Abstract
Activity-Based Protein Profiling (ABPP) is a chemical proteomics approach that utilizes small-molecule probes to determine the functional state of enzymes directly in native systems. ABPP probes selectively label active enzymes, but not their inactive forms, facilitating the characterization of changes in enzyme activity that occur without alterations in protein levels. ABPP can be a tool superior to conventional gene expression and proteomic profiling methods to discover new enzymes active in adipocytes, and to detect differences in the activity of characterized enzymes that may be associated with disorders of adipose tissue function. ABPP probes have been developed that react selectively with most members of specific enzyme classes. Here, using as an example the serine hydrolase family that includes many enzymes with critical roles in adipocyte physiology, we describe methods to apply ABPP analysis to the study of adipocyte enzymatic pathways.
Keywords: ABPP, activity-based protein profiling, chemoproteomic methods, enzyme activity, serine hydrolases, ABPP probes, adipocyte enzymes, functional proteomics
1. Introduction
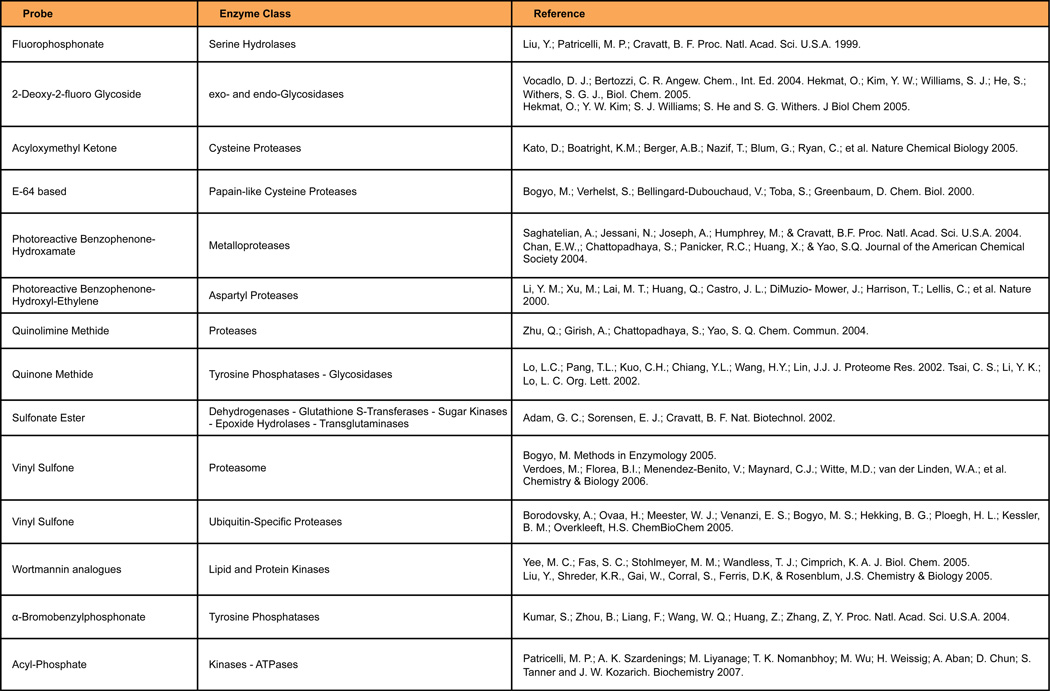
One of the greatest challenges of the postgenomic era is to identify and assign function to the large number of potential new enzymes that genome sequencing projects have unearthed. Adipocytes are cells that depend on multiple enzymatic pathways to carry out their function: to store and release energy as needed (white adipocytes), or to dissipate energy to generate heat (brown adipocytes). Although many of the enzymes involved in these processes are well known, gene expression and proteomic profiling studies have revealed that numerous poorly characterized enzymes are present in adipocytes. Recent advances in chemical proteomic methods have provided tools to establish the level of enzyme activity in cells and tissues. Application of these approaches to the study of adipogenesis and fat cell physiology is likely to provide significant insight into the enzymatic pathways active in adipocytes, and those that may be abnormally regulated in disease. Among the tools developed by chemical biologists to analyze enzyme activity, Activity-Based Protein Profiling (ABPP) stands out as a particularly powerful method to discover new enzymes involved in normal adipocyte physiology, as well as to uncover differences in activity of well-characterized enzymes in pathologic states involving adipose depots. ABPP is a chemical proteomics approach that utilizes small-molecule probes to determine the functional state of enzymes directly in native systems (Cravatt, Wright and Kozarich, 2008; Heal, Dang and Tate, 2011). An ABPP probe contains at least two key features: 1) a reactive group that binds and covalently modifies the active sites of a large number of enzymes that share conserved mechanistic and/or structural features, and 2) a reporter tag, such as a fluorophore or biotin, to enable detection, enrichment, and identification of probe-labeled enzymes by gel electrophoresis and in-gel fluorescence scanning (gel-based ABPP (Jessani, Liu, Humphrey and Cravatt, 2002)) or liquid chromatography-mass spectrometry (LC-MS, e.g., Multidimensional Protein Identification Technology, MudPIT; ABPP-MudPIT (Jessani, Niessen, Wei, Nicolau, Humphrey, Ji et al, 2005)) (Figure 1). ABPP probes have been generated for more than a dozen enzyme classes, including probes that react specifically with serine hydrolases (Liu, Patricelli and Cravatt, 1999), cysteine proteases (Greenbaum, Medzihradszky, Burlingame and Bogyo, 2000), kinases (Patricelli, Szardenings, Liyanage, Nomanbhoy, Wu, Weissig et al, 2007), and glycosidases (Hekmat, Kim, Williams, He and Withers, 2005) (Figure 2). These probes selectively label active enzymes, but not their inactive forms, facilitating the characterization of changes in enzyme activity that occur without alterations in protein levels (Kidd, Liu and Cravatt, 2001; Jessani, Humphrey, McDonald, Niessen, Masuda, Gangadharan et al, 2004).
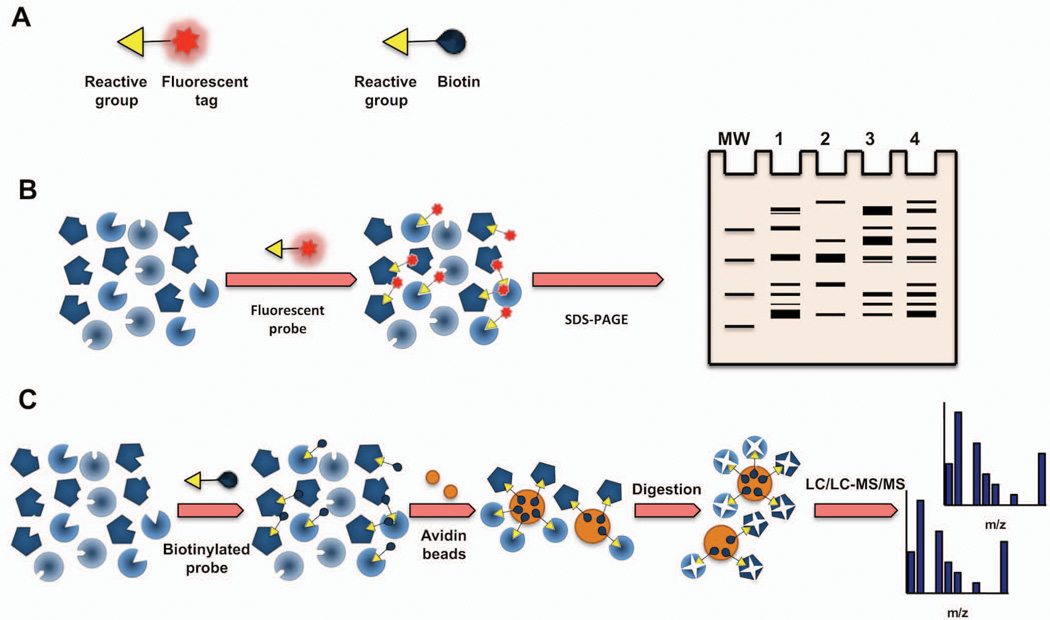
Figure 1. Basics of Activity-Based Protein Profiling (ABPP).
(a) Schematic of ABPP probes showing their basic features: 1) a reactive head group that targets a specific enzyme class, and 2) a reporter tag. (b) Gel-based ABPP. A fluorophosphonate-reactive group can be coupled to a flurophore tag (e.g., rhodamine) to covalently label and detect active serine hydrolases by SDS-PAGE and in-gel fluorescence scanning. (c) ABPP-MudPIT. Biotin-conjugated ABPP probes can be used to label, enrich, and identify by mass spectrometry active serine hydrolases (SH).
Figure 2. ABPP probes are available for a variety of enzyme classes.
A major application of ABPP technology is target discovery. Two or more proteomes are analyzed by ABPP in parallel to identify enzymes with different levels of activity. If the samples analyzed reflect different biological conditions (e.g., physiological vs. pathological), then the enzyme(s) showing differential activity can be hypothesized to be involved in the phenotype in question. The applications of ABPP methods to the study of adipocyte physiology are multiple. For example, ABPP may be used to profile and identify enzyme activities that vary between subcutaneous and visceral white adipose depots and that may explain the opposite relationship that these depots show to the development of insulin resistance and metabolic syndrome. Or it may be used to monitor enzyme activities that are altered in adipose tissue of diabetics relative to healthy individuals. ABPP can also be used to identify enzymes whose activity is modified upon exposure to chemical or environmental stimuli, as in the response to cold of brown adipose tissue. Because ABPP reports changes in enzyme activity, not gene or protein levels, it can be more powerful than traditional gene or proteomic profiling approaches in revealing those enzymes that play central molecular roles in the phenotypic differences under comparison. An additional advantage of ABPP relative to other approaches is that it can detect changes in activity of very low-abundance enzymes in highly complex samples.
When performed in a competitive mode, ABPP can also be used to screen for enzyme inhibitors or to identify the target(s) to which a small-molecule inhibitor binds (Li, Blankman and Cravatt, 2007; Bachovchin, Ji, Li, Simon, Blankman, Adibekian et al, 2010) directly in native cell and tissue proteomes (Bachovchin and Cravatt, 2012). In competitive ABPP, proteomes are first incubated with the small molecule of interest (e.g., a phenotypic hit that targets an unknown enzyme), and then labeled with a broad ABPP probe that reacts with most enzymes in the class under study (Figure 3). Competitive ABPP can be performed using irreversible or reversible inhibitors, although the identification of reversible inhibitors requires a kinetically controlled condition in which labeling with the ABPP probe has not reached saturation. In the case of activity-based irreversible enzyme inhibitors, the enzymes they target will not be subsequently labeled with the broad ABPP probe, and loss of probe labeling will reveal the identity of the inhibitor’s target(s). Enzyme activities present in the vehicle, but not the inhibitor-treated sample, represent the molecular target of the compound in question.
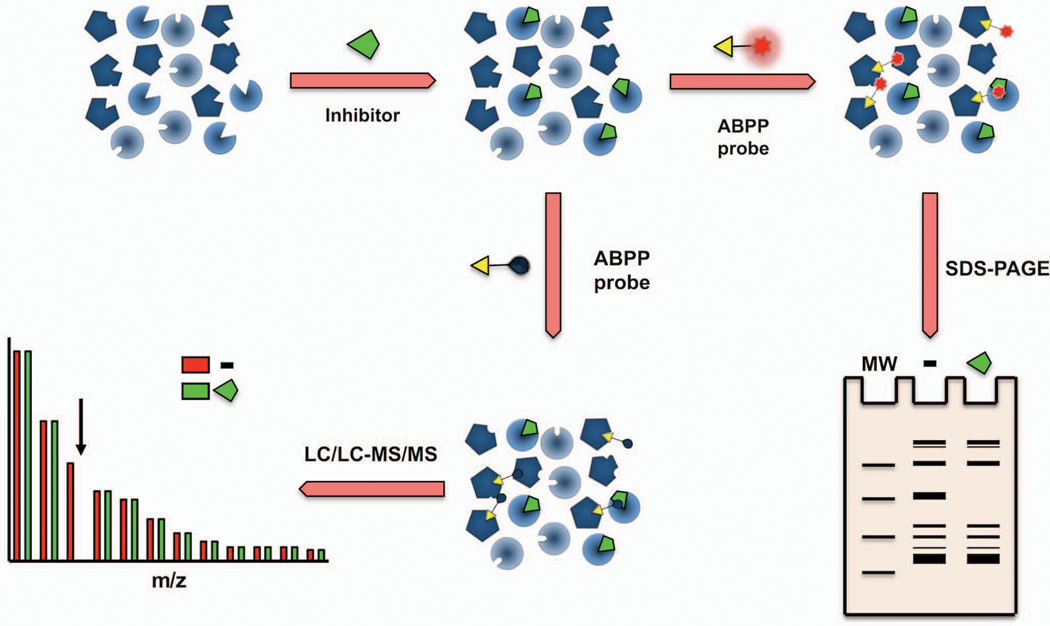
Figure 3. Competitive ABPP for enzyme and inhibitor discovery.
In a competitive Activity-Based Protein Profiling experiment, cells, animals, or prepared proteomes are first treated with either an inhibitor or vehicle and subsequently labeled with a broad ABPP probe. Gel-based ABPP or ABPP-MudPIT is then used to either visualize or identify active enzymes (i.e. labeled by the ABPP probe). An enzyme that is the target of an inhibitor will show reduced signals in the inhibitor-treated samples relative to vehicle controls.
To illustrate the application of ABPP methods to the study of adipocyte enzymes, we describe here several basic protocols that use activity-based probes that specifically label enzymes of the serine hydrolase family. Serine hydrolases (SHs) constitute one of the largest and most diverse enzyme class in mammals, where they perform crucial roles in many biological processes (Long and Cravatt, 2011, Bachovchin and Cravatt, 2012). There are ~240 human SHs, and these enzymes include lipases, peptidases/proteases, and (thio)esterases, all of which hydrolyze their substrates through a conserved mechanism involving an active-site serine nucleophile. A substantial fraction of human SHs (>50%) remain unannotated, with no described function or identified physiological substrates, and an even greater number (>80%) lack selective inhibitors to aid in their characterization (Bachovchin and Cravatt, 2012). SHs play a particularly prominent role in adipocytes, where they regulate lipolysis (hormone-sensitive lipase, HSL; adipose triglyceride lipase, ATGL; monoacylglycerol lipase, MGLL; PNPLA4) and lipid uptake (lipoprotein lipase, LPL) and synthesis (fatty acid synthase, FASN). Adipocytes also express many poorly characterized SHs that may play equally important roles in their cellular physiology (Soukas, Saatkamp, Novelli and Friedman, 2001).
ABPP can prove to be a powerful tool to discern the function of these unannotated SHs in adipocyte biology and to establish which may serve as therapeutic targets for metabolic disorders. ABPP probes bearing a fluorophosphonate (FP) electrophile that shows broad reactivity with SHs and negligible interaction with other enzymes have been generated (Liu et al, 1999; Kidd et al, 2001). These FP-based probes have been shown to label >80% of the 115+ predicted mouse metabolic SHs in tissue/cell proteomes (Bachovchin et al, 2010). An FP probe conjugated to a fluorophore (e.g., FP-Rhodamine) can be used to reveal differences in serine hydrolase activity by gel-based ABPP, while an FP-Biotin probe can be used to enrich and identify the specific serine hydrolases in question using ABPP-MudPIT. An example of data generated with these broad FP-based ABPP probes is shown in Figure 4.
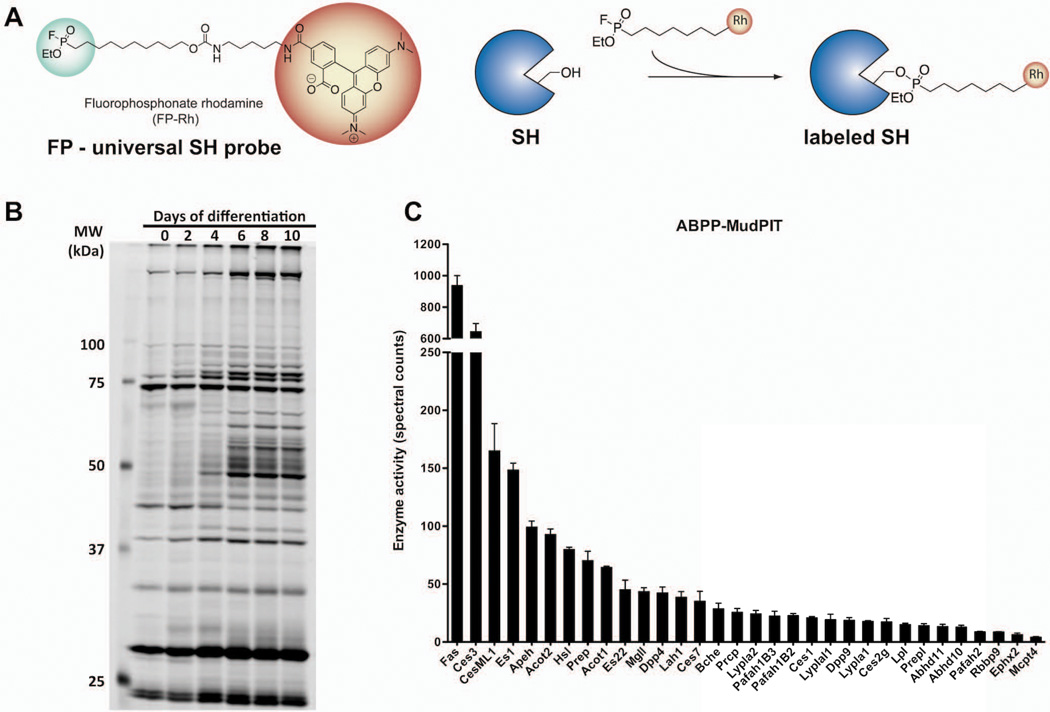
Figure 4. Sample ABPP data obtained with FP-based probes.
(a) Reporter-tagged fluorophosphonate ABPP probes that react covalently only with the active form of serine hydrolases can be used to (b) profile the pattern of serine hydrolase activity by gel-based ABPP during differentiation of 10T1/2 adipocytes or (c) identify active serine hydrolases in mouse white adipose tissue using ABPP-MudPIT.
2. Technical Aspects
2.1 Preparation of Proteomes for ABPP Analysis
Total proteome extracts
To preserve enzyme activity, only non-denaturing lysis buffers should be used in ABPP analysis. Mechanical disruption of cells in PBS is the best procedure to maintain proteome integrity. Protease inhibitors may be added to the PBS solution but they are not recommended, as they may inhibit the activity of some enzymes and interfere with labeling of enzymes with the broad activity probe. To preserve proteome integrity and minimize protein degradation, all solutions and collecting tubes must be pre-cooled and kept on ice, and all procedures must be carried out at 4°C.
Materials
Cell culture or tissue sample
Ice-cold Phosphate buffered saline without calcium or magnesium (PBS; e.g., Corning catalog number 21-031-CV)
Dounce homogenizer
TissueLyser (Qiagen) or similar mechanical tissue disruption equipment (e.g. polytron homogenizer)
Beads (for use with TissueLyser)
Probe sonicator
Ultracentrifuge (required only if fractionation of soluble and membrane-bound proteomes is desired)
In the case of cells, wash and scrape the cells of interest from the culture plate using PBS and centrifuge to pellet them. Start with enough cells to yield sufficient proteome for gel-based ABPP (50 µg/reaction) or ABPP-MudPIT (1 mg/reaction) as desired; bear in mind the low protein yield of adipocytes. Resuspend the cell pellet in 4 volumes of ice-cold PBS and homogenize on ice by performing 10–15 strokes with a Dounce homogenizer or by sonicating 5 times for 5 sec at 50% power. To prepare proteomes from tissues (a reasonable starting amount is 30–100 mg of adipose tissue), add 4 volumes of PBS to freshly isolated (preferred) or frozen tissue, and homogenize using a bead-based TissueLyser for 3 min at maximum shaking frequency. Repeat the procedure until the solution is homogeneous. Once the tissue is fully homogenized, sonicate the solution 5 times for 5 sec at 30% power. If a TissueLyser (or similar instrument) is not available, alternative methods of mechanical tissue disruption are also appropriate (e.g. manual polytron homogenizer) as long as the protein solution is not unduly heated.
Centrifuge proteome 30 min at 14,000×g at 4°C and collect the supernatant. Be careful to avoid the fat cake that is present on top of the solution.
Quantify protein concentration and adjust to desired concentration (1–2 mg/mL).
Aliquot samples and store at −80°C, or proceed with ABPP probe labeling if desired.
Prior to ABPP, thaw samples on ice and briefly allow them to warm to room temperature.
Fractionation of soluble and membrane-associated proteomes
To capture the greatest number of enzyme activities in ABPP experiments, and to simplify interpretation of enzyme activity profiles in gel-based ABPP or mass spectrometry (i.e. ABPP-MudPIT) analyses, it is often useful to separate the soluble from the membrane-bound proteome of cells/tissues, and to analyze them separately.
Mechanically homogenize cells or tissues as in step 1 above.
Centrifuge for 5 min at 1,000×g at 4°C to eliminate nuclei and unbroken cells or small pieces of tissue.
Transfer supernatant to a clean ultracentrifuge tube, being careful to avoid the fat cake. Centrifuge for 1 hr at 100,000 × g at 4°C to pellet the membrane-associated fraction and separate it from the soluble proteome.
Transfer supernatant to a clean tube. Gently wash the pellet once with ice-cold PBS. Discard wash and add 1–2 volumes of cold PBS to the pellet. Sonicate pellet 3 times at 30% power to resuspend membrane-associated proteome.
Quantify concentration of membrane-associated and soluble proteomes and adjust to desired concentration (1–2 mg/mL).
Aliquot samples and store at −80°C, or proceed with ABPP probe labeling if desired.
2.2 Gel-based ABPP
The following protocol is the standard procedure for labeling enzymes of the serine hydrolase family in proteomes prepared from cells or tissues (i.e. in vitro ABPP labeling) using Flurophosphonate (FP)-based ABPP probes (e.g., FP-Rhodamine), and visualizing differences in enzyme activity levels in SDS-PAGE gels. This basic protocol can be modified as necessary and used to profile the activity levels of enzymes in other families for which appropriate activity-based probes are available (see examples in Figure 2). An unlabeled control sample should be used for comparison to the experimental samples to confirm efficiency of the labeling reaction. Each enzyme class may have different ABPP analysis requirements. For example, serine hydrolases are often modified at the post-translational level (e.g., subject to glycosylation). This can result in altered migration patterns in electrophoresis gels and in the presence of multiple bands corresponding to the same enzyme. To reduce the number of bands visualized in gels and simplify interpretation of data, an optional deglycosylation step can be performed after the labeling procedure. An additional control sample labeled with the ABPP probe but not subjected to deglycosylation is required in this case (to verify efficiency of the procedure).
Materials
Fluorophore-conjugated Fluorophosphonate (FP) ABPP probe (e.g., FP-Rhodamine; FP-TAMRA, Thermo Fisher catalog number 88318)
Proteome (cell/tissue lysate prepared as described above)
Cold PBS without calcium or magnesium pH 7.4
50X FP fluorophore-conjugated ABPP probe (50 µM stock in DMSO; can be stored at −20°C for over a year)
DMSO
10% Nonidet P-40 (NP-40) (w/v) solution (New England Biolabs, catalog number B2704S)
10X Glycoprotein denaturing buffer (New England Biolabs, catalog number B1704S)
Reaction buffer G7 (New England Biolabs, catalog number B3704S)
PNGaseF (New England Biolabs, catalog number P0704S)
4X LDS sample buffer (Life Technologies, catalog number NP0007)
Sample Labeling
For each experimental and control sample, aliquot 49 µL of 1 mg/mL cell/tissue proteome into microcentrifuge tubes.
To the experimental samples add 1 µL of 50X ABPP probe stock (50 µM) for a final concentration of 1 µM FP probe. To the control sample add 1 µL of DMSO.
-
Vortex samples and allow the reaction to proceed for 1 hr at RT. The labeling reaction can also be carried out at 37°C, which can result in better visualization of slow-labeling enzymes.
Optional: Deglycosylation step. Go to step 8 if deglycosylation is not desired
Add 5 µL of 10X glycoprotein denaturing buffer.
Heat samples for 10 min at 95°C.
Add 5 µL of 10% NP-40 solution, 5 µL of reaction buffer G7, and 0.5 µL of PNGaseF to each sample. Do not add PNGaseF to the labeled control sample.
Vortex samples and incubate for 1 hr at 37°C.
Add 20 µL of 4X LDS sample buffer to quench the reaction.
Heat samples for 5 min at 95°C. At this point, samples can be stored at −20°C overnight.
Run 40 µL of labeled sample on a 10% SDS-PAGE gel (250 V constant for a large gel). To avoid overheating due to the high voltage, electrophoresis may be performed at 4°C, or cooling plates added to the outside of the gel box.
Visualize enzyme activities using an in-gel fluorescence flatbed scanner (e.g., Hitachi FMBio IIe). Inverse grey-scale images are typically most useful to analyze and present data.
2.3 ABPP-MudPIT
This technique allows the identification by mass spectrometry of specific enzyme activities, in these case serine hydrolases. It is highly recommended that all mass spectrometry experiments be run in four or more biological replicates.
Materials
Biotin-conjugated Fluorophosphonate ABPP probe (e.g., FP-Biotin; FP-Desthiobiotin Thermo Fisher catalog number 88317). Make a 1 mM stock solution in DMSO.
Proteome (cell/tissue lysate) prepared as described in 2.1
10% Triton X-100 (e.g., Sigma Aldrich, catalog number T-9284) solution in PBS
PBS without calcium or magnesium, pH 7.4
PD-10 separation columns (GE Healthcare, catalog number 17-0851-0)
10% SDS (e.g., Fluka, catalog number BP166-500) solution in water
Avidin beads (Avidin-agarose beads from egg white, Sigma Aldrich, catalog number A9207)
Water, LC-MS grade (e.g., Fluka, catalog number 39253)
Urea (e.g., Fluka, catalog number U15-500)
Tris (2-Carboxyethyl) phosphine hydrochloride (TCEP) (Sigma Aldrich, catalog number C4706)
Iodoacetamide (e.g., Sigma Aldrich, catalog number I6125)
Trypsin (Promega, catalog number V5111)
Formic acid (e.g., Fluka, catalog number O6440)
Sample Labeling
For each reaction, label 1 mg of proteome (as a 1 mg/mL solution) with FP-Biotin at 5 µM final concentration (5 µL of 1mM stock solution in DMSO) for 2 hr at room temperature.
Solubilize proteome by adding 100 µL of Triton X-100 (10% stock solution in PBS) for a final concentration of approximately 1%. The final volume of the reaction should now be 1.1 mL.
Rotate at 4°C for 1 hr.
Equilibrate PD-10 column with 25 mL of PBS.
To eliminate the excess of ABPP probe, add the 1.1 mL labeling reaction to the column and rinse the tubes where it was carried out twice with 700 µL of PBS, adding each wash to the column. The final volume of the reaction should now be 2.5 mL.
Elute the labeled proteome with 3.5 mL of PBS and collect the fraction. The final volume of the reaction should now be 6 mL.
Add 0.3 mL of SDS (10% stock solution in water) for a final concentration of about 0.5%.
Heat samples at 90°C for 8 min.
Cool samples to room temperature.
Add PBS to reach a final volume of 8.5 mL.
When working with avidin beads, use cut pipette tips to ensure integrity of beads. Wash avidin beads (100 µL of slurry per sample; 100 µL contains 50 µL of glycerol and 50 µL of beads) 3 times with 10 mL of PBS. Centrifuge 1 min at 400 × g between washes. Resuspend the washed beads pellet in 1 volume of PBS.
Mix labeled proteome and washed beads (100 µL/sample) in a 15 mL conical tube.
Rotate at room temperature for 1 hr.
Centrifuge labeled proteome/bead mixture at 400 × g for 3 min.
-
Discard supernatant.
From this point on, use only mass spectrometry grade water or its equivalent for all solutions
Wash pelleted beads twice with 8 mL of 1% SDS. For each wash, rotate tubes at room temperature for 5 min, prior to pelleting beads (400 × g for 3 min). This same washing procedure should be used for all subsequent washes described below.
Wash beads twice with 8 mL of 6 M urea.
Wash beads three times with 8 mL of PBS.
After the final wash, bring beads up to 1 mL in PBS and transfer to screw top microfuge tubes.
Spin in tabletop microcentrifuge at 1,400 rpm for 3 min.
Discard buffer and add 150 µL of 8 M urea to the pellet.
Add 10 µL of 100 mM TCEP for a final concentration of approximately 5 mM.
Add 5 µL of freshly made 500 mM iodoacetamide (approximately 12 mM final concentration) to each sample and rotate at room temperature in the dark for 30 min.
Spin on tabletop microcentrifuge at 400 × g for 3 min. Discard all but 50 µL of supernatant and add 250 µL of PBS to the pellet in order to reduce the concentration of urea to less than 2 M prior to tryptic digestion.
Add 4 µL of 0.5 µg/µL trypsin to each sample and incubate at 37°C overnight.
Spin in tabletop microcentrifuge at 400 × g for 4 min.
Transfer peptide supernatant to mass spectrometry grade tubes. Be careful not to aspirate beads that can clog the HPLC columns during mass spectrometry analysis.
Add 0.1 mL of PBS to the beads, gently shake and centrifuge at 4,000 rpm for 4 min. Add supernatant to that obtained in the previous step. Volume should now be ~400 µL.
Add formic acid to a final concentration of 5%.
Store samples at −80°C or apply to equilibrated capillary column for mass spectrometry analysis.
Protocols to carry out mass spectrometry analysis can vary widely. For an example of a MudPIT protocol, the reader is referred to Kline and Wu, 2009.
2.3 Competitive ABPP
As mentioned above, competitive ABPP can be performed using irreversible or reversible inhibitors, but the clear identification of reversible inhibitors requires a kinetically controlled condition in which labeling with the ABPP probe has not reached saturation. The protocol below is tailored for irreversible enzyme inhibitors, but incubation with the ABPP probe can be shortened to enable the isolation of reversible inhibitors.
For each compound to be tested, aliquot 50 µg in 49 µl (for gel-based ABPP) or 1 mg in 1 mL (for ABPP-MudPIT) of proteome into microcentrifuge tubes.
To each sample add 0.5 µL of each compound to be evaluated for gel-based ABPP, or 5 µL for ABPP-MudPIT. Compound stock solutions should be made to reflect the final concentration desired in the competitive ABPP analysis (e.g., if the compound is to be tested at a 10 µM dose, stock should be 1 mM). Add 0.5 µL of DMSO to the control sample for gel-based ABPP, or 5 µL for ABPP-MudPIT.
Vortex samples and incubate for 1 hr at room temperature.
Label samples by adding 0.5 µL of ABPP probe stock for gel-based ABPP or 5 µL for ABPP-MudPIT, keeping in mind that for gel-based ABPP a final probe concentration of 1 µM should be used, while for ABPP-MudPIT the final probe concentration should be 5 µM.
From this point on, the labeling reactions should be treated just as described in the protocols above for Sample labeling, picking up at the step immediately after ABPP probe addition.
3. Discussion
Critical Parameters and Troubleshooting
Protein degradation during the preparation of the proteomes may affect the quality of ABPP analysis. The use of protease inhibitors will help avoid this problem, but it may affect the ability of ABPP probes to label certain enzyme classes. This is a particular problem for serine hydrolases, some of which are themselves proteases (e.g., DPP-IV). If warranted, protease inhibitors should be avoided and all proteome preparation steps carried out at 4°C. Use of mass spectrometry grade water is critical in ABPP-MudPIT analysis to reduce background signals. To maximize tryptic digestion, the TCEP solution must be prepared fresh every time (TCEP powder should be stored at 4°C) in order to obtain complete protein denaturation. Similarly, to prevent disulfide bond regeneration, the iodoacetamide solution must be fresh and not be exposed to light. Significant non-specific protein binding is frequently observed with streptavidin beads (as used in ABPP-MudPIT). Until the investigator is familiar enough with the procedure to recognize common non-specific proteomic analysis artifacts, two controls are suggested. First, it is important to include a control sample that has not been labeled with the ABPP probe but that has been processed in parallel with labeled samples. Proteins identified by MudPIT in both datasets represent proteins that bind non-specifically to the streptavidin beads and should be filtered out in the analysis. Second, to obtain the complete peptide spectrum of the proteomes in question (i.e. the input condition), a control tryptic sample obtained by digesting the intact proteome prior to any enrichment should be performed. Peptides identified only in the streptavidin-enriched samples but absent in the “input” tryptic digest sample should be discarded as false positives. It is important to keep in mind for all ABPP experiments that once the labeling reaction has started, the procedure must be carried out to completion until a freezing step is indicated. Investigators are encouraged to plan ahead, particularly while performing ABPP-MudPIT experiments with multiple samples. Although ABPP probes label previously frozen proteomes well, greater labeling efficiency may be observed in fresh lysates; whenever possible, fresh proteomes are preferred for ABPP analysis. Finally, the lipid content of adipocytes results in the generation of a fat cake layer during cell/tissue homogenization that should be avoided while harvesting supernatants. Lipid contamination of proteomes may interfere with labeling by ABPP probes.
In addition to the basic ABPP techniques described above, there are several embodiments of ABPP that merit brief mention.
In situ ABPP
Because the majority of ABPP probes have limited cell permeability due to their bulky reporter tag, labeling of enzymes with tagged ABPP probes is performed in homogenized proteomes (in vitro ABPP as described in all protocols above), not intact cells or tissues. This limits the ability of in vitro ABPP to report on the activity of all endogenous enzymes, as some are likely to degrade during the proteome preparation steps. Moreover, some enzymes may work as part of complexes that need to remain intact to be active. To enable activity-based labeling of enzymes directly in living cells and organisms, cell-permeable ABPP probes have been developed in which the reporter tag is substituted with a small, latent chemical handle (an alkyne or an azide), which allows cell permeability. An orthogonally-functionalized reporter tag is then attached to the probe using click chemistry methods (Cu(I)-catalyzed stepwise azide-alkyne cycloaddiction of Huisgen) (Kolb and Sharpless, 2003). Following the click reaction that couples a reporter tag to the ABPP probe, labeled proteins can then be analyzed by gel-based ABPP or ABPP-MudPIT. Interested readers are referred to Speers and Cravatt, 2009 for protocols for in situ ABPP.
ABPP-SILAC
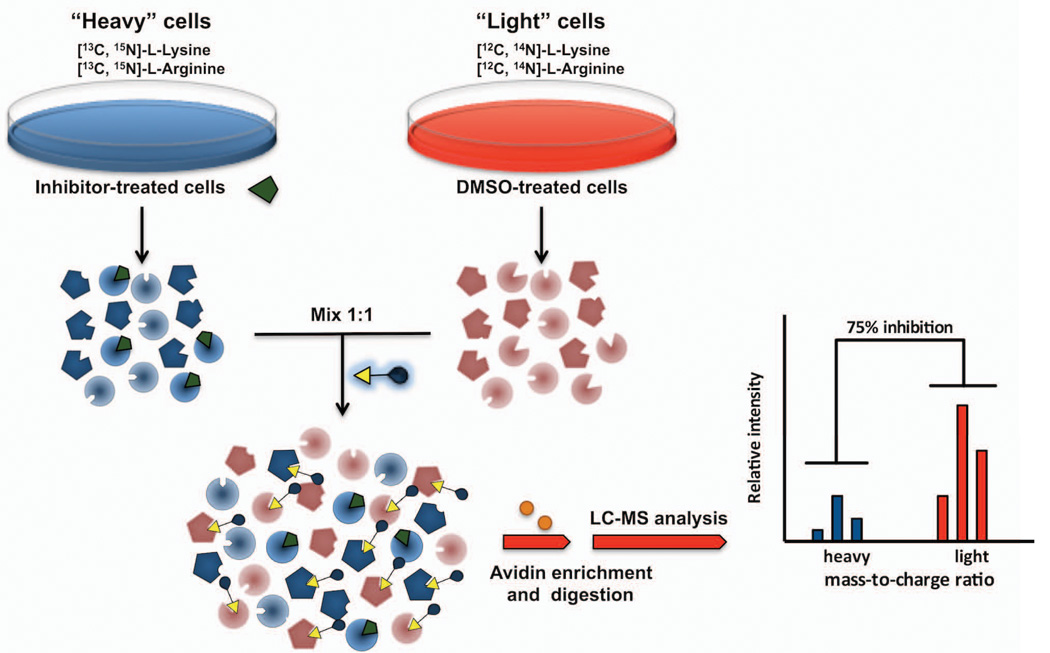
Stable isotope labeling by amino acids in cell culture (SILAC) enables in vivo incorporation of a label into proteins for mass spectrometry-based quantitative proteomic analysis (Ong, Blagoev, Kratchmarova, Kristensen, Steen, Pandey and Mann, 2002). SILAC relies on the metabolic incorporation of a “light” or “heavy” form of an amino acid (e.g., labeled with deuterium, 13C, or 15N) into all newly synthesized proteins. SILAC involves growing cells in isotopically “light” and “heavy” amino acid media for several population doublings (typically 5) to allow full incorporation of the heavy amino acid into the proteome. Because the “light” and “heavy” form of the amino acids used are virtually identical, “heavy” cells behave similarly to cells grown in “light” media. Incorporation of isotopically labeled amino acids into proteins results in a mass shift of the corresponding peptides that can be detected by a mass spectrometer. When “light” and “heavy” samples are combined, the ratio of peak intensities in the mass spectrum reflects the relative protein abundance (Figure 5). Because quantitative differences between samples can be established with greater confidence that with other methods, SILAC has been integrated with ABPP analysis (Adibekian, Martin, Wang, Hsu, Bachovchin, Niessen et al, 2011). A representative competitive ABPP-SILAC experiment to determine the target of a serine hydrolase inhibitor would involve growing cells in isotopically “light” and “heavy” amino acid media, which would then be treated with inhibitor or DMSO, respectively, for a given time (typically 1–4 hr), lysed, combined, and treated with an FP-Biotin probe. FP-labeled SHs would then be enriched by avidin chromatography and identified and quantified by LC-MS based on analysis of MS2 spectra and MS1 profiles using a mass spectrometer. ABPP-SILAC can also be used in combination with in situ ABPP (Adibekian, Martin, Chang, Hsu, Tsuboi, Bachovchin et al, 2012), thus maximizing the range of applications and quantitative power of ABPP.
Figure 5. Competitive ABPP-SILAC to identify the target of enzyme inhibitors.
Cells are grown in different media with isotopically stable amino acids. Proteomes prepared from compound-treated “heavy” and vehicle-treated “light” cells are labeled with a biotin-conjugated ABPP probe, and mixed in a 1:1 ratio. Following avidin enrichment and on-bead digestion, samples are analyzed by mass spectrometry and representative peptides belonging to treated or control cells are identified by mass-to-charge shift.
Acknowledgements
Work was supported by NIH grants CA179489 and DK099810 to E.S. and B.F.C. and the American Heart Association to A.G.
References
- Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nature Biotechnology. 2002;20:805–809. doi: 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Hodder PS, Rosen H, Cravatt BF. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J Am Chem Soc. 2012;134(25):10345–10348. doi: 10.1021/ja303400u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat Chem Biol. 2011;7(7):469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat Rev Drug Discov. 2012;11(1):52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010;107(49):20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogyo M, Verhelst S, Bellingard-Dubouchaud V, Toba S, Greenbaum D. Selective targeting of lysosomal cysteine proteases with radiolabeled electrophilic sub-strate analogs. Chemistry & Biology. 2000;7:27–38. doi: 10.1016/s1074-5521(00)00061-2. [DOI] [PubMed] [Google Scholar]
- Bogyo M. Screening for selective small molecule inhibitors of the proteasome using activity-based probes. Methods in Enzymolog. 2005;399:609–622. doi: 10.1016/S0076-6879(05)99040-X. [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Ovaa H, Meester WJ, Venanzi ES, Bogyo MS, Hekking BG, et al. Small-molecule inhibitors and probes for ubiquitin- and ubiquitin-like-specific proteases. ChemBioChem. 2005;6:287–291. doi: 10.1002/cbic.200400236. [DOI] [PubMed] [Google Scholar]
- Chan EW, Chattopadhaya S, Panicker RC, Huang X, Yao SQ. Developing photoactive affinity probes for proteomic profiling: hydroxamate-based probes for metalloproteases. Journal of the American Chemical Society. 2004;126(44):14435–14446. doi: 10.1021/ja047044i. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- Heal WP, Dang TH, Tate EW. Activity-based probes: discovering new biology and new drug targets. Chem Soc Rev. 2011;40(1):246–257. doi: 10.1039/c0cs00004c. [DOI] [PubMed] [Google Scholar]
- Hekmat O, Kim YW, Williams SJ, He S, Withers SG. Active-site peptide "fingerprinting" of glycosidases in complex mixtures by mass spectrometry. Discovery of a novel retaining beta-1,4-glycanase in Cellulomonas fimi. J Biol Chem. 2005;280(42):35126–35135. doi: 10.1074/jbc.M508434200. [DOI] [PubMed] [Google Scholar]
- Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR, 3rd, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A. 2004;101(38):13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer invasiveness. Proc Natl Acad Sci U.S.A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2(9):691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, et al. Activity-based probes that target diverse cysteine protease families. Nature Chemical Biology. 2005;1(1):33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40(13):4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- Kline KG, Wu CC. MudPIT analysis: application to human heart tissue. Methods Mol Biol. 2009;528:281–293. doi: 10.1007/978-1-60327-310-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov Today. 2003;8(24):1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- Kumar S, Zhou B, Liang F, Wang WQ, Huang Z, Zhang ZY. Activity- based probes for protein tyrosine phosphatases. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7943–7948. doi: 10.1073/pnas.0402323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J Am Chem Soc. 2007;129(31):9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio- Mower J, et al. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96(26):14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shreder KR, Gai W, Corral S, Ferris DK, Rosenblum JS. Wortmannin, a widely used phosphoinositide 3-kinase inhibitor, also potently inhibits mammalian polo-like kinase. Chemistry & Biology. 2005;12(1):99–107. doi: 10.1016/j.chembiol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Lo LC, Pang TL, Kuo CH, Chiang YL, Wang HY, Lin JJ. Design and synthesis of class-selective activity probes for protein tyrosine phosphatases. Journal of Proteome Research. 2002;1:35–40. doi: 10.1021/pr015506a. [DOI] [PubMed] [Google Scholar]
- Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111(10):6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46(2):350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proceedings of the National Academy of Sciences of the United States. 2004;101(27):10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem. 2001;276(36):34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF. Activity-Based Protein Profiling (ABPP) and Click Chemistry (CC)-ABPP by MudPIT Mass Spectrometry. Curr Protoc Chem Biol. 2009;1:29–41. doi: 10.1002/9780470559277.ch090138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CS, Li YK, Lo LC, Tsai CS, Li YK, Lo LC. Design and synthesis of activity probes for glycosidases. Organic Letters. 2002;4(3607):3610. doi: 10.1021/ol0265315. [DOI] [PubMed] [Google Scholar]
- Verdoes M, Florea BI, Menendez-Benito V, Maynard CJ, Witte MD, van der Linden WA, et al. A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chemistry & Biology. 2006;13(11):1217–1226. doi: 10.1016/j.chembiol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Vocadlo DJ, Bertozzi CR. A strategy for functional proteomic analysis of gly- cosidase activity from cell lysates. Angewandte Chemie, International Edition. 2004;43:5338–5342. doi: 10.1002/anie.200454235. [DOI] [PubMed] [Google Scholar]
- Yee MC, Fas SC, Stohlmeyer MM, Wandless TJ, Cimprich KA. A cell- permeable, activity-based probe for protein and lipid kinases. The Journal of Biological Chemistry. 2005;280:2905–2909. doi: 10.1074/jbc.M504730200. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Girish A, Chattopadhaya S, Yao SQ. Developing novel activity- based fluorescent probes that target different classes of proteases. Chemical Communica- tions. 2004;13:1512–1513. doi: 10.1039/b404471a. [DOI] [PubMed] [Google Scholar]