Abstract
Diagnosis of neurologic and neuropsychiatric disorders typically involves considerable assessment including clinical observation, neuroimaging, and biological and neuropsychological measurements. While it is reasonable to expect that the integration of neuroimaging data and complementary non-imaging measures is likely to improve early diagnosis on individual basis, due to technical challenges associated with the task of combining different data types, medical image pattern recognition analysis has been largely focusing solely on neuroimaging evaluations. In this paper, we explore the potential of integrating neuroimaging and clinical information within a pattern classification framework, and propose that the multi-kernel learning (MKL) paradigm may be suitable for building a multimodal classifier of a disorder, as well as for automatic identification of the relevance of each information type. We apply our approach to the problem of detecting cognitive decline in healthy older adults from single-visit evaluations, and show that the performance of a classifier can be improved when nouroimaging and clinical evaluations are used simultaneously within a MKL-based classification framework.
Keywords: Multi-Kernel Learning (MKL), Normal aging, MRI
1 Introduction
Image-based high-dimensional pattern classification has gained considerable attention, and has begun to provide tests of high sensitivity and specificity on an individual patient basis, in addition to characterizing group differences [7, 6, 11]. One of the major advantages of pattern recognition methods is their ability to capture multivariate relationships among various anatomical regions for more effective characterization of group differences.
Despite the advances of image-based pattern recognition, clinical observations still form the basis for the diagnosis in neurologic and neuropsychiatric disorders. Moreover, besides imaging evaluations, there are a number of non-imaging measures that are of significance in a variety of studies. For example, the potential alternative markers associated with aging can be biological [2, 12], genetic [10], or cognitive [5]. While the potential of a computational approach that would integrate disparate types of information (e.g., structural, functional, clinical, genetic, etc.) is obvious, technical challenges associated with such an approach often prevented joint use of the available alternative measures.
Several attempts have been made recently in the direction of integrating different types of imaging or non-imaging information for pattern classification in the studies of aging. Zhang et al. [20] integrate MRI, PET modalities with CSF markers via a weighted combination of multiple kernels, which provided improvements in the problem of discriminating Alzheimer’s disease (AD) (or Mild Cognitive Impairment (MCI)) and healthy controls. In a similar problem of classifying AD and healthy subjects, Hinrichs et al. [9] employed a multi-kernel learning (MKL) approach to integrate different imaging and non-imaging data. In [4], a computational imaging marker of AD-like atrophy was combined with CSF to predict MCI to AD conversion. At the same time, studies that would investigate possible benefits of integrating imaging and clinical evaluations for prediction of cognitive decline in healthy subjects are absent.
The prospect of finding potential treatments of AD makes it critical to identify biomarkers for very early stages of cognitive decline. As the diagnosis of MCI and AD involves comprehensive clinical observations, it is tempting to use cognitive evaluations when predicting aging-related cognitive decline. There are, however, several challenges associated with the reliability of cognitive measures. While cognitive measures may be significantly different in cognitively declined populations as compared to healthy controls, these measures may not be able to predict cognitive decline at baseline when the decline is not yet evident. As the result of the low predictive power of cognitive measures at baseline, as well as due to the associated significant noise, a number of followup evaluations are typically needed to detect cognitive decline. Considering that brain structure may actually precede reduction in cognitive function by at least several years [13], it is reasonable to expect that single-visit imaging evaluations together with the respective cognitive measures can jointly provide richer information for the detection of cognitive decline.
In this paper, we describe a general MKL-based framework that integrates imaging and clinical information for classification. Our approach consists of an image processing pipeline and a MKL component that combines disparate data for classification. The application focus of this paper is in predicting cognitive decline in healthy older adults by combining MRI and a cognitive assessment test, where our method allows inference about longitudinal outcomes based on the analysis of single-visit imaging and cognitive evaluations.
2 Background
Support Vector Machines (SVM) [19] have been shown to provide high classification accuracy, and are among the most widely used classification algorithms in the brain MRI classification studies [7, 11]. SVM project the data into a high-dimensional space, and find the classification function as the separating hyperplane with the largest margin, where the margin is the distance from the separating hyperplane to the closest training examples.
Multi-Kernel Learning (MKL) [18] extends the theory of SVM by allowing different kernel functions to represent subsets of features (e.g., MRI, cognitive evaluations). Given a set of points χ = {x1,…,xn}, and their respective labels {y1,…, yn}, the MKL problem for K kernels can be formulated as follows:
| (1) |
where the slack variables ξi are introduced to allow some amount of misclassification in the case of non-separable classes, constant C implicitly controls the tolerable misclassification error, the kernel functions φk(x) map the original data into a high-dimensional, possibly infinitely-dimensional, space, and β = (β1,…,βK) are the subkernel weights. The sparsity of the kernel combinations is controlled by a constraint on the subkernel weights, where the commonly used sparse constraint is ‖β‖1 = 1, and the typical non-sparse constraint is ‖β‖2 = 1. The task of the MKL optimization problem is then to find subkernel weights while simultaneously maximizing the margin for the training data.
3 Classification of Imaging and Clinical Evaluations
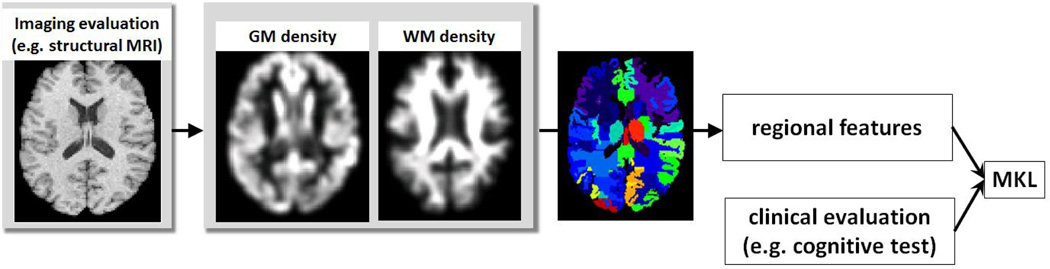
Figure 1 presents the diagram of our approach, and uses the task of integrating structural MRI and cognitive evaluations as an example. The main components of our approach are: (1) tissue density estimation; (2) ROI features extraction; and (3) integration of image measurements and clinical evaluations via MKL.
Fig. 1.
Diagram of our approach (example of integrating structural MRI and cognitive evaluations). The main steps include tissue density estimation, ROI features extraction, and integration of imaging and cognitive evaluations via MKL.
Tissue density estimation
All MR images are preprocessed following a mass-preserving shape transformation framework [3]. Gray matter (GM) and white matter (WM) tissues are segmented out from each skull-stripped MR brain image by a brain tissue segmentation method [14]. Each tissue-segmented brain image is then spatially normalized into a template space, by using a high-dimensional image warping method [16]. The total tissue mass is preserved in each region during the image warping, and tissue density maps are generated in the template space. These tissue density maps give a quantitative representation of the spatial distribution of tissue in a brain, with brightness being proportional to the amount of local tissue volume before warping.
Extracting regional features
The original brain images, and the respective tissue density maps, have very high dimensionality which makes it difficult to discover patterns in the resulting high-dimensional space. By registering brain images to a common template with a predefined number of labeled anatomical regions of interest (ROIs), and by calculating mean tissue density at each ROI, the dimensionality of the original data can be reduced to a manageable size. We use a template image with 101 manually labeled ROIs, and compute average GM and WM tissue densities at ROIs as the image features.
MKL classification
For the purpose of integrating imaging and clinical evaluations we use one kernel for imaging features, and another kernel for the respective clinical evaluations. We use the publicly available implementation of ℓ2-norm MKL [17], and consider Gaussian kernels with σi and σc representing kernel widths for imaging features and clinical measures, respectively.
4 Results
4.1 Materials and evaluation
Dataset
We analyzed a population of 127 healthy individuals from the Baltimore Longitudinal Study of Aging (BLSA) [15] which has been following a set of older adults with annual and semi-annual imaging and clinical evaluations. In this paper we focus on MRI evaluations of the BLSA as the imaging component of our analysis. In conjunction with each imaging evaluation, every individual’s cognitive performance was evaluated on tests of mental status and memory. We selected the following three measures for our analysis: the immediate free recall score (sum of five immediate recall trials) on the California Verbal Learning Test (CVLT) [5], the total number of errors from the Benton Visual Retention Test (BVRT) [1], and the total score from the Mini-Mental State Exam (MMSE) [8].
Defining cognitively stable and declining groups
While the individual cognitive evaluations are often noisy and unreliable, one can identify trends of cognitive decline by considering rates of change in cognitive evaluations over time. We formed the cognitively stable labeled subset from 25 subjects who had the highest slopes of CVLT, and 25 subjects with the lowest CVLT slope values were assigned into the labeled cognitively declining subset. The slope of the CVLT score represents the rate of cognitive decline, and lower slopes of the score indicate higher rates of decline. The remaining 77 subjects were unlabeled.
Evaluation
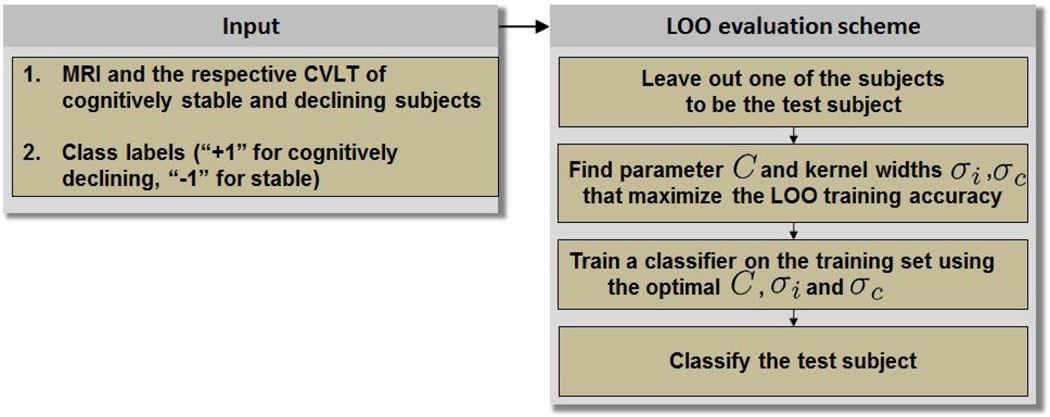
In order to assess the classification performance of our approach, we adopted a leave-one-out (LOO) evaluation scheme (Figure 2). At each run of the LOO evaluation we removed one subject from the labeled set as the test subject. The remaining subjects formed the training set and the classifier was trained on the training data. The free parameters of the classifier (e.g., C, kernel widths) were identified as the ones that yielded the highest LOO classification accuracy on the training set. After the classifier was trained on the training set, it was applied to the test subject to obtain the subject’s test label.
Fig. 2.
LOO evaluation scheme (example of integrating structural MRI and CVLT).
4.2 Classification of single-visit evaluations
In our first experiment, we classified baseline (i.e., first-visit) evaluations following the leave-one-out scheme in Figure 2. The distributions of age in cognitively stable and cognitively declining subpopulations had means 65.8 ± 6.3 years and 70.4 ± 7.0 years, respectively, with the age of cognitively declining individuals being slightly higher than in the stable subjects (p = 0.02). The MKL classifier with a single kernel is equivalent to the SVM classifier, and yielded classification LOO accuracy of 58.0% when using only the MRI information, and 66.0% when using only CVLT at baseline. At the same time, by integrating MRI and CVLT at baseline, we were able to achieve classification accuracy of 74.0%. Table 1 summarizes the classification performance at baseline using imaging evaluations, cognitive evaluations, and the combination of both. The subkernel weights β estimated by MKL using all labeled subjects were 0.884 and 0.467 for the MRI and CVLT kernels, respectively.
Table 1.
Classification of first-visit evaluations
| Kernels | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| MRI | 58.0% | 52.0% | 64.0% |
| CVLT | 66.0% | 68.0% | 64.0% |
| MRI+CVLT | 74.0% | 76.0% | 72.0% |
Similarly, we assessed the ability of our approach to detect cognitive decline using last-visit evaluations. The mean ages in cognitively stable and cognitively declined populations during the last visits were 73.4±7.6 and 78.2±6.4, respectively, with age in cognitively declining individuals being slightly higher than in the stable subjects (p = 0.02). Table 2 summarizes the classification performance for last-visit evaluations. The CVLT-only classifier at last visits performed on a par with the MKL classifier that integrated MRI and CVLT. The subkernel weights estimated by MKL using the last-visit evaluations of all labeled subjects were 0.162 and 0.987 for the MRI and CVLT kernels, respectively.
Table 2.
Classification of last-visit evaluations
| Kernels | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| MRI | 70.0% | 76.0% | 64.0% |
| CVLT | 88.0% | 88.0% | 88.0% |
| MRI+CVLT | 88.0% | 88.0% | 88.0% |
As expected, the performance of the MKL classifier in the task of discriminating cognitively stable and cognitively declining individuals was significantly better for relatively older individuals (i.e., 88.0% during last visits vs. 74.0% during first visits).
4.3 Biomarker of cognitive decline
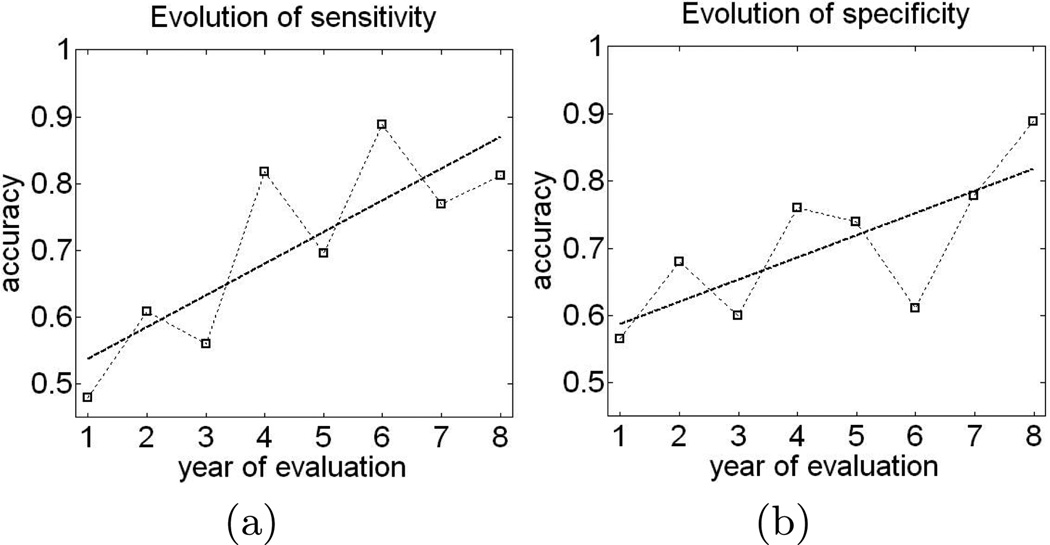
Next, we investigated whether the classifier trained on the last visit scans can predict cognitive decline during earlier evaluations. For a given test subject with a set of longitudinal evaluations x1,…,xt, and the MKL classifier estimated from the last visit evaluations of the training subjects, we obtained the values of the classification function ℱ(x1),…,ℱ(xt), where . The value of the classification function for the subject’s evaluation x reflects the presence of brain phenotypic as well as cognitive pattern inherent to cognitive decline, with larger values of the classification function indicating higher similarity with “imaging-cognitive” pattern of decline. The plot in Figure 3 shows sensitivity and specificity of the classifier tested at every given year of evaluation, where the year of evaluation for an individual is defined relative to the year of the individual’s first visit. Note, that not all subjects may have underwent evaluation at any given year. Indeed, apart from the first and the third year of evaluation, none of the evaluation years witnessed evaluations performed for all 50 labeled subjects. Moreover, some subjects had as few as four evaluations, while some had as many as eleven. Consequently, classification results shown in the plot in Figure 3 were obtained for different, although overlapping, sets of subjects. For example, only 34 subjects had evaluation during their 8-th year. As a result, the classification performance of the classifier trained on the last-visit evaluations of the training subjects and applied to all evaluations of the test subject may not be directly comparable for any two years of evaluation. Nonetheless, the trends in Figure 3 suggest that the classification performance of the classifier noticeably improves with subjects’ age. The performance of the MKL classifier trained on the last-visit evaluations and applied to the very early evaluations is somewhat surprising. In particular, the sensitivity and specificity of the classifier trained on the last-visit evaluations is very low when applied to the first-visit evaluations (i.e., sensitivity and specificity at year one in Figure 3(a,b)). At the same time, the results in Section 4.2 show that the classifier that is specifically trained on the first-visit evaluations can predict cognitive decline based on first-visit evaluations with significantly higher accuracy. This may suggest that different classifiers may be needed for prediction of cognitive decline from the evaluations of different age groups.
Fig. 3.
Sensitivity (a) and specificity (b) of the classifier trained on last-visit evaluations and applied to earlier visits.
4.4 Analysis of individuals with uncertain trends of decline
As we described in the Materials, the trends of cognitive decline in 77 out of 127 individuals were not clear, and the subjects could not have been reliably assigned into one of the two labeled groups. We analyzed the ability of the MKL classifier trained on the last-visit evaluations of the labeled subjects to detect cognitive decline in the subjects with weak trends of decline. After the MKL classifier was trained on the last-visit evaluations to discriminate between cognitively stable and cognitively declining labeled sets, we obtained values of the classification function for every evaluation of the 77 unlabeled individuals.
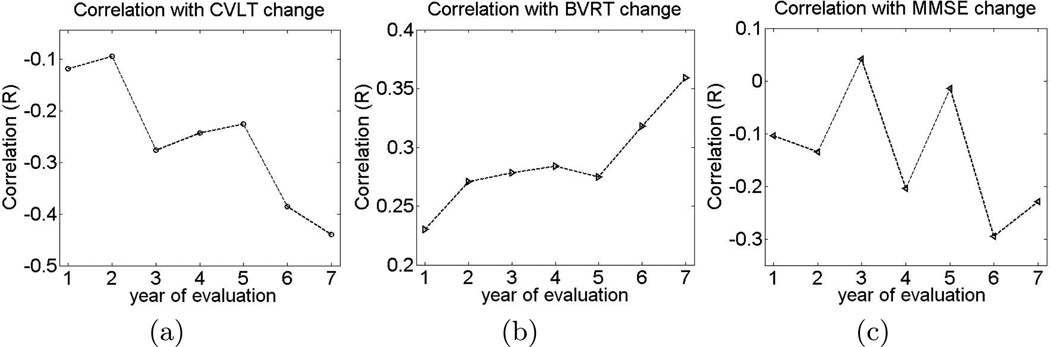
Figure 4(a) shows correlation between the classification values and the rate of change in CVLT for different years of evaluation In general, correlation between the classification values and rate of change in cognitive performance increases with age, which is expected given that the MKL classifier was trained on the last-visit evaluations of the labeled individuals. Additionally, Figures 4(b) and 4(c) show evolution of correlation between the classification values and BVRT and MMSE, respectively. While the increase in correlation with BVRT is evident, increase in correlation with MMSE during later evaluations is less obvious, which reects the fact that MMSE is typically more noisy than CVLT and BVRT.
Fig. 4.
Correlation between the value of the classification function and the rate of change in cognitive evaluations for specific evaluation years. (Lower values of CVLT and MMSE, as well as higher values of BVRT, indicate worse cognitive performance.)
5 Conclusion
In this paper, we presented a pattern classification framework for integration of imaging and non-imaging evaluations. Our method involves an image preprocessing and feature extraction protocol, and employs MKL methodology to integrate imaging and non-imaging features. The application focus of our approach was in prediction of cognitive decline in healthy older adults, where we used MKL to integrate single-visit structural neuroimaging and cognitive evaluations. Our results suggest that, while neither MRI nor CVLT individually carry sufficient information to predict cognitive decline based on a single evaluation, they allow us to achieve promising prediction accuracy when considered jointly. Our proposed approach is general and can potentially be used for integration of other types of neuroimaging and non-imaging data. In particular, we are planning to further explore the problem of predicting cognitive decline in older adults by integrating structural MRI, PET, and other cognitive, as well as genetic, evaluations.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (NIA), and R01-AG14971, N01-AG-3-2124, N01-AG-3-2124.
References
- 1.Benton A. Revised Visual Retention Test. New York: The Psych. Corp.; 1974. [Google Scholar]
- 2.Bouwman FH, van der Flier WM, Schoonenboom NSM, van Elk EJ, Kok A, Rijmen F, Blankenstein MA, Scheltens P. Longitudinal changes of CSF biomarkers in memory clinic patients. Neurology. 2007;69(10):1006–1011. doi: 10.1212/01.wnl.0000271375.37131.04. [DOI] [PubMed] [Google Scholar]
- 3.Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the ravens maps: methods and validation using simulated longitudinal atrophy. Neuroimage. 2001;14(6):1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- 4.Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of mci to ad conversion, via mri, csf biomarkers, and pattern classification. Neurobiology of Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.05.023. In Press, Corrected Proof, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delis D, Kramer J, Kaplan E, Ober B. California Verbal Learning Test - Research Edition. New York: The Psychological Corporation; 1987. [Google Scholar]
- 6.Duchesne S, Bocti C, De Sousa K, Frisoni GB, Chertkow H, Collins DL. Amnestic mci future clinical status prediction using baseline mri features. Neurobiol Aging. 2010;31(9):1606–1617. doi: 10.1016/j.neurobiolaging.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Fan Y, Batmanghelich N, Clark CM, Davatzikos C. Spatial patterns of brain atrophy in mci patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. NeuroImage. 2008;39(4):1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folstein MF, Folstein SE, McHugh PR. ”mini-mental state”. a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975 Nov;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 9.Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for ad in a multi-modality framework: An analysis of mci progression in the adni population. NeuroImage. 2011;55(2):574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji Y, Permanne B, Sigurdsson EM, Holtzman DM, Wisniewski T. Amyloid beta40/42 clearance across the blood-brain barrier following intra-ventricular injections in wild-type, apoe knock-out and human apoe3 or e4 expressing transgenic mice. J Alzheimers Dis. 2001;3(1):23–30. doi: 10.3233/jad-2001-3105. [DOI] [PubMed] [Google Scholar]
- 11.Kloppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RSJ. Automatic classification of mr scans in Alzheimer’s disease. Brain. 2008 Mar;131(3):681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Leon M, Mosconi L, Li J, De Santi S, Yao Y, Tsui W, Pirraglia E, Rich K, Javier E, Brys M, Glodzik L, Switalski R, Saint Louis L, Pratico D. Longitudinal csf isoprostane and mri atrophy in the progression to ad. Journal of Neurology. 2007;254:1666–1675. doi: 10.1007/s00415-007-0610-z. [DOI] [PubMed] [Google Scholar]
- 13.Petersen R, Jack C., Jr Imaging and biomarkers in early Alzheimer’s disease and mild cognitive impairment. Clin Pharmacol Ther. 2009;84(4):438–441. doi: 10.1038/clpt.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pham DL, Prince JL. Adaptive fuzzy segmentation of magnetic resonance images. IEEE Trans. Med. Imaging. 1999;18(9):737–752. doi: 10.1109/42.802752. [DOI] [PubMed] [Google Scholar]
- 15.Resnick SM, Pham DL, Kraut MA, Zonderman AB, Davatzikos C. Longitudinal magnetic resonance imaging studies of older adults: A shrinking brain. J. Neurosci. 2003;23(8):3295–3301. doi: 10.1523/JNEUROSCI.23-08-03295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen D, Davatzikos C. Hammer: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imag. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 17.Sonnenburg S, Rätsch G, Henschel S, Widmer C, Behr J, Zien A, Bona Fd, Binder A, Gehl C, Franc V. The shogun machine learning toolbox. J. Mach. Learn. Res. 2010 Aug;99:1799–1802. [Google Scholar]
- 18.Sonnenburg S, Räatsch G, Schäafer C, Schäolkopf B. Large scale multiple kernel learning. J. Mach. Learn. Res. 2006 Dec;7:1531–1565. [Google Scholar]
- 19.Vapnik VN. The nature of statistical learning theory. New York, NY, USA: Springer-Verlag New York, Inc.; 1995. [Google Scholar]
- 20.Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]