Summary
Trimethylation of Histone H3 at Lysine 4 (H3K4me3) is a chromatin modification known to mark the transcription start sites of active genes. Here we show that H3K4me3 domains that spread more broadly over genes in a given cell type preferentially mark genes essential for the identity and function of that cell type. Using the broadest H3K4me3 domains as a discovery tool in neural progenitor cells, we identify novel regulators of these cells. Machine learning models reveal that the broadest H3K4me3 domains represent a distinct entity, characterized by increased marks of elongation. Broadest H3K4me3 domains also have more paused polymerase at their promoters, suggesting a unique transcriptional output. Indeed, genes marked by broadest H3K4me3 domains exhibit enhanced transcriptional consistency rather than increased transcriptional levels, and perturbation of H3K4me3 breadth leads to changes in transcriptional consistency. Thus, H3K4me3 breadth contains information that could ensure transcriptional precision at key cell identity/function genes.
Introduction
Diverse cell types within multi-cellular organisms are characterized by specific transcriptional profiles. Chromatin states influence some aspects of transcription, such as expression levels or alternative splicing, and may play a role in the establishment and maintenance of gene expression programs (Bernstein et al., 2005; Dunham et al., 2012). For example, subtypes of enhancers direct the high expression of cell identity genes (Parker et al., 2013; Rada-Iglesias et al., 2011; Whyte et al., 2013). Whether other aspects of transcription are linked to cell identity and can be predicted by chromatin states is unknown.
Trimethylation of Histone H3 Lysine 4 (H3K4me3) is a major chromatin modification in eukaryotes (Santos-Rosa et al., 2002; Strahl et al., 1999). Modifiers of H3K4me3 play roles in fundamental biological processes, including embryonic development (Ingham, 1998) and stem cell biology (Ang et al., 2011; Schmitz et al., 2011). Perturbations in H3K4me3-modifying complexes lead to cancer in mammals (Shilatifard, 2012)and lifespan changes in invertebrates (Greer et al., 2010; Siebold et al., 2010). The H3K4me3 modification is associated with the promoters of actively transcribed genes (Barski et al., 2007; Guenther et al., 2007; Santos-Rosa et al., 2002), and is thought to serve as a transcriptional on/off switch (Dong et al., 2012). However, H3K4me3 can also mark poised genes (Bernstein et al., 2006), and transcription can occur in the absence of H3K4me3 (Hodl and Basler, 2012). Thus, how this mark affects specific transcriptional outputs to influence diverse cellular functions is still largely unclear.
Important information regarding specific transcriptional outputs could be contained in the spread of epigenetic modifications over a genomic locus. Repressive chromatin marks, such as H3K9me3, are deposited over broad genomic regions (~megabases) (Shah et al., 2013; Soufi et al., 2012; Zhu et al., 2013). Active chromatin marks are usually restricted to specific genomic loci, but have also been observed in broader deposits (~kilobases) (Parker et al., 2013). For example, broad depositions of H3K4me3 have been reported in embryonic stem cells (ESCs), Wilms tumor cells, hematopoietic stem cells, and hair follicle stem cells at some key regulators in these cells (Adli et al., 2010; Aiden et al., 2010; Lien et al., 2011). However, the overall biological significance of H3K4me3 breadth is unexplored.
Here we performed a meta-analysis of the H3K4me3 mark, which revealed that extremely broad H3K4me3 domains in one cell type mark cell identity/function genes in that cell type across species. Using the broadest H3K4me3 domains, we discovered novel regulators of neural progenitor cells and propose that these domains could be used to identify regulators of a particular cell type. Remarkably, genes marked by the broadest H3K4me3 domains showed increased transcriptional consistency (i.e. low transcriptional variability), and perturbation of H3K4me3 breadth led to changes in transcriptional consistency. Our study identifies a new chromatin signature linked to transcriptional consistency and cell identity, and highlights that breadth is a key component of chromatin states.
Results
Broad H3K4me3 domains mark subsets of genes in all organisms, but do not predict expression levels
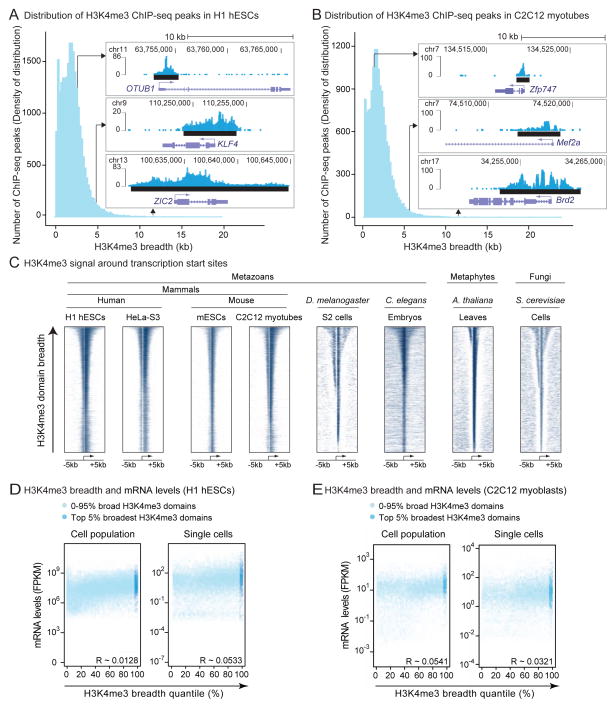
To investigate the importance of H3K4me3 breadth, we analyzed the landscape of H3K4me3 domains in >200 datasets of H3K4me3 chromatin-immunoprecipitation followed by sequencing (ChIP-seq) or microarray hybridization (ChIP-chip) in stem, differentiated, or cancer cells from 9 species (Table S1). Consistent with previous reports, H3K4me3 was mostly present in 1–2kb regions around transcription start sites (TSSs) (Figure 1A–1C). However, as previously noted in mammalian stem cells (Adli et al., 2010; Aiden et al., 2010; Lien et al., 2011), broader domains of H3K4me3 spanning up to 60kb were present in all cell types and organisms (Figure 1A–1C and S1A). Broad H3K4me3 domains were mostly found close to genes, extending both 5′ and 3′ of TSSs (Figure 1C). The genes marked by these regions were different between cell types (Figure S1B). Broad H3K4me3 domains were not associated with higher H3K4me3 ChIP intensities (Figure S1C and S1D), and were observed regardless of sequencing depth, method of chromatin fragmentation, peak-calling algorithm (Figure S1E), or H3K4me3 antibody.
Figure 1. H3K4me3 breadth is an evolutionarily conserved feature that is not predictive of expression levels.
A–B) Breadth distributions of H3K4me3 ChIP-seq peaks in H1 hESCs (A) and C2C12-derived myotubes (B) display ‘heavy right tails’, indicative of broader H3K4me3 domains than expected. Inserts: Example H3K4me3 regions in H1 hESCs or C2C12 myotubes. Black bar: ChIP-seq peaks called by MACS2.
C) H3K4me3 ChIP signal sorted by breadth at −5kb, +5kb around transcription start sites (TSSs).
D–E) mRNA levels is not a function of H3K4me3 breadth quantile at the population (left panels) or single cell (right panels) level by RNA-seq in H1 hESCs (D) or C2C12 myoblasts (E). Insert: Pearson correlation coefficients. See also Figure S1J.
We asked if broader H3K4me3 domains may be explained by underlying promoter or gene structure, or by expression levels. Broader H3K4me3 regions did not mark gene cluster regions (Figure S1F). There was no correlation between H3K4me3 domain breadth and gene length (Figure S1G), nor with the numbers of used TSSs (e.g. alternative promoters) (Figure S1H and S1I). H3K4me3 breadth did not correlate with mRNA levels (Figure 1D and 1E), even when comparing the most extreme examples (the top 5% broadest H3K4me3 domains) to the rest of the breadth distribution (Figure S1J). Thus, broad H3K4me3 domains are present in many cell types across taxa but cannot be explained as simple readouts of promoter complexity or high expression levels. These observations prompted us to investigate the biological relevance of broad H3K4me3 domains.
Broad H3K4me3 domains preferentially mark cell identity and function genes in a given cell type
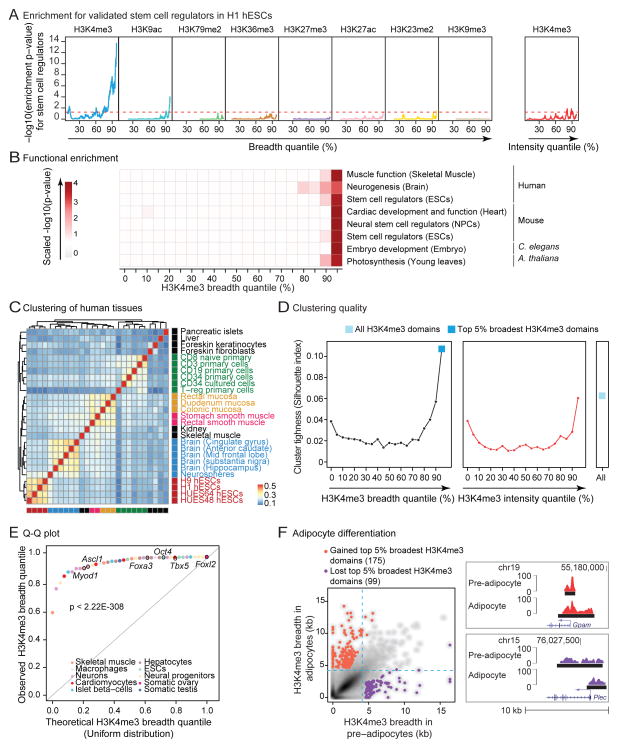
To assess if broad H3K4me3 domains mark specific gene sets, we analyzed H3K4me3 ChIP-seq datasets from >20 different cell and tissue types in mice and humans. When compared to all H3K4me3 domains, the top 5% broadest H3K4me3 domains in a particular cell/tissue type enriched for annotations linked to the ‘function’ (e.g. specialized cytoskeleton for contractile cells) and the ‘identity’ (e.g. factors required to establish that cell lineage) (Figure S2A and S2B). In human embryonic stem cells (hESCs), the set of 5% broadest H3K4me3 domains enriched the most, along the H3K4me3 breadth continuum, for validated embryonic stem cell regulators (Figure 2A and S2C; Table S2). In contrast, the broadest domains of other histone marks (e.g. H3K27ac) or the top 5% most ‘intense’ H3K4me3 domains (i.e. with the highest ChIP-seq signal normalized to peak breadth) did not strongly enrich for stem cell regulators (Figure 2A). More generally, along the breadth continuum, the top 5% broadest H3K4me3 domains in a given cell/tissue type most enriched for genes with important functions for that particular cell/tissue type across cell types and taxa (Figure 2B).
Figure 2. H3K4me3 breadth enriches for genes that are important for cell identity and function.
A) The top 5% broadest H3K4me3 domains enrich for stem cell regulators in H1 hESCs. Enrichment expressed as −log10(p-value) in Fisher’s exact test. Red dashed line: p = 0.05. See also Figure S2C.
B) The top 5% broadest H3K4me3 domains enriches for genes involved in cell/tissue function. Significance as scaled −log10(p-value) in Fisher’s exact test (see Extended Experimental Procedures and Table S2). The NPC dataset is described in Figure 3A–3C.
C) Hierarchical clustering of the top 5% broadest H3K4me3 domains from human tissues and cells based on Jaccard Index similarity. See also Figure S2D.
D) Measure of cluster tightness (Silhouette index) from different sets of H3K4me3 domains in human tissues. See also Figure S2E–S2G.
E) Quantile by quantile (Q-Q) plot of the quantile ranks of H3K4me3 domains marking known cell identity or reprogramming genes in tissue of relevance (Table S3). Significance in Kolmogorov-Smirnov test.
F) H3K4me3 breadth is remodeled at a subset of loci during differentiation. Left panel: Scatterplots of H3K4me3 breadth for adipogenesis (3T3L1 in pre- vs. mature adipocyte). Right panel: Remodeled top 5% broadest H3K4me3 domains between pre- and mature adipocytes. See also Figure S2J.
We next asked if the broadest H3K4me3 domains could separate cells or tissues by lineage better than the complete set of H3K4me3 domains (Zhu et al., 2013). The top 5% H3K4me3 broadest domains indeed discriminated cells or tissues according to their lineage in human and mouse (Figure 2C and S2D). Clustering quality measures showed that the set of the 5% broadest H3K4me3 domains outperformed all other 5% subsets of H3K4me3 domains (binned by breadth or intensity) as well as the complete set of H3K4me3 domains (Figure 2D and S2E–S2G). The broadest H3K4me3 domains could also distinguish fibroblasts from fully and even partially reprogrammed iPSCs (Figure S2H and S2I). Reciprocally, genes encoding factors known to be critical for cell identity/fate (e.g. Myod1 in skeletal muscle) had significantly broader H3K4me3 domains in the relevant tissue than expected by chance (p < 2.22×10−308 in Kolmogorov-Smirnov test; Figure 2E and Table S3).
A prediction from these observations is that a subset of the broadest H3K4me3 domains should be remodeled as cells differentiate along a lineage. Indeed, some top 5% broadest H3K4me3 domains are remodeled between progenitors and derived progeny (e.g. adipocyte progenitors and adipocytes) (Figure 2F). Genes that gained top 5% broadest H3K4me3 domains during differentiation were enriched for differentiated cell functions (e.g. lipid homeostasis), whereas genes that lost top 5% broadest H3K4me3 domains were enriched for progenitor cell functions (e.g. cell proliferation) (Figure S2J). A subset of genes was marked by broad H3K4me3 domains in most cells (e.g. Foxo3), and those marked genes tended to be shared between mouse and human (p < 1.4×10−75 in Fisher’s exact test), perhaps representing genes that are key for basic cell function across tissues.
Next, we compared the top 5% broadest H3K4me3 domains to another signature linked to cell identity, ‘super enhancers’ (Whyte et al., 2013). Genes marked by the top 5% broadest H3K4me3 domains were distinct from genes assigned to super enhancers (Figure S2K). The top 5% broadest H3K4me3 domains and super enhancers were similarly effective at predicting validated stem cell regulators in mESCs (Figure S2L), but the broadest H3K4me3 domains discriminated better between lineages (Figure S2M). Additionally, the breadth of H3K4me3 domains provides a ranking system to further enrich for key regulators (Figure S2L). Thus, the top 5% broadest H3K4me3 domains have the potential to identify genes with functions in a given cell type or lineage.
Broad H3K4me3 domains as a discovery tool for novel regulators of neural progenitor cells
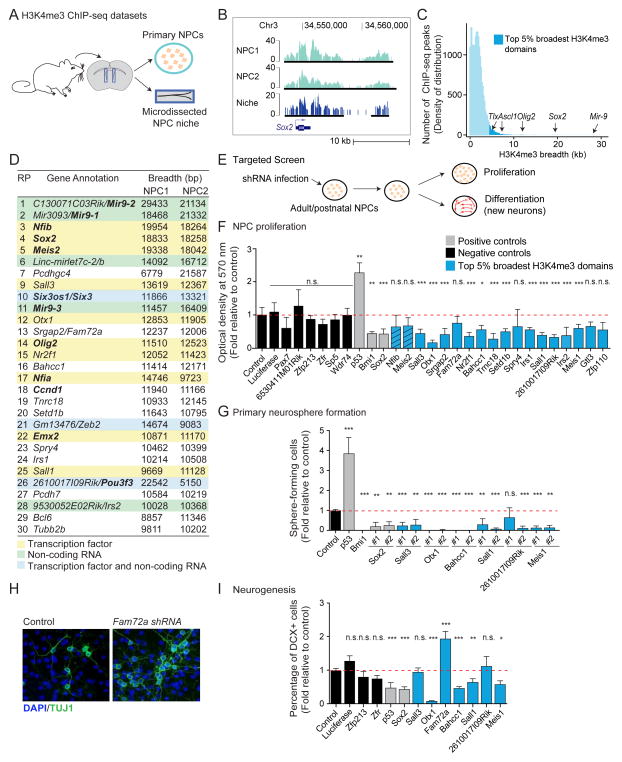
We tested whether the top 5% broadest H3K4me3 domains could be used to discover novel regulators in a particular cell type. We chose adult neural progenitor cells (NPCs) because these cells give rise to all brain cell types and may be a source of cells for regenerative therapies (Bonaguidi et al., 2011; Doetsch et al., 1999; Lujan et al., 2012). We generated H3K4me3 ChIP-seq datasets from NPC cultures isolated from adult mice and from the microdissected brain region that hosts these cells in vivo (Figure 3A–3C and S3A).
Figure 3. The top 5% broadest H3K4me3 domains can be used as a discovery tool to identify new regulators of neural progenitor cells.
In all panels: n.s. not significant; * p < 0.05; ** p < 0.01; *** p < 0.005 in a Wilcoxon test against control with Bonferroni correction for multiple testing.
A) Experimental design for H3K4me3 ChIP-seq datasets in primary cultures of neural progenitors (NPCs) and microdissected niche (subventricular zone).
B) H3K4me3 ChIP-seq peaks at a known NPC regulator in independent NPC primary cultures and in the NPC niche. Black bars: ChIP-seq peaks called by MACS2.
C) Distribution of H3K4me3 ChIP-seq peaks as a function of their breadth in NPCs reveals that known NPC regulators are marked by broad H3K4me3 domains.
D) Genes associated to top 30 broadest H3K4me3 domains in NPCs. Domains ranked by decreasing H3K4me3 breadth. Known regulators of NPCs in bold. RP: rank of rank product.
E) Experimental design to test the role of genes marked by top 5% broadest H3K4me3 domains in NPC proliferation and neurogenesis.
F) Proliferation capacity as normalized MTT optical density relative to control. Mean + SD of 2 independent experiments conducted in triplicate. Hashed blue bars: genes whose role in NPCs was discovered while this study was in preparation(Agoston et al., 2014; Ninkovic et al., 2013). See also Figure S3D.
G) Proliferation capacity as percentage of infected cells that formed primary neurospheres relative to control. Mean + SD of at least 2 independent experiments conducted in triplicate.
H) Images of new neurons upon Fam72a knock-down. Green: TUJ1 (neurons). Blue: DAPI (nuclei).
I) Neurogenesis measured by percentage of DCX+ cells (new neurons) normalized to control. Mean + SEM of at least 2 independent experiments conducted in triplicate. See also Figure S3F.
As seen in other cell types, genes marked by the top 5% broadest H3K4me3 domains in adult NPCs were enriched for known regulators based on a literature-curated list (Figure 2B, 3C and S3B; Table S2). A large fraction of broad H3K4me3 domains in NPCs mark genes encoding transcription factors, non-coding RNAs or both (Figure 3D). Interestingly, the broadest H3K4me3 domains in NPCs marked genes that had not been previously implicated in the maintenance of the neuronal lineage (e.g. Bahcc1, Fam72a, 2610017I09Rik; Figure 3D; Table S4) and genes that, while involved in brain development, have not been studied in adult/postnatal NPCs (e.g. Otx1)(Sakurai et al., 2010).
Using a lentiviral-based RNA interference (shRNA) approach, we knocked-down candidate genes marked by the broadest H3K4me3 domains in primary NPCs, and we quantified the ability of NPCs to proliferate and generate new neurons (neurogenesis) (Figure 3E and S3C). Knock-down of known regulators p53 and Sox2 had the expected effect on NPC proliferation and neurogenesis (Figure 3F, 3G, and 3I) (Wang et al., 2011). Knocking-down two-thirds of genes marked by broad H3K4me3 domains (12/18) significantly decreased NPC proliferation (Figure 3F, 3G, S3D, and S3E), whereas knock-down of control genes (i.e. genes with shorter or no H3K4me3 domains) did not significantly affect NPC proliferation. Knock-down of a subset of genes marked by the broadest H3K4me3 domains also decreased neurogenesis, suggesting that they are necessary for the differentiation ability of NPCs (Figure 3H, 3I, and S3F). These include the genes encoding the transcription factor SALL1, the homeobox transcription factor OTX1, and BAHCC1, a protein with a bromo-adjacent homology domain that can bind acetylated histones (Kuo et al., 2012). In contrast, knock-down of Fam72a, a gene misregulated in an Alzheimer’s mouse model (Nehar et al., 2009), led to an increase in neurogenesis, suggesting FAM72A may function to restrain differentiation in the progenitor state (Figure 3H, 3I, and S3F). Thus, this targeted screen allowed us to identify previously uncharacterized genes involved in NPC self-renewal and/or neurogenesis and to confirm that the top 5% broadest H3K4me3 domains can be used to discover genes that regulate the biology of a given cell type. To facilitate the use of H3K4me3 breadth as a discovery tool in other systems, we created a searchable database accessible at http://bddb.stanford.edu (Figure S3G and Table S5).
The top 5% broadest H3K4me3 domains represent a distinct subclass of H3K4me3 domains
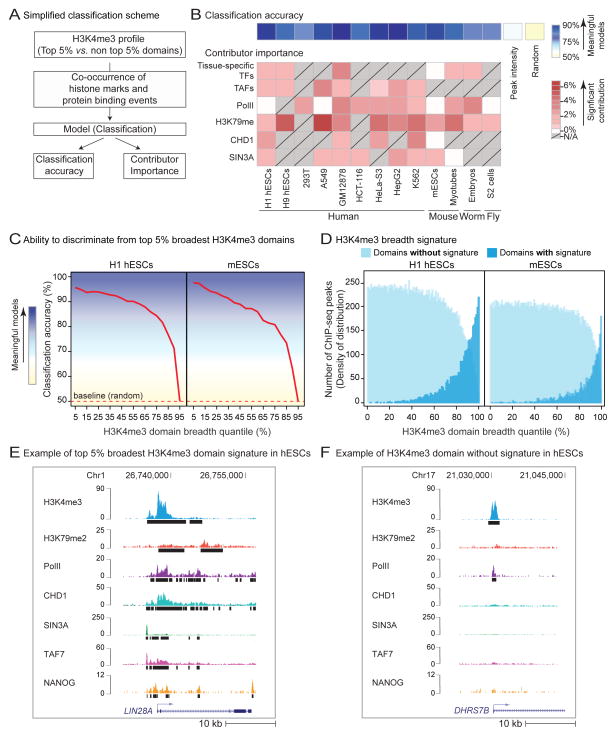
Considering their association with biological function, we asked whether the top 5% broadest H3K4me3 domains constitute a distinct entity with specific (epi-)genomic characteristics. We used 4 different classification algorithms – a family of machine-learning algorithms that can learn to separate entities based on discriminating features. We built classification models based on the co-occurrence of protein binding profiles and chromatin modifications with H3K4me3 domains in 13 cell types and organisms (Figure 4A, S4A, and Table S1; see Extended Experimental Procedures). All 4 algorithms were able to discriminate with high accuracy (>75%) the top 5% broadest H3K4me3 domains apart from random sets of the same number of H3K4me3 domains from the rest of the distribution (Figure 4B and S4B–S4D). In contrast, models built from the top 5% most intense H3K4me3 peaks lacked discriminative power (Figure 4B and S4D). The discriminative power of the classification models remained high when the top 5% H3K4me3 broadest domains were compared to domains up to the 85th percentile of the breadth distribution, and then sharply dropped (Figure 4C). The presence of this inflexion point supports the notion that a specific set of features distinguishes the broadest H3K4me3 domains from the rest of the distribution. Reciprocally, H3K4me3 domains sharing the discriminating (epi-)genomic characteristics of the broadest H3K4me3 domains were broader than expected by chance (Figure 4D). Together, these results suggest that specific combinations of histone marks and protein binding distinguish the top 5% broadest H3K4me3 domains from the rest of the breadth continuum.
Figure 4. The broadest H3K4me3 domains are characterized by a specific epigenomic signature.
A) Simplified scheme of computational modeling. See also Figure S4A.
B) Average classification accuracy and most important contributors associated to top 5% broadest H3K4me3 domains identified by Random Forest models in 13 cell types and organisms. Contributors for which no data was available in grey with diagonal lines. Tissue-specific transcription factors (TFs) refer to: NANOG (H1 hESCs), SMAD2/3 (H9 hESCs), STAT5 (GM12878), NANOG (mESCs), MYOG/MYOD (Myotubes), LIN-13 (C. elegans embryos). See also Figure S4B–S4E.
C) Accuracy of progressive classifications in H1 hESCs and mESCs. Classifications performed between top 5% broadest H3K4me3 domains and other 5% quantile subsets along the breadth continuum. The accuracy of progressive classifications reflects the ability to discriminate domains of that quantile from top 5% broadest H3K4me3 domains.
D) Breadth of H3K4me3 domains ‘with/without the top 5% broadest H3K4me3 domain signature’ in H1 hESCs and in mESCs.
E–F) Example domains with/without signature in H1 hESCs. Black bars: peaks called by MACS2.
We next asked which (epi-)genomic characteristics most contributed to the ability of models to accurately discriminate the top 5% broadest H3K4me3 domains from the rest of the distribution (Figure S4E). Consistent with the tissue-specific genomic deposition pattern of H3K4me3 breadth, we found that lineage-specific transcription factors (TFs), such as MYOD/MYOG in myotubes, were recurrent important contributors to the models (Figure 4B). The enrichment of lineage-specific TFs at broad H3K4me3 domains was observed in multiple other cell types (Figure S4G). Other important recurrent contributors included the transcription initiation complex (e.g. TAFs and RNA Polymerase II (PolII)), marks of transcriptional elongation (e.g. H3K79me2), the H3K4me3 reader CHD1, which is involved in nucleosome repositioning at transcribed genes (Smolle et al., 2012), and the transcriptional repressor SIN3A, which constrains spurious transcription in yeast (Carrozza et al., 2005) (Figure 4B, 4E, 4F, S4E, and S4F). These contributors point towards a link between extreme H3K4me3 breadth and specific regulation of transcriptional initiation and elongation – a surprising finding given that H3K4me3 breadth is not correlated with expression level (Figure 1E and S1H) and that the models were built to discriminate the top 5% broadest H3K4me3 domains from the rest of H3K4me3 domains, all of which are thought to be transcriptionally active.
The top 5% broadest H3K4me3 domains exhibit unique regulation of PolII pausing and elongation
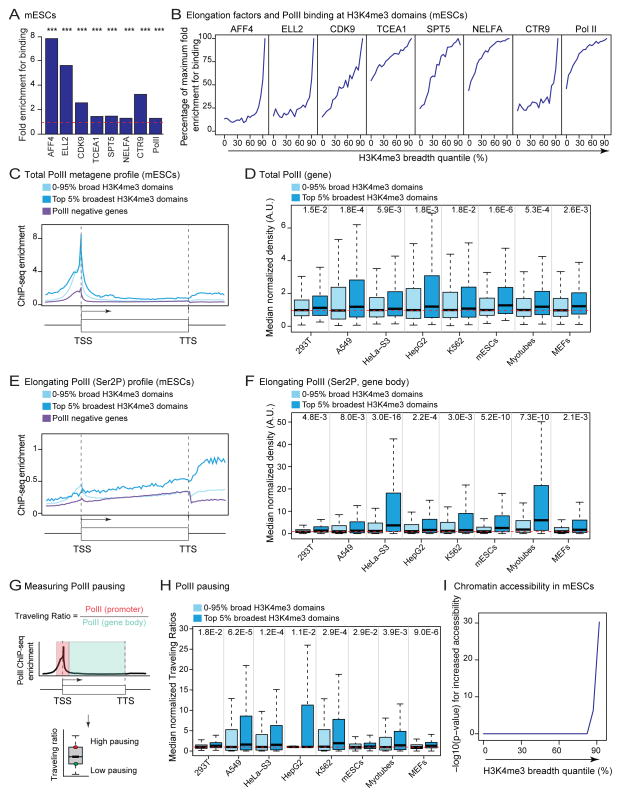
We examined whether the top 5% broadest H3K4me3 domains had unique features of transcriptional regulation. Increasing H3K4me3 domain breadth was associated with enriched binding of positive regulators of transcription elongation (e.g. Super Elongation Complex, P-TEFb) (p < 1×10−4 in permutation test; Figure 5A, 5B, and S5A–S5E) (Luo et al., 2012; Sims et al., 2004). The top 5% broadest H3K4me3 domains were also the most likely to be bound by PolII (Figure 5A, 5B, and S5B–S5E), despite similar numbers of PolII binding sites (Figure S1H). The top 5% broadest H3K4me3 domains are associated with overall higher levels of PolII (p < 4.7×10−2 in one-sample Wilcoxon tests; Figure 5D and S5G). Serine 2-phosphorylated PolII (Ser2P), which is characteristic of productive elongation (Sims et al., 2004), was significantly higher at genes marked by top 5% broadest H3K4me3 domains (p < 8.0×10−3 in one-sample Wilcoxon tests; Figure 5E–5F). Somewhat surprisingly, Serine 5-phosphorylated PolII (Ser5P), which is associated with transcriptional initiation, was also increased at genes marked by top 5% broadest H3K4me3 domains (p < 8.0×10−3 in one-sample Wilcoxon tests; Figure S5H–S5I). This observation, coupled with the association of the top 5% broadest H3K4me3 domains to factors that promote PolII pausing (e.g. NELFA) (Sims et al., 2004) (Figure 5A, 5B and S5A), suggests that broad H3K4me3 domains are associated with PolII pausing. Indeed, genes marked by the top 5% broadest H3K4me3 domains had significantly higher Traveling Ratios, a measure of PolII pausing (p < 2.9×10−2 in one-sample Wilcoxon tests; Figure 5G, 5H and S5J). While it may seem contradictory that the same class of domains would associate to increased elongation and PolII pausing, this phenomenon may represent a footprint of steady PolII release from a heavily pre-loaded promoter. Increased PolII pausing at proximal promoters has been suggested to promote chromatin accessibility (Gilchrist et al., 2010). Indeed, promoters of genes coated by the top 5% broadest H3K4me3 domains were more accessible than promoters marked by shorter H3K4me3 domains, as assessed by DNAse-seq (p < 4.2×10−20 in one-sample Wilcoxon tests; Figure 5I and S5K) or ATAC-seq (Buenrostro et al., 2013) (p = 4.9×10−156 in one-sample Wilcoxon tests; Figure S5L). Thus, the top 5% broadest H3K4me3 domains are associated with unique regulation of PolII initiation and elongation and increased chromatin accessibility, suggesting a specific transcriptional output.
Figure 5. The top 5% broadest H3K4me3 domains are associated with marks of transcriptional elongation and PolII pausing.
Indicated p-values for top 5% broadest H3K4me3 domain associated-genes calculated in one-sided one-sample Wilcoxon tests against expected genome-wide value from 10,000 random samplings (red dashed line).
A) Differential binding of components of the elongation machinery to top 5% vs. non top 5% broadest H3K4me3 domains in mESCs. p-values from permutation test.
B) Enrichments for components of the elongation machinery in mESCs expressed as a percentage of the maximal binding enrichment that can be observed along the H3K4me3 breadth continuum (see A for enrichment at top 5% broadest H3K4me3 domains). See also Figure S5A–S5E.
C) Mean ChIP-seq enrichment of Total PolII in mESCs. TSS: transcription start site; TTS: transcription termination site.
D) Normalized PolII ChIP-seq density over the proximal promoter and gene body. Comparisons of top 5% broadest H3K4me3 domains against the rest of the distribution also significant in one-sided Wilcoxon tests (9.6×10−10 < p < 5.4×10−3) (continued in Figure S5G).
E) Mean ChIP-seq enrichment of elongating PolII (Ser2P) in mESCs. TSS: transcription start site; TTS: transcription termination site.
F) Normalized elongating PolII (Ser2P) ChIP-seq density over gene bodies. Comparisons of top 5% broadest H3K4me3 domains against the rest of the distribution also significant in one-sided Wilcoxon tests (7.3×10−23 < p < 2.6×10−5) (continued in Figure S5J).
G) Measure of PolII pausing. Traveling Ratio is defined as background subtracted ChIP-seq density value of PolII at the promoter vs. gene body.
H) Normalized Traveling Ratios. Comparisons of top 5% broadest H3K4me3 domains against the rest of the distribution also significant in one-sided Wilcoxon tests (1.8×10−9 < p < 4.7×10−2) (continued in Figure S5K).
I) Significance for increased chromatin accessibility in mESCs against expected genome-wide value shown as −log10(p-value) in one-sided Wilcoxon tests.
The top 5% broadest H3K4me3 domains are associated with increased transcriptional consistency
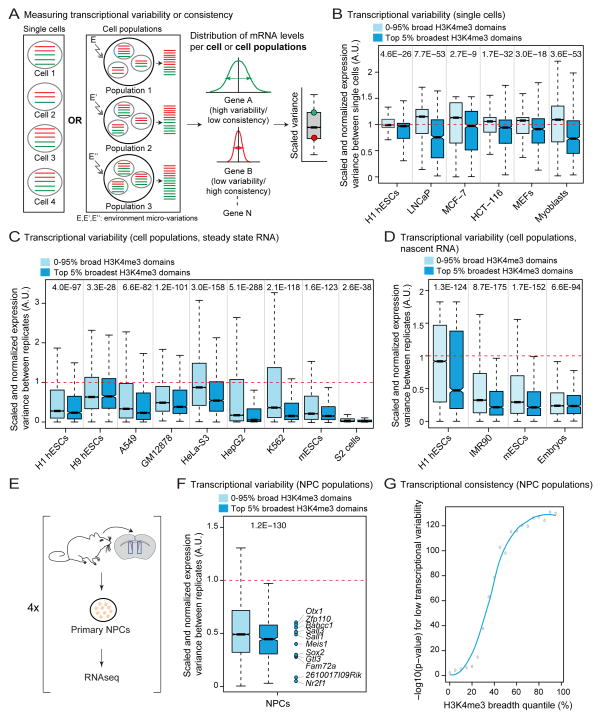
Increased PolII pausing and elongation have been linked to transcriptional consistency (e.g. lower transcriptional variability, or “transcriptional noise” in single cells) (Boettiger et al., 2011; Lagha et al., 2013). In addition, increased chromatin accessibility at promoters may facilitate transcriptional consistency, in part by minimizing transcriptional bursting (Field et al., 2008). We asked whether the top 5% broadest H3K4me3 domains are associated with transcriptional consistency (Figure 6A). To measure transcriptional consistency in single cells, we calculated the variance in expression relative to expression level for each gene in single cell RNA-seq datasets (Table S6). Remarkably, genes marked by the top 5% broadest H3K4me3 domains had reduced transcriptional variability in single cell RNA-seq experiments (p < 4.6×10−26 in one-sample Wilcoxon tests; Figure 6B). Transcriptional consistency significantly increased with H3K4me3 breadth, but not with peak intensity (Figure S6A).
Figure 6. H3K4me3 breadth is associated with transcriptional consistency.
Indicated p-values for top 5% broadest H3K4me3 domain associated-genes calculated using one-sided one-sample Wilcoxon tests against expected transcriptome-wide value from 10,000 random samplings (red dashed line).
A) Transcriptional consistency/variability at the level of single cells or cell populations is defined as variance of expression levels scaled to expression levels (i.e. scaled variance).
B) Transcriptional variability at the single cell level (steady state mRNA). Comparisons of top 5% broadest H3K4me3 domains against the rest of the distribution also significant in Wilcoxon tests (2.9×10−93 < p < 2.4×10−16). See also Figure S6A.
C) Transcriptional variability at the cell population level (steady state mRNA). Comparisons of top 5% broadest H3K4me3 domains against the rest of the distribution also significant in Wilcoxon tests (1.5×10−170 < p < 3.9×10−3)(Continued in Figure S6B).
D) Transcriptional variability at the cell population level (nascent mRNA by GRO-seq). Comparisons of top 5% broadest H3K4me3 domains against the rest of the distribution also significant in Wilcoxon tests (1.8×10−51 < p < 4.4×10−20). See also Figure S6C.
E) Experimental design for RNA-seq datasets in primary NPCs cultures.
F) Transcriptional variability at the cell population level in adult NPCs (steady state mRNA). Comparison of top 5% broadest H3K4me3 domains against the rest of the distribution also significant in a Wilcoxon test (p = 3.7×10−12).
G) Significance for lower transcriptional variability in adult NPCs against expected transcriptome-wide value expressed as −log10(p-value) in one-sided Wilcoxon tests.
To expand our analysis, we tested transcriptional consistency in datasets generated from cell populations. A gene with stable transcription levels will tend to have consistent expression values between biological replicates of cell populations despite microvariations in the environment (Figure 6A) (Dong et al., 2011). Similar to our observations at the single cell level, analysis of 15 different cell population transcriptome datasets (Table S6) showed that genes marked by the top 5% broadest H3K4me3 domains had reduced transcriptional variability (p < 4×10−3 in one-sample Wilcoxon tests; Figure 6C and S6B–S6D). The broadest H3K4me3 domains were also associated with increased transcriptional consistency when considering only nascent RNA levels in cell populations by GRO-seq (General Run On sequencing) (p < 6.6×10−94 in one-sample Wilcoxon tests; Figure 6D, S6E and Table S6), indicating that transcriptional consistency at the broadest H3K4me3 domains does not result from increased mRNA stability. By performing RNA-seq, we confirmed that genes marked by the top 5% broadest H3K4me3 domains in primary NPC cultures were more transcriptionally consistent in these cells (p = 1.2×10−130 in a one-sample Wilcoxon test; Figure 6E, 6F and 7E). Transcriptional consistency increased as a function of H3K4me3 domain breadth in NPCs (Figure 6G). Together, our results indicate that the broadest H3K4me3 domains are linked to transcriptional consistency.
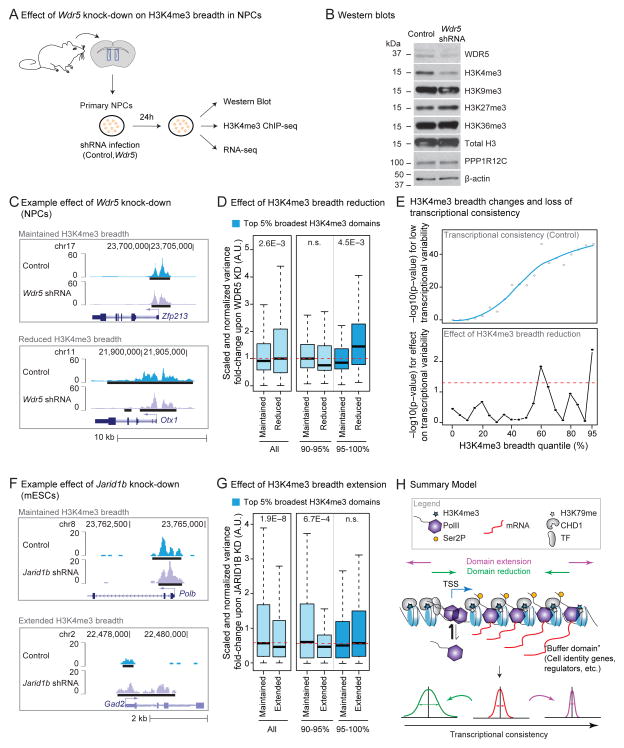
Figure 7. Experimental perturbation of H3K4me3 breadth results in changes to transcriptional consistency.
A) Experimental design to study the effect of knocking-down Wdr5 in primary NPC cultures.
B) Western Blot analysis of NPCs treated in control (empty vector) or Wdr5 knock-down after 24h of infection. See also Figure S7B.
C) Examples of H3K4me3 peaks in control (empty vector) and Wdr5 knock-down in NPCs after 24h of infection.
D) Reduction of H3K4me3 breadth upon 24h Wdr5 knock-down is linked to increased transcriptional variability in NPCs. Variability was measured between 3 biological replicates at genes whose H3K4me3 domains were maintained or reduced upon Wdr5 knock-down. Red dashed line: expected transcriptome-wide value. p-values between genes with maintained vs. reduced breadth in Wilcoxon tests.
E) H3K4me3 breadth remodeling upon Wdr5 knock-down and loss of transcriptional consistency in NPCs. Upper panel: −log10(p-value) in one-sided Wilcoxon test for lower transcriptional variability in control infected NPCs than expected transcriptome-wide value. Lower panel: −log10(p-value) in one-sided Wilcoxon tests for increased variability of genes losing H3K4me3 breadth vs. genes of the same original H3K4me3 quantile with maintained breadth. Red dashed line: p = 0.05.
F) Examples of H3K4me3 peaks in control (scramble) and Jarid1b knock-down in mESCs after 48h of infection.
G) Gain of H3K4me3 breadth upon Jarid1b knock-down is linked to increased transcriptional consistency in mESCs after 48h of infection. Variability between 3 biological replicates at genes whose H3K4me3 domains were maintained vs. extended upon Jarid1b knock-down. Red dashed line: expected transcriptome-wide value. p-values between genes with maintained or extended breadth using Wilcoxon tests.
H) Summary model. Broad H3K4me3 domains extend 5′ and 3′ of TSSs and mark genes important for cell identity/function and genes with increased transcriptional consistency. Broad H3K4me3 domains may promote chromatin accessibility, thereby allowing efficient PolII loading and elongation. The mechanism responsible for the deposition of broad H3K4me3 domains is unknown but may involve tissue-specific transcription factors and the elongation machinery in a positive feedback loop. These domains may help ‘buffer’ important cell lineage/function genes against environmental fluctuation and can serve as discovery tool for such genes.
Perturbation of H3K4me3 breadth leads to changes in transcriptional consistency
We next tested whether perturbation of H3K4me3 breadth could result in changes in transcriptional consistency. H3K4me3 is deposited by members of the COMPASS/Trithorax/Trithorax-related family of methyltransferase complexes and removed by the JARID1 family of demethylases (Black et al., 2012). Components of the H3K4me3 regulatory machinery were enriched to bind loci marked by the broadest H3K4me3 domains (Figure S7A), suggesting that these regions may require increased presence of regulators for their deposition/maintenance.
We used the adult NPC model to test if decreasing H3K4me3 breadth could affect transcriptional consistency (Figure 7A). Because WDR5 is an essential scaffolding subunit shared by all COMPASS/Trithorax-like complexes (Trievel and Shilatifard, 2009), we reasoned that its knock-down might be more efficient at reducing H3K4me3 breadth than that of individual methyltransferases. We tested the effect of short term (24h) Wdr5 knock-down on H3K4me3 levels and breadth in NPCs (Figure 7A). By 24h of knock-down, WDR5 RNA and protein levels were reduced (Figure 7B, S7B and S7C) without major consequences on cell viability (Figure S7D). Short-term WDR5 depletion led to a global reduction of H3K4me3 levels in NPCs, but did not significantly affect methylation of other histone residues (Figure 7B). In line with the global decrease of H3K4me3, there was a decrease in the signal-to-noise ratio of H3K4me3 ChIP-seq from Wdr5 shRNA treated cells compared to control cells (Figure S7E and S7F). After accounting for the overall loss of H3K4me3 ChIP-seq intensity (see Extended Experimental Procedures; Figure S7E and S7F), we observed that H3K4me3 breadth decreased genome-wide in response to WDR5 depletion (Figure 7C and S7G). The mean proportion of H3K4me3 domains maintaining or losing breadth was fairly constant above the 30th percentile of breadth (Figure S7G). Thus, a short-term Wdr5 knock-down provides a way to compare genes at which H3K4me3 breadth is reduced or maintained (Figure 7C). To evaluate the impact of H3K4me3 breadth reduction on transcriptional consistency, we generated RNA-seq datasets from replicates of NPC cultures infected with control or Wdr5 shRNA (Figure 7A and S7I). In line with our results in uninfected NPCs (Figure 6G), H3K4me3 breadth enriched for increased transcriptional consistency in control infected NPCs (Figure 7E, top). Upon short-term Wdr5 knock-down, only a few genes were differentially expressed (Figure S7H). In contrast, genes with reduced H3K4me3 breadth (<50% of original breadth) upon Wdr5 knock-down exhibited increased transcriptional variability compared to genes with maintained breadth (Figure 7D). Reduction of H3K4me3 breadth was most significantly associated with loss of transcriptional consistency at the top 5% broadest H3K4me3 domains (Figure 7D and 7E, bottom). Thus, reduction of H3K4me3 breadth is associated with a decrease in transcriptional consistency.
To test the effect of H3K4me3 breadth extension on transcriptional consistency, we used H3K4me3 ChIP-seq datasets and expression microarrays obtained in mESCs following short term (48h) knock-down of the H3K4me3 demethylase Jarid1b/Kdm5b (Schmitz et al., 2011). Depletion of JARID1B induces a substantial increase in H3K4me3 levels in mESCs (Schmitz et al., 2011). After accounting for changes in H3K4me3 ChIP-seq intensity, we observed that H3K4me3 breadth tended to increase genome-wide in response to Jarid1b knock-down (Figure 7F, S7K and S7L). Few genes were differentially expressed after short-term knock-down of Jarid1b (Figure S7K and S7M). However, genes that gained H3K4me3 breadth (>2x their original breadth) upon Jarid1b knock-down had a significant decrease in transcriptional variability compared against those that maintained H3K4me3 breadth (Figure 7G and S7L). There was no further gain of transcriptional consistency upon Jarid1b knock-down for genes marked by the top 5% broadest domains (Figure 7G), maybe because genes marked by these domains have already reached their lowest level of variability. The decreased transcriptional variability associated with the extension of H3K4me3 breadth in mESCs mirrors our findings that reduced H3K4me3 breadth leads to increased transcriptional variability in NPCs. Together, these results are consistent with the idea that the broadest H3K4me3 domains may promote transcriptional consistency at key cell identity/function genes (Figure 7H).
Discussion
Through meta-analysis of high-throughput genomics data, construction of machine-learning models and experimental validation of predictions, we uncover the existence of a subclass of H3K4me3 domains that preferentially marks genes important for cell identity and function. Our study also identifies for the first time a genome-wide link between a specific chromatin landscape, H3K4me3 breadth, and low transcriptional variability. We propose to refer to this subclass of broad H3K4me3 domains as ‘buffer domains’, as they may help prevent spurious bursts of transcription.
Buffer domains as a cell identity signature and discovery tool
Several epigenetic signatures have been linked to cell identity or developmental genes: active enhancers (regions of enrichment for H3K4me1, H3K27ac and p300 binding) (Heintzman et al., 2009; Rada-Iglesias et al., 2011), super enhancers (regions intensely bound by mediator and cell-specific transcription factors) (Whyte et al., 2013), stretch enhancers (extended enhancer regions) (Parker et al., 2013) and DNA methylation canyons or valleys (large hypomethylated regions) (Jeong et al., 2014; Xie et al., 2013). While there is some overlap in the genes marked by buffer domains and other signatures, many genes are uniquely captured by each of these signatures (Figure S2K; BAB, DU and AB, unpublished observations). Thus, complementary strategies of transcriptional regulation may control cell identity, with buffer domains tuning transcriptional consistency and super/stretch enhancers promoting high expression (Parker et al., 2013; Whyte et al., 2013).
Here we test the potential of the buffer domain signature to assist in the discovery of new cell identity/function genes in NPCs. We identify new regulators of NPC proliferation and neurogenesis, such as the putative chromatin reader Bahcc1 or the non-coding RNA 2610017I09Rik. Although we have not assessed the function of all genes marked by buffer domains in NPCs, our study illustrates the potential of this signature to identify new regulators and annotate poorly characterized genes. As reprogramming factors tend to be marked by buffer domains, this signature may also help find candidates for reprogramming cells into a cell type of interest, a task facilitated by our web-accessible database (http://bddb.stanford.edu). The predictive value of buffer domains could also be used to identify genes in contexts for which few functional genes have been identified so far, such as aging or neurological disorders.
A specific transcriptional output linked to H3K4me3 breadth
There is a general consensus that H3K4me3 plays a role in transcriptional initiation (Lauberth et al., 2013). However, recent work in Drosophila suggests that H3K4me3 may be dispensable for the expression of some genes (Hodl and Basler, 2012). Here we find that short-term manipulation of H3K4me3 breadth impacts transcriptional consistency, which suggests that H3K4me3 may ensure the robustness of transcriptional outputs. Interestingly, deficiencies in histone acetyltransferases or chromatin remodelers lead to increased transcriptional noise in yeast (Hansen and O’Shea, 2013; Raser and O’Shea, 2004; Weinberger et al., 2012), supporting the idea that chromatin states are critical for transcriptional precision.
The mechanisms leading to the deposition/maintenance of broad H3K4me3 domains and their connection with PolII regulation and transcriptional consistency are still unknown. Subunits of H3K4me3-depositing complexes are required for PolII priming (Perez-Lluch et al., 2011) and may promote PolII release into productive elongation (Ardehali et al., 2011). Conversely, PAF1, which associates with elongating PolII, can recruit methyltransferase complexes responsible for the deposition of both H3K4me3 and H3K79me2 (Krogan et al., 2003), an elongation mark enriched at the broadest H3K4me3 domains. Based on these previous findings and our own, we propose a model where, at some key regulatory genes, a positive feedback mechanism may link robust initiation and elongation of PolII with sustained H3K4me3 deposition over broad regions (Figure 7H). Sustained elongation may first allow increased recruitment of H3K4me3-depositing complexes to specific genes. In turn, this recruitment would promote the broadening of H3K4me3 deposits and subsequent recruitment of the H3K4me3 reader CHD1, which facilitates the passage of elongating PolII (Smolle et al., 2012). This loop could work towards ensuring transcriptional consistency at specific loci. How these loci get selected is unclear, though tissue-specific transcription factors, which are enriched at the broadest H3K4me3 domains, might be involved. As transcription factor dynamics has been associated with transcriptional noise (Hansen and O’Shea, 2013), there may also exist an interplay between transcription factor binding and H3K4me3 breadth in tuning transcriptional consistency.
Biological implications of regulating transcriptional consistency
Control of transcriptional noise, through PolII pausing or nucleosome positioning, has emerged as a new layer of complexity in biological processes (Levine, 2011; Raser and O’Shea, 2004). In metazoans, a dual requirement for transcriptional consistency has been observed. Reduction of transcription stochasticity is required for embryo patterning (Lagha et al., 2013; Raj et al., 2010), yet increased transcriptional variability may be permissive for cell fate decisions (Balazsi et al., 2011). While the importance of transcriptional consistency post-development is not fully understood, transcriptional noise increases with age in cardiomyocytes (Bahar et al., 2006), but not in hematopoietic stem cells (Warren et al., 2007). The prevalence of buffer domains suggests that control of transcriptional consistency is critical in most cells and organisms. This conserved signature may ensure consistent expression of key genes involved in identity maintenance against variations of a fluctuating environment, a feature that may be lost during aging or disease.
Experimental procedures
H3K4me3 ChIP-seq analysis
Publicly available datasets were obtained from ENCODE (Dunham et al., 2012), Roadmap Epigenomics (Zhu et al., 2013), GEO datasets, ArrayExpress/EBI or Sequence Read Archive (SRA) (Table S1). ChIP-seq experiments in NPCs were performed as previously described (Webb et al., 2013), using H3K4me3 antibody (Active Motif, antibody 39159). Libraries were generated according to Illumina instructions and sequenced on an Illumina GAII sequencer. Reads were mapped to reference genomes using bowtie0.12.7 (Langmead et al., 2009). ChIP-seq peaks were called using MACS2.08 (Feng et al., 2012) with the “—broad” option for histone marks. Peaks were assigned to the gene with the closest TSS.
Proliferation and neurogenesis in primary NPC culture
Adult/post-natal mouse NPCs were isolated by microdissection of the subventricular zone and maintained as non-adherent neurospheres as described previously (Webb et al., 2013). For proliferation or neurogenesis assays, adherent adult (proliferation) or postnatal NPCs (neurogenesis) were transduced with a 30% lentiviral dilution and selected using 0.5 μg/ml of puromycin (Invivogen). For proliferation, cell growth was quantified by MTT (Molecular Probes) 4 days following infection. For neurogenesis, cells were plated to account for proliferation differences, then switched to NPC differentiation media for 4 days. The number of new neurons was assessed by Doublecortin (DCX) staining (Santa Cruz, sc-8066).
Transcriptional variability
Transcriptional variability was assessed using microarray or RNA-seq datasets with ≥3 replicates generated in conditions matching H3K4me3 ChIP datasets. The expression variance per gene across replicates was scaled to the expression level of the gene (i.e. normalized to maximum level observed for that gene). Details are in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Broad H3K4me3 domains mark cell identity genes and can be used as a discovery tool
Broad H3K4me3 domains are a distinct entity defined by specific PolII regulation
Genes marked by broad H3K4me3 domains have increased transcriptional consistency
Perturbation of H3K4me3 breadth leads to changes in transcriptional consistency
Acknowledgments
We especially thank Joanna Wysocka and Michael Bassik for stimulating scientific discussion on this research. We thank Sivan Bercovici, Andreas Beyer, Will Greenleaf, Eric Greer, Courtney Hodges, Jonathan Pritchard, Julien Sage, Dario Valenzano, Anton Valouev, Reiner Veitia and members of the Brunet lab for critical discussion and feedback on the manuscript. This work was supported by NIH DP1 AG044848 (A.B.), P01 GM099130 (A.B. and M.P.S), P01 AG036695 (A.B. and T.A.R.), R01 GM103787 (J.C.B.), the Glenn Laboratories for the Biology of Aging (A.B. and T.A.R.), the Dean’s fellowship at Stanford University (B.A.B.), NSF GRFP and NIH F31 AG043232 graduate fellowship (E.A.P.), the Department of Genetics at Stanford University (D.U.) and EMBO long-term post-doctoral fellowship and the Wenner-Gren Fellowhip (S.M.).
Footnotes
Accession numbers
Previously published datasets are reported in Table S1 and S6. Sequencing data have been deposited to the Sequence Read Archive (SRA) under accession number SRP024294.
B.A.B., E.A.P, D.U. and A.B. designed the study. B.A.B and D.U. performed the computational analysis with help from E.A.P. E.A.P. initiated the project and performed experiments with help from B.A.B. S.M. constructed the RNA-seq libraries. B.A.B. created the MySQL database and E.D.W. and K.K. wrote the web interface with supervision from B.C.H. and J.M.C. A.C.D. did chromatin accessibility analyses. A.B.K. advised on the pipeline for breadth change analysis. E.M. and T.A.R. provided intellectual contributions. K.D. assisted in experiments. R.G. and J.C.B. gave advanced access to frog H3K4me3 datasets and M.P.S. to unpublished ENCODE data. B.A.B., D.U., E.A.P. and A.B. wrote the paper. All authors commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adli M, Zhu J, Bernstein BE. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat Methods. 2010;7:615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoston Z, Heine P, Brill MS, Grebbin BM, Hau AC, Kallenborn-Gerhardt W, Schramm J, Gotz M, Schulte D. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development. 2014;141:28–38. doi: 10.1242/dev.097295. [DOI] [PubMed] [Google Scholar]
- Aiden AP, Rivera MN, Rheinbay E, Ku M, Coffman EJ, Truong TT, Vargas SO, Lander ES, Haber DA, Bernstein BE. Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell. 2010;6:591–602. doi: 10.1016/j.stem.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. Embo J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Ralph PL, Evans SN. Transcriptional regulation: effects of promoter proximal pausing on speed, synchrony and reliability. PLoS Comput Biol. 2011;7:e1001136. doi: 10.1371/journal.pcbi.1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Dong D, Shao X, Deng N, Zhang Z. Gene expression variations are predictive for stochastic noise. Nucleic Acids Res. 2011;39:403–413. doi: 10.1093/nar/gkq844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Greven MC, Kundaje A, Djebali S, Brown JB, Cheng C, Gingeras TR, Gerstein M, Guigo R, Birney E, et al. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol. 2012;13:R53. doi: 10.1186/gb-2012-13-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, O’Shea EK. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol Syst Biol. 2013;9:704. doi: 10.1038/msb.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl M, Basler K. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol. 2012;22:2253–2257. doi: 10.1016/j.cub.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Ingham PW. trithorax and the regulation of homeotic gene expression in Drosophila: a historical perspective. Int J Dev Biol. 1998;42:423–429. [PubMed] [Google Scholar]
- Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet. 2014;46:17–23. doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153:976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- Nehar S, Mishra M, Heese K. Identification and characterisation of the novel amyloid-beta peptide-induced protein p17. FEBS Lett. 2009;583:3247–3253. doi: 10.1016/j.febslet.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Ninkovic J, Steiner-Mezzadri A, Jawerka M, Akinci U, Masserdotti G, Petricca S, Fischer J, von Holst A, Beckers J, Lie CD, et al. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Black BL, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lluch S, Blanco E, Carbonell A, Raha D, Snyder M, Serras F, Corominas M. Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res. 2011;39:4628–4639. doi: 10.1093/nar/gkq1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai Y, Kurokawa D, Kiyonari H, Kajikawa E, Suda Y, Aizawa S. Otx2 and Otx1 protect diencephalon and mesencephalon from caudalization into metencephalon during early brain regionalization. Dev Biol. 2010;347:392–403. doi: 10.1016/j.ydbio.2010.08.028. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K. Jarid1b targets genes regulating development and is involved in neural differentiation. Embo J. 2011;30:4586–4600. doi: 10.1038/emboj.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013;27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci U S A. 2010;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci U S A. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- Wang YZ, Plane JM, Jiang P, Zhou CJ, Deng W. Concise review: Quiescent and active states of endogenous adult neural stem cells: identification and characterization. Stem Cells. 2011;29:907–912. doi: 10.1002/stem.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren LA, Rossi DJ, Schiebinger GR, Weissman IL, Kim SK, Quake SR. Transcriptional instability is not a universal attribute of aging. Aging Cell. 2007;6:775–782. doi: 10.1111/j.1474-9726.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Webb AE, Pollina EA, Vierbuchen T, Urban N, Ucar D, Leeman DS, Martynoga B, Sewak M, Rando TA, Guillemot F, et al. FOXO3 Shares Common Targets with ASCL1 Genome-wide and Inhibits ASCL1-Dependent Neurogenesis. Cell Rep. 2013;4:477–491. doi: 10.1016/j.celrep.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L, Voichek Y, Tirosh I, Hornung G, Amit I, Barkai N. Expression noise and acetylation profiles distinguish HDAC functions. Mol Cell. 2012;47:193–202. doi: 10.1016/j.molcel.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.