2142–50. doi: 10.1002/pmic.200700199. [DOI] [PubMed] [Google Scholar]
- [62].Schröder C, Jacob A, Tonack S, Radon TP, Sill M, Zucknick M, Rüffer S, Costello E, Neoptolemos JP, Crnogorac-Jurcevic T, Bauer A, Fellenberg K, Hoheisel JD. Dual-color proteomic profiling of complex samples with a microarray of 810 cancer-related antibodies. Molecular & cellular proteomics. 2010;9:1271–80. doi: 10.1074/mcp.M900419-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fang C, Wang Y, Vu NT, Lin W-Y, Hsieh Y-T, Rubbi L, Phelps ME, Müschen M, Kim Y-M, Chatziioannou AF, Tseng H-R, Graeber TG. Integrated microfluidic and imaging platform for a kinase activity radioassay to analyze minute patient cancer samples. Cancer research. 2010;70:8299–308. doi: 10.1158/0008-5472.CAN-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Göttert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. Journal of the American Chemical Society. 2008;130:8633–41. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Du Z, Colls N, Cheng KH, Vaughn MW, Gollahon L. Microfluidic-based diagnostics for cervical cancer cells. Biosensors & bioelectronics. 2006;21:1991–5. doi: 10.1016/j.bios.2005.09.005. [DOI] [PubMed] [Google Scholar]
- [66].Kim MS, Kwon S, Kim T, Lee ES, Park J-K. Quantitative proteomic profiling of breast cancers using a multiplexed microfluidic platform for immunohistochemistry and immunocytochemistry. Biomaterials. 2011;32:1396–403. doi: 10.1016/j.biomaterials.2010.10.040. [DOI] [PubMed] [Google Scholar]
- [67].Henry OY, Fragoso A, Beni V, Laboria N, Sánchez JLA, Latta D, Von Germar F, Drese K, Katakis I, O'Sullivan CK. Design and testing of a packaged microfluidic cell for the multiplexed electrochemical detection of cancer markers. Electrophoresis. 2009;30:3398–405. doi: 10.1002/elps.200900368. [DOI] [PubMed] [Google Scholar]
- [68].Sukhanova A, Nabiev I. Fluorescent nanocrystal-encoded microbeads for multiplexed cancer imaging and diagnosis. Critical reviews in oncology/hematology. 2008;68:39–59. doi: 10.1016/j.critrevonc.2008.05.006. [DOI] [PubMed] [Google Scholar]
- [69].Xie C, Wang G. Development of simultaneous detection of total prostate-specific antigen (tPSA) and free PSA with rapid bead-based immunoassay. Journal of clinical laboratory analysis. 2011;25:37–42. doi: 10.1002/jcla.20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Long Y, Zhang Z, Yan X, Xing J, Zhang K, Huang J, Zheng J, Li W. Multiplex immunodetection of tumor markers with a suspension array built upon core-shell structured functional fluorescence-encoded microspheres. Analytica chimica acta. 2010;665:63–8. doi: 10.1016/j.aca.2010.03.009. [DOI] [PubMed] [Google Scholar]
- [71].Shin K-S, Lee SW, Han K-C, Kim SK, Yang EK, Park JH, Ju B-K, Kang JY, Kim TS. Amplification of fluorescence with packed beads to enhance the sensitivity of miniaturized detection in microfluidic chip. Biosensors & bioelectronics. 2007;22:2261–7. doi: 10.1016/j.bios.2006.11.011. [DOI] [PubMed] [Google Scholar]
- [72].Fu Z, Hao C, Fei X, Ju H. Flow-injection chemiluminescent immunoassay for alpha-fetoprotein based on epoxysilane modified glass microbeads. Journal of immunological methods. 2006;312:61–7. doi: 10.1016/j.jim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- [73].Retnakumari A, Jayasimhan J, Chandran P, Menon D, Nair S, Mony U, Koyakutty M. CD33 monoclonal antibody conjugated Au cluster nano-bioprobe for targeted flow-cytometric detection of acute myeloid leukaemia. Nanotechnology. 2011;22:285102. doi: 10.1088/0957-4484/22/28/285102. [DOI] [PubMed] [Google Scholar]
- [74].Ray S, Reddy PJ, Choudhary S, Raghu D, Srivastava S. Emerging nanoproteomics approaches for disease biomarker detection: a current perspective. Journal of proteomics. 2011;74:2660–81. doi: 10.1016/j.jprot.2011.04.027. [DOI] [PubMed] [Google Scholar]
- [75].Kah JCY, Kho KW, Lee CGL, James C, et al. Early diagnosis of oral cancer based on the surface plasmon resonance of gold nanoparticles. International journal of nanomedicine. 2007;2:785–98. [PMC free article] [PubMed] [Google Scholar]
- [76].Yu X, Munge B, Patel V, Jensen G, Bhirde A, Gong JD, Kim SN, Gillespie J, Gutkind JS, Papadimitrakopoulos F, Rusling JF. Carbon nanotube amplification strategies for highly sensitive immunodetection of cancer biomarkers. Journal of the American Chemical Society. 2006;128:11199–205. doi: 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lin J, He C, Zhang L, Zhang S. Sensitive amperometric immunosensor for alpha-fetoprotein based on carbon nanotube/gold nanoparticle doped chitosan film. Analytical biochemistry. 2009;384:130–5. doi: 10.1016/j.ab.2008.09.033. [DOI] [PubMed] [Google Scholar]
- [78].Ji NY, Kang YH, Park M-Y, Lee CI, Kim MK, Kim DG, Kim JW, Song EY. Development of a fluorescent microsphere immunoassay for cystatin B (CSTB) in serum of patients with hepatocellular carcinoma. Clinical chemistry and laboratory medicine. 2011;49:151–5. doi: 10.1515/CCLM.2011.008. [DOI] [PubMed] [Google Scholar]
- [79].Xie C, Kim HJ, Haw JG, Kalbasi A, Gardner BK, Li G, Rao J, Chia D, Liong M, Punzalan RR, Marks LS, Pantuck AJ, de la Taille A, Wang G, Mukouyama H, Zeng G. A novel multiplex assay combining autoantibodies plus PSA has potential implications for classification of prostate cancer from non-malignant cases. Journal of translational medicine. 2011;9:43. doi: 10.1186/1479-5876-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Dekking E, van der Velden VHJ, Böttcher S, Brüggemann M, Sonneveld E, Koning-Goedheer A, Boeckx N, Lucio P, Sedek L, Szczepański T, Kalina T, Kovac M, Evans P, Hoogeveen PG, Flores-Montero J, Orfao A, Comans-Bitter WM, Staal FJT, van Dongen JJM. Detection of fusion genes at the protein level in leukemia patients via the flow cytometric immunobead assay. Best practice & research. Clinical haematology. 2010;23:333–45. doi: 10.1016/j.beha.2010.09.010. [DOI] [PubMed] [Google Scholar]
- [81].Jokerst JV, Raamanathan A, Christodoulides N, Floriano PN, Pollard A, Simmons GW, Wong J, Gage C, Furmaga WB, Redding SW, McDevitt JT. Nano-bio-chips for high performance multiplexed protein detection: determinations of cancer biomarkers in serum and saliva using quantum dot bioconjugate labels. Biosensors & bioelectronics. 2009;24:3622–9. doi: 10.1016/j.bios.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lee KH, Galloway JF, Park J, Dvoracek CM, Dallas M, Konstantopoulos K, Maitra A, Searson PC. Quantitative molecular profiling of biomarkers for pancreatic cancer with functionalized quantum dots. Nanomedicine : nanotechnology, biology, and medicine. 2012 doi: 10.1016/j.nano.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Barat B, Sirk SJ, McCabe KE, Li J, Lepin EJ, Remenyi, Roland Koh AL, Olafsen T, Gambhir SS, Weiss S, Wu AM. Cys-diabody quantum dot conjugates (immunoQdots) for cancer marker detection. Bioconjugate chemistry. 2009;20:1474–81. doi: 10.1021/bc800421f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zheng H, Chen G, DeLouise LA, Lou Z. Detection of the cancer marker CD146 expression in melanoma cells with semiconductor quantum dot label. Journal of biomedical nanotechnology. 2010;6:303–11. doi: 10.1166/jbn.2010.1136. [DOI] [PubMed] [Google Scholar]
- [85].Hu M, Yan J, He Y, Lu H, Weng L, Song S, Fan C, Wang L. Ultrasensitive, multiplexed detection of cancer biomarkers directly in serum by using a quantum dot-based microfluidic protein chip. ACS nano. 2010;4:488–94. doi: 10.1021/nn901404h. [DOI] [PubMed] [Google Scholar]
- [86].Kah JCY, Olivo MC, Lee CGL, Sheppard CJR. Molecular contrast of EGFR expression using gold nanoparticles as a reflectance-based imaging probe. Molecular and cellular probes. 2008;22:14–23. doi: 10.1016/j.mcp.2007.06.010. [DOI] [PubMed] [Google Scholar]
- [87].Liu X, Dai Q, Austin L, Coutts J, Knowles G, Zou J, Chen H, Huo Q. A one-step homogeneous immunoassay for cancer biomarker detection using gold nanoparticle probes coupled with dynamic light scattering. Journal of the American Chemical Society. 2008;130:2780–2. doi: 10.1021/ja711298b. [DOI] [PubMed] [Google Scholar]
- [88].Lu W, Arumugam SR, Senapati D, Singh AK, Arbneshi T, Khan SA, Yu H, Ray PC. Multifunctional Oval-Shaped Gold-Nanoparticle-Based Selective Detection of Breast Cancer Cells Using Simple Colorimetric and Highly Sensitive Two- Photon Scattering Assay. ACS nanoano. 2010;4:1739–1749. doi: 10.1021/nn901742q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ladd J, Lu H, Taylor AD, Goodell V, Disis ML, Jiang S. Direct detection of carcinoembryonic antigen autoantibodies in clinical human serum samples using a surface plasmon resonance sensor. Colloids and surfaces. B, Biointerfaces. 2009;70:1–6. doi: 10.1016/j.colsurfb.2008.11.032. [DOI] [PubMed] [Google Scholar]
- [90].Choi S, Chae J. A microfluidic biosensor based on competitive protein adsorption for thyroglobulin detection. Biosensors & bioelectronics. 2009;25:118–23. doi: 10.1016/j.bios.2009.06.017. [DOI] [PubMed] [Google Scholar]
- [91].Lee K, Drachev VP, Irudayaraj J. DNA-gold nanoparticle reversible networks grown on cell surface marker sites: application in diagnostics. ACS nano. 2011;5:2109–17. doi: 10.1021/nn1030862. [DOI] [PubMed] [Google Scholar]
- [92].Law W-C, Yong K-T, Baev A, Prasad PN. Sensitivity improved surface plasmon resonance biosensor for cancer biomarker detection based on plasmonic enhancement. ACS nano. 2011;5:4858–64. doi: 10.1021/nn2009485. [DOI] [PubMed] [Google Scholar]
- [93].Yu C, Nakshatri H, Irudayaraj J. Identity profiling of cell surface markers by multiplex gold nanorod probes. Nano letters. 2007;7:2300–6. doi: 10.1021/nl070894m. [DOI] [PubMed] [Google Scholar]
- [94].Piliarik M, Bocková M, Homola J. Surface plasmon resonance biosensor for parallelized detection of protein biomarkers in diluted blood plasma. Biosensors & bioelectronics. 2010;26:1656–61. doi: 10.1016/j.bios.2010.08.063. [DOI] [PubMed] [Google Scholar]
- [95].Kazuno S, Fujimura T, Arai T, Ueno T, Nagao K, Fujime M, Murayama K. Multi-sequential surface plasmon resonance analysis of haptoglobin-lectin complex in sera of patients with malignant and benign prostate diseases. Analytical biochemistry. 2011;419:241–9. doi: 10.1016/j.ab.2011.08.029. [DOI] [PubMed] [Google Scholar]
- [96].Mallidi S, Larson T, Tam J, Joshi PP, Karpiouk A, Sokolov K, Emelianov S. Multiwavelength photoacoustic imaging and plasmon resonance coupling of gold nanoparticles for selective detection of cancer. Nano letters. 2009;9:2825–31. doi: 10.1021/nl802929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Qian X, Peng X-H, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nature biotechnology. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- [98].Lutz B, Dentinger C, Sun L, Nguyen L, Zhang J, Chmura A, Allen A, Chan S, Knudsen B. Raman nanoparticle probes for antibody-based protein detection in tissues. The journal of histochemistry and cytochemistry. 2008;56:371–9. doi: 10.1369/jhc.7A7313.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wang G, Lipert RJ, Jain M, Kaur S, Chakraboty S, Torres MP, Batra SK, Brand RE, Porter MD. Detection of the potential pancreatic cancer marker MUC4 in serum using surface-enhanced Raman scattering. Analytical chemistry. 2011;83:2554–61. doi: 10.1021/ac102829b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Maiti KK, Dinish US, Fu CY, Lee J-J, Soh K.-Seng., Yun S-W, Bhuvaneswari R, Olivo M, Chang Y-T. Development of biocompatible SERS nanotag with increased stability by chemisorption of reporter molecule for in vivo cancer detection. Biosensors & bioelectronics. 2010;26:398–403. doi: 10.1016/j.bios.2010.07.123. [DOI] [PubMed] [Google Scholar]
- [101].Aydin O, Altaş M, Kahraman M, Bayrak OF, Culha M. Differentiation of healthy brain tissue and tumors using surface-enhanced Raman scattering. Applied spectroscopy. 2009;63:1095–100. doi: 10.1366/000370209789553219. [DOI] [PubMed] [Google Scholar]
- [102].Maiti KK, Samanta A, Vendrell M, Soh K-S, Olivo M, Chang Y-T. Multiplex cancer cell detection by SERS nanotags with cyanine and triphenylmethine Raman reporters. Chemical communications. 2011;47:3514–6. doi: 10.1039/c0cc05265e. [DOI] [PubMed] [Google Scholar]
- [103].Chon H, Lee S, Yoon S-Y, Chang S-I, Lim DW, Choo J. Simultaneous immunoassay for the detection of two lung cancer markers using functionalized SERS nanoprobes. Chemical communications. 2011;47:12515–7. doi: 10.1039/c1cc15707h. [DOI] [PubMed] [Google Scholar]
- [104].Sha MY, Xu H, Natan MJ, Cromer R. Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood. Journal of the American Chemical Society. 2008;130:17214–5. doi: 10.1021/ja804494m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Domenici F, Bizzarri AR, Cannistraro S. SERS-based nanobiosensing for ultrasensitive detection of the p53 tumor suppressor. International journal of nanomedicine. 2011;6:2033–42. doi: 10.2147/IJN.S23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chikkaveeraiah BV, Bhirde A, Malhotra R, Patel V, Gutkind JS, Rusling JF. Single-wall carbon nanotube forest arrays for immunoelectrochemical measurement of four protein biomarkers for prostate cancer. Analytical chemistry. 2009;81:9129–34. doi: 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Malhotra R, Patel V, Vaqué JP, Gutkind JS, Rusling JF. Ultrasensitive electrochemical immunosensor for oral cancer biomarker IL-6 using carbon nanotube forest electrodes and multilabel amplification. Analytical chemistry. 2010;82:3118–23. doi: 10.1021/ac902802b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Huang K-J, Niu D-J, Xie W-Z, Wang W. A disposable electrochemical immunosensor for carcinoembryonic antigen based on nano-Au/multi-walled carbon nanotubes-chitosans nanocomposite film modified glassy carbon electrode. Analytica chimica acta. 2010;659:102–8. doi: 10.1016/j.aca.2009.11.023. [DOI] [PubMed] [Google Scholar]
- [109].Yeung D, Hughes T. Therapeutic targeting of BCR-ABL: prognostic markers of response and resistance mechanism in chronic myeloid leukaemia. Critical reviews in oncology. 2012 doi: 10.1615/critrevoncog.v17.i1.30. [DOI] [PubMed] [Google Scholar]
- [110].Saintigny P, Burger JA. Recent Advances in Non-small Cell Lung Cancer Biology and Clinical Management. Discovery Medicine. 13:287–297. n.d. [PubMed] [Google Scholar]
- [111].Ghosh G, Yan X, Lee AG, Kron SJ, Palecek SP. Quantifying the sensitivities of EGF receptor (EGFR) tyrosine kinase inhibitors in drug resistant non-small cell lung cancer (NSCLC) cells using hydrogel-based peptide array. Biosensors & bioelectronics. 2010;26:424–31. doi: 10.1016/j.bios.2010.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Mizutani T, Kondo T, Darmanin S, Tsuda M, Tanaka S, Tobiume M, Asaka M, Ohba Y. A novel FRET-based biosensor for the measurement of BCR-ABL activity and its response to drugs in living cells. Clinical cancer research. 2010;16:3964–75. doi: 10.1158/1078-0432.CCR-10-0548. [DOI] [PubMed] [Google Scholar]
- [113].Jarvius M, Paulsson J, Weibrecht I, Leuchowius K-J, Andersson A-C, Wählby C, Gullberg M, Botling J, Sjöblom T, Markova B, Ostman A, Landegren U, Söderberg O. In situ detection of phosphorylated platelet-derived growth factor receptor beta using a generalized proximity ligation method. Molecular & cellular proteomics. 2007;6:1500–9. doi: 10.1074/mcp.M700166-MCP200. [DOI] [PubMed] [Google Scholar]
- [114].Ghosh G, Lee AG, Palecek SP. Hydrogel-based protein array for quantifying epidermal growth factor receptor activity in cell lysates. Analytical biochemistry. 2009;393:205–14. doi: 10.1016/j.ab.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Lee AG, Beebe DJ, Palecek SP. Quantification of kinase activity in cell lysates via photopatterned macroporous poly(ethylene glycol) hydrogel arrays in microfluidic channels. Biomedical microdevices. 2012;14:247–57. doi: 10.1007/s10544-011-9602-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kim P, Liu X, Lee T, Liu L, Barham R, Kirkland R, Leesman G, Kuller A, Ybarrondo B, Ng S-C, Singh S. Highly sensitive proximity mediated immunoassay reveals HER2 status conversion in the circulating tumor cells of metastatic breast cancer patients. Proteome science. 2011;9:75. doi: 10.1186/1477-5956-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Khan IH, Zhao J, Ghosh P, Ziman M, Sweeney C, Kung H-J, Luciw P. Microbead arrays for the analysis of ErbB receptor tyrosine kinase activation and dimerization in breast cancer cells. Assay and drug development technologies. 2010;8:27–36. doi: 10.1089/adt.2009.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lu S, Wang Y. Fluorescence resonance energy transfer biosensors for cancer detection and evaluation of drug efficacy. Clinical cancer research. 2010;16:3822–4. doi: 10.1158/1078-0432.CCR-10-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Lowe SB, Dick J. a G., Cohen BE, Stevens MM. Multiplex sensing of protease and kinase enzyme activity via orthogonal coupling of quantum dot-peptide conjugates. ACS nano. 2012;6:851–7. doi: 10.1021/nn204361s. [DOI] [PubMed] [Google Scholar]
- [120].Toita R, Mori T, Naritomi Y, Kang J-H, Shiosaki S, Niidome T, Katayama Y. Fluorometric detection of protein kinase Cα activity based on phosphorylation-induced dissociation of a polyion complex. Analytical biochemistry. 2012;424:130–6. doi: 10.1016/j.ab.2012.01.036. [DOI] [PubMed] [Google Scholar]
- [121].Fujiwara A, Shibata E, Terashima H, Shishido A, Nishiki J, Yoshida K, Miyauchi K, Madachi A, Matsuura N. Evaluation of matrix metalloproteinase-2 (MMP-2) activity with film in situ zymography for improved cytological diagnosis of breast tumors. Breast cancer. 2006;13:272–8. doi: 10.2325/jbcs.13.272. [DOI] [PubMed] [Google Scholar]
- [122].Darragh MR, Schneider EL, Lou J, Phojanakong PJ, Farady CJ, Marks JD, Hann BC, Craik CS. Tumor detection by imaging proteolytic activity. Cancer research. 2010;70:1505–12. doi: 10.1158/0008-5472.CAN-09-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Ryu JH, Lee A, Lee S, Ahn C-H, Park JW, Leary JF, Park S, Kim K, Kwon IC, Youn I-C, Choi K. “One-step” detection of matrix metalloproteinase activity using a fluorogenic peptide probe-immobilized diagnostic kit. Bioconjugate chemistry. 2010;21:1378–84. doi: 10.1021/bc100008b. [DOI] [PubMed] [Google Scholar]
- [124].Liu GL, Chen FF, Ellman J. a, Lee LP. Peptide-nanoparticle hybrid SERS probe for dynamic detection of active cancer biomarker enzymes. Conference proceedings : … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Conference; 2006. pp. 795–8. [DOI] [PubMed] [Google Scholar]
- [125].Fajkus J. Detection of telomerase activity by the TRAP assay and its variants and alternatives. Clinica chimica acta. 2006;371:25–31. doi: 10.1016/j.cca.2006.02.039. [DOI] [PubMed] [Google Scholar]
- [126].Rothacker J, Ramsay RG, Ciznadija D, Gras E, Neylon CB, Elwood NJ, Bouchier-Hayes D, Gibbs P, Rosenthal M, Nice EC. A novel magnetic bead-based assay with high sensitivity and selectivity for analysis of telomerase in exfoliated cells from patients with bladder and colon cancer. Electrophoresis. 2007;28:4435–46. doi: 10.1002/elps.200600829. [DOI] [PubMed] [Google Scholar]
- [127].Fizazi K, Morat L, Chauveinc L, Prapotnich D, De Crevoisier R, Escudier B, Cathelineau X, Rozet F, Vallancien G, Sabatier L, Soria JC. High detection rate of circulating tumor cells in blood of patients with prostate cancer using telomerase activity. Annals of oncology. 2007;18:518–21. doi: 10.1093/annonc/mdl419. [DOI] [PubMed] [Google Scholar]
- [128].Xu T, Lu B, Tai Y-C, Goldkorn A. A cancer detection platform which measures telomerase activity from live circulating tumor cells captured on a microfilter. Cancer research. 2010;70:6420–6. doi: 10.1158/0008-5472.CAN-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Zuo X, Xia F, Patterson A, Soh HT, Xiao Y, Plaxco KW. Two-step, PCR-free telomerase detection by using exonuclease III-aided target recycling. Chembiochem. 2011;12:2745–7. doi: 10.1002/cbic.201100592. [DOI] [PubMed] [Google Scholar]
- [130].Zhou X, Xing D, Zhu D, Jia L. Magnetic bead and nanoparticle based electrochemiluminescence amplification assay for direct and sensitive measuring of telomerase activity. Analytical chemistry. 2009;81:255–61. doi: 10.1021/ac801914b. [DOI] [PubMed] [Google Scholar]


