Abstract
Inhibiting protein-protein interactions (PPIs) with synthetic molecules remains a frontier of chemical biology. Many PPIs have been successfully targeted by mimicking α-helices at interfaces, but most PPIs are mediated by non-helical, non-strand peptide loops. We sought to comprehensively identify and analyze these loop-mediated PPIs by writing and implementing LoopFinder, a customizable program that can identify loop-mediated PPIs within all protein-protein complexes in the Protein Data Bank. Comprehensive analysis of the entire set of 25,005 interface loops revealed common structural motifs and unique features that distinguish loop-mediated PPIs from other PPIs. “Hot loops,” named in analogy to protein hot spots, were identified as loops with favorable properties for mimicry using synthetic molecules. The hot loops and their binding partners represent new and promising PPIs for the development of macrocycle and constrained peptide inhibitors.
INTRODUCTION
Specific interactions between proteins are responsible for a wide range of signaling processes in the cell. As a result, targeting protein-protein interactions (PPIs) is a growing field of drug discovery.2 Recent work using macrocyclic molecules has demonstrated that such compounds are particularly adept at inhibiting PPIs.3 Macrocyclic natural products such as polyketides and non-ribosomally-synthesized peptides can have exquisite potency and selectivity, and recent approaches have developed synthetic macrocycles that approach their sophistication. Decades of work with selection techniques such as phage display and RNA display have revealed that cyclization almost universally augments the affinities and selectivities of the selected molecules. Macrocyclic linkers and other conformational constraints can endow even large peptides and proteins with surprising bioavailability, and these effects have been observed in areas as diverse as the optimization of peptide hormones and the engineering of highly disulfide-bonded natural products.4 Macrocycles are theoretically capable of inhibiting nearly any PPI, but PPIs mediated by short peptide loops provide the most direct starting point for designing macrocyclic inhibitors.
Designing inhibitors is not straightforward, but peptides and peptidomimetics offer the advantage of being able to directly mimic specific secondary structures. β-turns and β-strands are readily mimicked by a diverse collection of small-molecule and peptide scaffolds.5,6 Numerous strategies are also available for mimicking or structurally stabilizing α-helices, including side-chain-to-side-chain crosslinks, backbone-to-backbone crosslinks, backbone replacements using unnatural residues such as β-amino acids, and non-peptidic scaffolds such as terphenyls or macrocycles.7 These and other classes of molecules have been used to target important helix-mediated PPIs, such as the p53-MDM2 and Bcl-xL-BH3 interactions. While campaigns focusing on inhibiting helix-mediated PPIs have been successful, a survey of the Protein Data Bank showed that only 26% of interface residues have α-helical secondary structure, with 24% having β-strand secondary structure and the remaining 50% having non-regular secondary structure.1
Some systematic methods have been developed to use structures of PPIs to identify druggable pockets and to design potential inhibitors. HotSprint searches for conserved residues at PPIs that satisfy requirements for solvent accessibility.8 Another approach searches PPI interfaces for regions with maximal changes in solvent-accessible surface area upon complexation, then uses these “anchor residues” as pharmacophores to design inhibitors.9 The relative accessibility of α-helix mimetics prompted a systematic survey of hot spots located within α-helices at protein-protein interfaces.10,11 In addition, an algorithm called PeptiDerive was used to search a set of 151 pre-selected PPIs for short segments that contain multiple hot spots, regardless of structure.12 The EphB4-ephrin B2 interaction was cited as a proof-of-concept result for PeptiDerive, specifically residues 116 to 128 on ephrin B2.12 A similar sequence discovered via phage display was found to be antagonistic for EphB4 with an IC50 of 15 nM, validating the approach.13 However, this kind of analysis has never been integrated for batch processing of the entire PDB, nor has it been done with customizable parameters that allow loops of interest to be defined by structure.
To explore loop-mediated PPIs and to facilitate the design of macrocyclic inhibitors, we sought to comprehensively identify all known protein complexes that are mediated by short peptide loops. Herein, we describe LoopFinder, an original program for comprehensively searching structural databases for peptide loops at PPIs. We used LoopFinder to identify a set of loops that contribute significantly to binding interactions – in analogy to hot spots, we chose to call these “hot loops”. These hot loops identify novel targets for inhibition and provide starting points for the rational design of macrocycles as PPI inhibitors.
RESULTS
Workflow for LoopFinder is depicted in Supplementary Results (Supplementary Figure 1). We acquired 19,657 multi-chain structures from the PDB in August 2013, representing all multi-chain structures with ≤ 4Å resolution and < 90% sequence identity. PDB files were manipulated with a C++ script to remove headers and to define each binary protein-protein interface within multi-protein complexes (for NMR structures, only the first structure in the file was analyzed). These interfaces were then inputted in bulk to LoopFinder, which identified peptide loops at protein-protein interfaces as defined by several parameters. First, loops were limited to segments of 4 to 8 consecutive amino acids, in order to conform to molecular mass ranges typical for useful peptide and macrocycle ligands. Another parameter required at least 80% of residues within the loop to reside near the protein-protein interface (having at least one atom within 6.5 Å of the binding partner). We also included a 6.2 Å cutoff between the alpha carbons of the loop termini, to ensure a loop-like conformation and to exclude repeating secondary structures such as β-strands and α-helices. This distance was further restricted to a maximum of 4.67 Å for four-amino-acid loops and 5.83 Å for five-amino-acid loops, in order to eliminate non-loop structures. All of these specific parameters were designed to identify loops that might be amenable to mimicry by small cyclic peptides and other macrocycles. With these parameters, LoopFinder identified 121,086 total loops in 9,388 different structures, including numerous redundancies such as overlapping loops, nested loops, and homologous loops on different chains of homomultimeric protein complexes. These redundant loops were retained for computational alanine scanning because they would not significantly increase the computational burden, and would allow us to select the most critical loops from among them after interaction energies were assigned to each residue.
The complete set of 121,086 loops was then analyzed by computational alanine scanning with PyRosetta v2.012 and Rosetta 3.0 using a modified version of the previously reported scoring function that lacked environment-dependent hydrogen-bonding terms.14–17 The computational alanine scan produced data that is interpreted as the relative, predicted ΔΔGresidue for each residue for that particular PPI. At this point, the loop set was consolidated to eliminate redundant loops, producing a master list of 25,005 interface loops. These interface loops were then sorted by the presence and relative location of hot spot residues, with hot spots defined as ΔΔGresidue ≥ 1 kcal/mol. To generate the set of “hot loops,” we identified loops with two consecutive hot spots, loops with at least three hot spots, and loops for which the average ΔΔGresidue was greater than 1 kcal/mol (Figure 1). This yielded a set of 1,407 hot loops, covering 1,242 multi-chain PDB structures (Supplementary Data Set 1). This represents only 5.6% of the interface loops, highlighting that this process identified those loops that are most critical for mediating PPIs. Further, for each hot loop, the total predicted energy associated with the hot loop was compared to the total predicted energy for the corresponding interface. This analysis revealed that 36% of hot loops are responsible more than half of the predicted binding energy for the associated interface, and 67% of hot loops are responsible for more than a quarter of the predicted binding energy (Supplementary Figure 2). Overall, hot loops represent a significant percentage of the total interface energy, making them ideal starting points for identifying novel targets for macrocycles and constrained peptides.
Figure 1.
Identification of hot loops. Hot loops are identified as those loops that satisfy one or more of three criteria: the average ΔΔGresidue over the entire loop is greater than 1 kcal/mol, the loop has three or more hot spot residues (ΔΔGresidue ≥ 1 kcal/mol), and the loop has two or more consecutive hot spot residues. Representative loops that satisfy each of these criteria are shown within the blue, red and yellow circles (structures from 1AXI, 1GK9, and 1L2U, respectively). Some hot loops satisfy two of these criteria, with representative loops from these categories shown in the purple, orange and green boxes (2QNR, 1GK9 and 2FPF, respectively). In addition, 67 hot loops satisfy all three criteria, an example of which is shown in the gray box to the left (2AST). All structures, rendered in Pymol,42 show the chain at the interface in blue, the binding partner as a gray surface, the hot loop in green, and hot spots in orange (ΔΔGresidue ≥ 1 kcal/mol) and yellow (ΔΔGresidue ≥ 2 kcal/mol). Representative hot loops display a wide range of loop structures and modes of interaction with the partner surface.
Structural classification of interface loops
The sets of 25,005 interface loops and 1,407 hot loops warrant closer characterization in order to better understand loop-mediated PPIs. First, we analyzed the hot loops with respect to loop structure by identifying secondary structures flanking the loops and canonical turns within the loops. Canonical turn motifs were identified by measuring φ and ψ angles and comparing them with motifs from the PDBeMotif database (see Supplementary Note for definitions).18 A breakdown of loop structures is shown in Figure 2. 61% of the hot loops possess specific turn motifs with characteristic backbone torsions and intramolecular hydrogen bonds. Loops with one helical turn made up 11% of the hot loops, and these were typically at the N-terminal or C-terminal cap of an α-helix. This shows that the parameters of LoopFinder were defined conservatively enough to exclude regular α-helices. The specific α-turns identified by LoopFinder have very little overlap with previously identified α-helix-mediated PPIs (see below).10,11
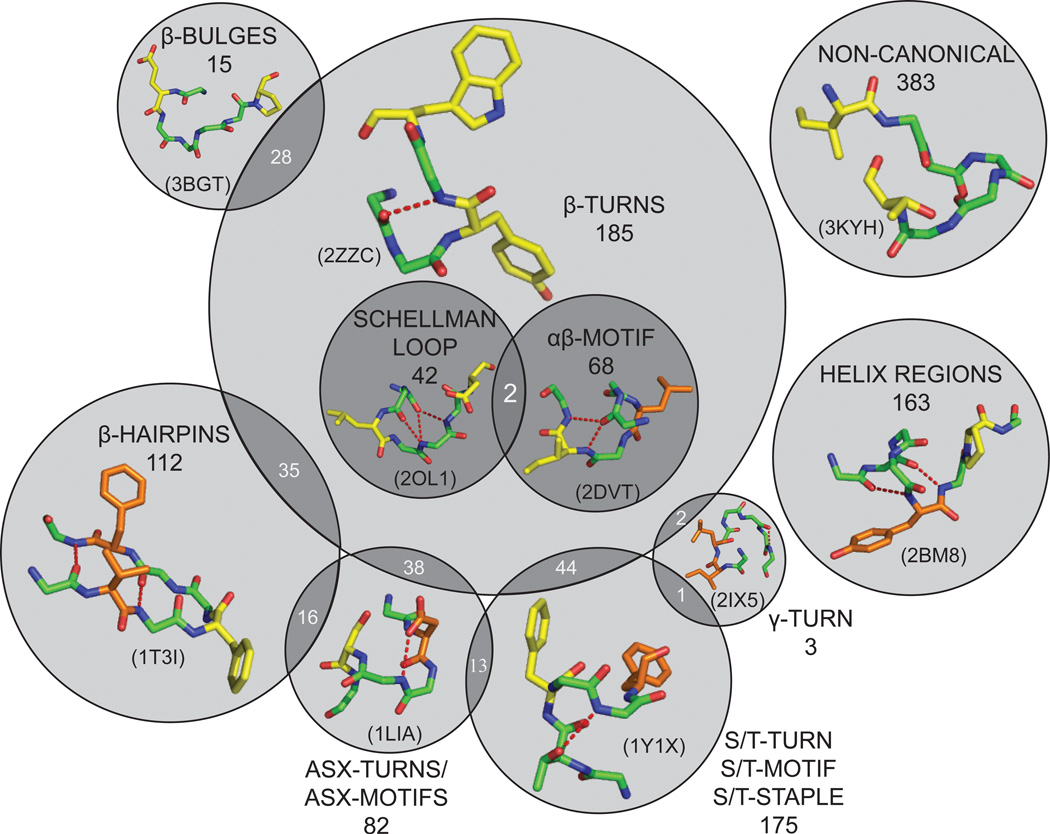
Figure 2.
Visualization of different loop structures observed among the hot loops. Representative examples of each type of loop are shown within each circle, including: β-turns (2ZZC), Schellman loops (2OL1), αβ-motifs (2DVT), β-bulges (3GBT), β-hairpins (1T3I), Asx-turns and motifs (1LIA), S/T-turns, motifs and staples (1Y1X), and γ-turns (2IX5). The remaining two categories shown above are α-helical regions identified by their backbone torsional angles (2BM8), and loops lacking canonical structural motifs (3KYH). All structures, rendered in Pymol,42 show the hot loop in green and hot spots in orange (ΔΔGresidue ≥ 1 kcal/mol) or yellow (ΔΔGresidue ≥ 2 kcal/mol).
Unsurprisingly, β-turns are the most common turns, present in 31% of all hot loops. Specific subcategories of β-turns that are prominent in the hot loop set include αβ-motifs and Schellmann loops. We also found that other structural motifs commonly overlap β-turn regions within hot loops (Figure 2). Another common motif within hot loops is a turn-like motif in which serine or threonine makes a side-chain-to-backbone hydrogen bond. There are three classes of such motifs, S/T-turns, S/T-motifs and S/T-staples, which together appear in 16% of all hot loops. β-hairpins, defined as any loop that connects two antiparallel β-strands regardless of the presence of a β-turn, are present in 11% of hot loops. β-hairpins have been thoroughly studied and successfully mimicked using peptides and peptidomimetics.6 Loops in which aspartate or asparagine form side-chain-to-backbone hydrogen bonds (Asx-turns and motifs) appear in 11% of hot loops. One additional small subset of structured hot loops are β-bulges, which are short breaks within β-strands. Structures of representative hot loops from each of these categories are shown in Figure 2, and demonstrate the wide diversity of loop structures identified by LoopFinder. No immediate correlation was observed among the relative three-dimensional orientations of hot spot residues within each structural class, indicating that, while common structural motifs were observed, different molecular scaffolds may need to be developed to target different structural classes or even different loops within each structural class.
An important finding from the hot loop set is that canonical turn motifs are not essential for loop-mediated PPIs. The loops categorized as “non-canonical” in Figure 2 have unique structures that are nonetheless still excellent starting points for inhibitor design (Supplementary Figure 3). Interestingly, some of these loops act as N-terminal or C-terminal caps of α-helices. There are a wide variety of torsional angles and intramolecular hydrogen bonds within these loops, giving each a unique topology that may promote high selectivity for their respective binding partners. Cyclization of these loops with flexible or rigidified linkers should produce constrained peptides with unique folded structures. These would represent novel three-dimensional scaffolds for targeting these and other PPIs.
How loops use individual residues to mediate PPIs
To quantify the relative energetic contributions of each amino acid to loop-mediated PPIs, we compiled the average ΔΔGresidue for each amino acid for both the interface loop set and the hot loop set (Table 1). The overall trends for both sets are similar, indicating that the hot loops are similar to all interface loops with respect to amino acid usage. The amino acids that have the highest average ΔΔGresidue within all interface loops are tryptophan, phenylalanine, histidine, aspartate, tyrosine, leucine, glutamate, isoleucine, and valine. These amino acids span charged, hydrophobic, and aromatic residues, and contain several striking features. Phenylalanine, which is disfavored for PPI hot-spots in general,19 has a higher average ΔΔGresidue than tyrosine, and almost as high as tryptophan. Thus, whatever causes phenylalanine to be disfavored as a hot spot for PPIs in general does not affect loop-mediated PPIs, making phenylalanine as important for loop-mediated PPIs as tryptophan. Another surprising result is that histidine is a major contributor to the binding energy of hot loops, whereas histidine is not observed at higher proportions as a hot-spot residue for PPIs in general.19 Arginine is not a major contributor to binding energy of loop-mediated PPIs, which is a stark contrast to its major role as one of the most common PPI hot spot residues. Finally, leucine and isoleucine have nearly equal average ΔΔGresidue within the interface loop dataset, and are similarly enriched in hot spots within those loops. Thus, while isoleucine is ten times more likely to be a hot spot than leucine within all PPIs,19 leucine and isoleucine contribute nearly equally to loop-mediated PPIs.
Table 1.
Amino acid bias within all 25,005 interface loops and within the 1,407 hot loops. The average ΔΔGresidue, percent abundance, and fold enrichment for hot spots compared to non-hot-spot positions were calculated for each amino acid within each loop set. See Supplementary Tables 1 and 2 for complete data.
| Residue | Interface Loops | Hot Loops | ||||
|---|---|---|---|---|---|---|
| Avg. ΔΔGresidue |
Percent Abundance |
Fold Enrichment in Hot Spots |
Avg. ΔΔGresidue |
Percent Abundance |
Fold Enrichment in Hot Spots |
|
| Trp | 0.30 | 1.3 | 3.4 | 1.6 | 2.1 | 1.8 |
| Phe | 0.28 | 4.0 | 3.1 | 1.3 | 5.6 | 1.9 |
| His | 0.27 | 2.5 | 2.5 | 1.1 | 3.7 | 1.6 |
| Asp | 0.21 | 5.9 | 1.5 | 1.0 | 6.9 | 1.4 |
| Tyr | 0.17 | 3.7 | 2.5 | 1.1 | 4.7 | 1.6 |
| Leu | 0.15 | 7.7 | 1.4 | 0.83 | 7.6 | 1.3 |
| Glu | 0.15 | 5.9 | 1.3 | 0.86 | 7.1 | 1.2 |
| Ile | 0.14 | 4.2 | 1.5 | 0.95 | 4.2 | 1.3 |
| Val | 0.12 | 5.2 | 0.73 | 0.74 | 4.8 | 0.82 |
| Ser | 0.09 | 6.8 | 0.61 | 0.59 | 6.8 | 0.75 |
| Pro | 0.09 | 4.7 | 0.89 | 1.3 | 5.3 | 1.1 |
| Thr | 0.09 | 5.6 | 0.69 | 0.75 | 5.4 | 0.86 |
| Asn | 0.08 | 4.9 | 0.74 | 0.56 | 4.8 | 0.68 |
| Arg | 0.07 | 5.3 | 1.4 | 0.83 | 6.3 | 1.2 |
| Lys | 0.03 | 4.8 | 0.67 | 0.50 | 4.0 | 0.82 |
| Ala | 0.01 | 8.0 | 0.13 | 0.30 | 5.4 | 0.22 |
| Met | 0.01 | 1.9 | 0.63 | 0.55 | 1.4 | 0.70 |
| Gly | 0.00 | 13 | 0.00 | 0.00 | 9.9 | 0.00 |
| Gln | −0.02 | 3.5 | 0.49 | 0.53 | 2.8 | 0.71 |
| Cys | −0.03 | 1.5 | 0.19 | 0.37 | 1.1 | 0.33 |
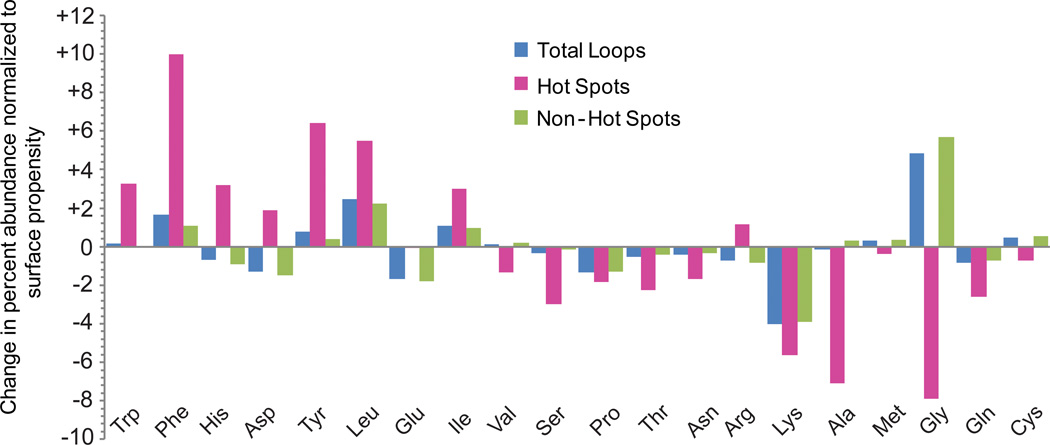
To examine the related question of amino acid abundance within interface loops, we normalized the percent abundance of each amino acid within the interface loop set to the propensity of each amino acid to reside at the protein surface. Then, we broke down these data into change in percent abundance (relative to surface propensity)20 for hot spot positions and non-hot spot positions. Overall, these values were similar for the interface loop set and the hot loops; data for all interface loops are shown in Figure 3. Some of the results of this analysis are not particularly surprising. For instance, glycine residues are highly enriched in the interface loop set, which is to be expected for loop regions and for some of the specific loop architectures identified in Figure 2. Another expected result is the prominence of large and hydrophobic amino acids, since they commonly mediate PPIs and have high average ΔΔGresidue values within loops. The large overabundance of phenylalanine at hot spots within loops agrees with its high average ΔΔGresidue, confirming that hot loops commonly use phenylalanine hot spots to recognize protein targets.
Figure 3.
Interface loops use a unique set of amino acids to recognize their binding partners. The percent abundances of each amino acid were normalized relative to propensity to reside on a protein surface.20 These normalized values were further broken down into all residues (blue), hot spot residues (red) and non-hot spot residues (green).
Proline might also be expected to be prominent in loops, but it is not over-represented in interface loops or hot loops. This is despite the fact that, when it is present, it (on average) contributes significantly to the binding energy (Table 1). Closer examination of the subset of hot loops containing a proline hot spot (179 loops, 13%) further elucidated the roles of “hot prolines” within these loops. For 70% of these loops, the hot proline is the residue that contributes the most to the interaction, and in 11% of these loops it is the only hot spot. In the majority of loops containing a hot proline, the proline sits at the boundary between the loop and an α-helix or β-strand. This suggests that prolines that act as secondary structure breakers can also play prominent roles in intermolecular interactions.
Within the interface loop set, charged amino acids contribute relatively large ΔΔGresidue. However, these are also generally prominent on protein surfaces, and are therefore not over-represented within loops (Figure 3). Thus, charged amino acids play similar roles in loop-mediated PPIs as they do in all PPIs. However, loop-mediated PPIs use arginine, aspartic acid, and glutamic acid in equal amounts, while PPIs in general use arginine more often than other charged residues. Strikingly, lysine is among the most abundant amino acids at protein surfaces and one of the most abundant amino acids in flexible loop regions,19–21 but is greatly under-represented within the non-redundant loop set, both overall and at hot spots. This may be because arginine, aspartate, and glutamate can more readily facilitate hydrogen-bond networks with high cooperativity and stability. The extensive underrepresentation of lysine residues within interface loops, both at hot spots and at non-hot spot positions, distinguishes the interface loops found by LoopFinder from other loops located at the protein surface. This indicates that underrepresentation of lysine (along with the other biases noted above) may be a method for identifying interface loops solely from primary sequence data and secondary structure predictions.
Finally, histidine is over-represented at hot spots within loops (Figure 3), and contributes a very high ΔΔGresidue on average (Table 1). This overall analysis does not distinguish whether histidine is contributing via polar, aromatic, or hydrophobic interactions. Visual inspection of “hot histidine” residues in the hot loop set indicates that histidine acts mainly as a hydrogen bond donor and acceptor, making specific polar contacts both within the loop and to the binding partner. Hydrophobic, Van der Waals, or π-stacking interactions involving the imizadole appear to play a less important role for these histidines.
Comparing loop-mediated PPIs to helix-mediated PPIs
Prior work has comprehensively analyzed PPIs mediated by α-helices in order to provide novel starting points for designing PPI inhibitors; these were recently compiled in a web-accessible database called HippDB.11 To evaluate the degree of overlap between this dataset and the loop-mediated interactions identified by LoopFinder, we cross-referenced the interface loop set to the collection of helical segments from HippDB. We found only 463 complexes were identified by both processes, indicating interfaces made up of surface loops and α-helical regions. Focusing on just the hot loops, we identified only 90 protein structures that were also in HippDB, and only 17 of these contained overlapping sequences between the hot loop and the helix (Supplementary Table 5). Even for these, LoopFinder identified a loop containing at least one additional amino acid outside the helical region for all but one. This reveals that the structural spaces identified by the two methods are essentially orthogonal.
Established and novel targets for inhibitor design
Overall, more than half the hot loops contained consecutive hot spots (Figure 1). Since this subset may be amenable to targeting with small molecules and established β-turn mimetics,5 we chose instead to examine more closely the set of 364 hot loops not containing consecutive hot spots (Supplementary Data Set 2). This set encompasses a diverse set of loop architectures which display hot spot side chains in wide diversity of three-dimensional orientations that may merit the development of new macrocyclic scaffolds. Included in this subset were several established and emerging drug targets that are discussed further below.
A classic PPI, hGH•hGHbp, contains a hot loop
One of the most thoroughly-studied PPIs has been the interaction between human growth hormone (hGH) and the soluble portion of its receptor, human growth hormone binding protein (hGHbp).22–24 We identified two hGH•hGHbp structures among our hot loops. The first consists of native hGH and hGHbp in a 1:2 complex (1HWG), and the second is a mutant interface that was re-optimized by phage display (1AXI).24 LoopFinder identified two hot loops within these structures, P61-E66 of hGH and I165-M170 of hGHbp. The most critical known hot spot within the hGH loop, R64, was accurately identified by our computational alanine scan, and contributed to the inclusion of hGH P61-E66 among the hot loops (Figure 4a). Likewise, the most critical known hot spots within the hGHbp I165-M170 loop, I165 and W169, were also identified by the computational alanine scan and contributed to its inclusion among the hot loops. Overall, the ability to compare our results to such a well-understood PPI speaks to the robustness and predictive power of Rosetta-based computational alanine scans and of LoopFinder.
Figure 4.
Established and novel targets for inhibitor design. a) LoopFinder identified a hot loop on the surface of hGH that is known to be essential for binding of hGHbp (1HWG).24 b) Hot loop within the transcription factor Nrf2 that binds its repressor, Keap1 (2FLU).26 c) The sC-connector loop of TIMP-3 is a hot loop that binds to the S2 pocket of TACE (3CKI).43 d) The interaction between Skp2 and Cks1 is essential for the formation of the SCFSkp2 complex and its ubiquitin E3 ligase activity (2AST).31 e) Inhibition of the histone acetyltransferase (HAT) MSL complex is a novel target identified by LoopFinder (2Y0N).37 All structures, rendered in Pymol,42 show the hot loop in green, and hot spots in orange (ΔΔGresidue ≥ 1 kcal/mol) or yellow (ΔΔGresidue ≥ 2 kcal/mol).
A transcription factor•repressor complex: Nrf2•Keap1
Cellular oxidative stress is associated with many disease states, including inflammation, cardiovascular disease, cancer, and neurodegenerative diseases. A coordinated program of protection from oxidative stressors called the antioxidant response is regulated by the transcription factor Nrf2 (Nuclear factor erythroid-derived-related factor 2). Under normal cellular conditions, Nrf2 remains at low levels through its PPI with the Kelch-like ECH-associated protein 1 (Keap1), which targets Nrf2 for ubiquitin-mediated degradation. The critical role of the Nrf2•Keap1 complex in the cell’s antioxidant response makes it a therapeutically relevant target. However, a lack of specificity associated with this strategy proved to be a major drawback. Inhibition of Nrf2 degradation by blocking the Nrf2•Keap1 PPI has the potential to be a more selective method of Nrf2 activation compared to prior work using nonspecific Michael acceptors to modify cysteines on Keap1.25 LoopFinder identified a six-residue sequence, D77-E82 (DEETGE), from the crystal structure of Keap1 bound to a Nrf2-derived peptide (2FLU; Figure 4b).26 This β-hairpin loop was previously identified as critical for this PPI, and a 16-residue peptide containing this loop (A69-L85) binds Keap1 with a Kd of 24 nM, retaining much of the affinity of full-length Nrf2 (Kd of 5 to 9 nM).27 Shorter peptides within this loop have binding affinity for Keap1 in the 100–350 nM range.27,28 Macrocyclic peptides derived from this hot loop have also been developed – these have IC50 values in the 15 nM range.29 Thus, in the case of Nrf2•Keap1, LoopFinder identified a key β-hairpin loop that directly translated into peptide inhibitors with low nanomolar affinity. This example illustrates the utility of LoopFinder as a resource for identifying hot loops as starting points for developing PPI inhibitors.
A protease•protease inhibitor complex: TACE•TIMP-3
Another hot loop identified by LoopFinder is the complex between tissue inhibitor of metalloproteinases 3 (TIMP-3) and TNF-α converting enzyme (TACE). Numerous peptide and small molecule inhibitors of TACE, many of which are broad-spectrum matrix metalloproteinase inhibitors, have been identified by academic and industrial groups.29 All of these molecules have targeted the catalytic site of TACE, most using a hydroxymate moiety to bind the catalytic zinc. The most successful small molecule dropped out of Phase II clinical trials due to concerns about specificity.29
TIMP-3 is an extracellular protein that binds TACE with sub-nanomolar affinity, inhibiting proteinase activity. The TIMP-3•TACE interaction has been extensively studied both in vitro and in vivo. The TIMP-3•TACE interaction is facilitated by three interface epitopes within TIMP-3 that bind TACE (PDB 3CKI).29 LoopFinder identified one of these epitopes, called the sC-connector loop, as a hot loop for the TIMP-3•TACE interaction (Figure 4c). The hot loop consists of a six-residue stretch from S64 to G69 (SESLCG), in which S64 and L67 are identified as hot spots (ΔΔGresidue = 3.62 kcal/mol and 1.73 kcal/mol respectively). Notably, S64 and L67 are unique residues within TIMP-3 compared to TIMP-1, TIMP-2, and TIMP-4, which show drastically reduced binding to TACE.29 In addition, this region is structurally constrained within TIMP-3 via a disulfide bond from C68 to Cys1. The binding pocket for the hot loop has been identified an “alternative pocket” on TACE that could be as important as the actual catalytic zinc active site,29 but this loop within TIMP-3 has never been used as a starting point for designing TACE inhibitors. Thus, identification of S64-G69 as a hot loop suggests specific designs for non-zinc-chelating inhibitors of TACE. Such inhibitors would have immense therapeutic potential as highly selective TACE inhibitors that do not bind other ADAMs or matrix metalloproteinases.
An E3 ligase complex: Skp2•Cks1
p27Kip1 is a G1-checkpoint protein that directly and indirectly regulates many components of the eukaryotic cell cycle, and enhanced degradation of p27Kip1 is associated with many common cancers. p27Kip1 is targeted for proteolysis by the ubiquitin E3 ligase SCFSkp2, which is a complex of Skp1, Cul1, Rbx1, and Skp2.30 Ubiquitination of p27Kip1 also requires the binding of an accessory protein, Cdc kinase subunit 1 (Cks1), to the SCFSkp2 complex. In crystal structures of the SCFSkp2 complex (2ASS) and SCFSkp2 bound to p27Kip1 (2AST),31 LoopFinder identified a six-residue loop on the surface of Cks1 comprising M38 to W43 (MSESEW). This loop contains four consecutive hot spot residues from S39 to E42 (ΔΔGresidue = 1.11 kcal/mol, 1.93 kcal/mol, 2.07 kcal/mol, and 2.94 kcal/mol respectively) located at the N-terminal cap of an α-helix (Figure 4d). Experimental mutagenesis has shown that S41 is essential for SCFSkp2 complex activity, confirming the importance of this hot loop.32 The S39-E42 hot loop represents a novel starting point for designing SCFSkp2 inhibitors using α-turn-mimicking scaffolds. Because prior small-molecule screening efforts yielded only weak inhibitors,33–35 it is likely that the shallow Skp2•Cks1interface is better targeted by constrained peptides or macrocycles.
A histone acetyltransferase complex: Msl1•Msl3
MOF (males-absent on the first) is a histone acetyltransferase (HAT) that exclusively catalyzes the acetylation of histone 4 lysine 16 (H4K16). Only a handful of HAT inhibitors have been discovered, and all target the catalytic site. MOF requires complexation with three regulatory proteins (MSL1, MSL2 and MSL3) for activity.36,37 LoopFinder identified two hot loops as critical for the formation of the MSL complex (4DNC).36 One of these is V575 to P580 (VAFGRP) on MSL1, with Phe577 and Arg579 as hot spots (ΔΔGresidue = 2.74 kcal/mol and 4.5 kcal/mol respectively; Figure 4e). This loop is highly conserved across eukaryotes (Supplementary Figure 7) and forms a β-hairpin-like structure that binds a shallow pocket on MSL3 (2Y0N).37 Co-immunoprecipitation experiments showed that variants of MSL1 with mutations in this loop have substantially lower MSL3 affinity, and that a residue that makes up the binding pocket on MSL3, F484, is essential for recognition of MSL1.37 The other hot loop in the MOF-MSL1/2/3 complex is H183 to G188 (HIGNYE) of MOF, which binds MSL1 using hot spot residues Asn186 and Glu188 (ΔΔGresidue = 1.54 kcal/mol and 3.23 kcal/mol respectively; Supplementary Figure 8). To our knowledge, there are no known inhibitors of any member of the MSL complex. LoopFinder has thus identified a hot loop and corresponding binding pocket that may represent a novel druggable interface for targeting cellular HAT activity.
DISCUSSION
Despite the large interfaces of most PPIs, it has long been shown that residues at the interface do not contribute equally to the binding interaction.19–21 “Hot spots” have been defined as individual residues that contribute a significant portion of binding free energy to the overall interaction (often, ΔΔG e 1 kcal/mol).38,39–41A computational alanine scanning engine based on Rosetta has been developed to computationally predict PPI interface hot spots, typically in 79% agreement with experimental values.14,15,39–41 Using this approach, computational alanine scans of protein-protein complexes have yielded a wide range of information about the different roles specific residues play at PPI interfaces. In this work, we extend this methodology to short peptide loops, and identify unique structures and properties of “hot loops” that play key roles in diverse PPIs.
LoopFinder is a useful tool for searching structure databases for peptide loops at protein-protein interfaces. LoopFinder identified all PPI interface loops in the PDB, 25,005 in total. Parameters within LoopFinder can be tailored to perform custom searches within our dataset or the entire PDB for loops of specific size or geometry. Three criteria (average ΔΔGresidue, presence of three hot spot residues, and presence of two consecutive hot spot residues) were used to identify “hot loops” as those loops that contribute maximally to the PPI. Other criteria can be readily added in the future. We speculate that hot loops with two consecutive hot spots may be unique loops that could be mimicked by traditional small molecules, while those with broader interaction surfaces represent starting points for the design of constrained peptides and other macrocycles. While the design of constrained peptides and small molecule macrocycles is inherently complex, the identification of many potentially fruitful starting points and targets will accelerate this growing field. The LoopFinder algorithm, the set of 25,005 interface loops, and our culled list of 1,407 hot loops are valuable resources for the development of constrained peptides and macrocycles, since these typically have well-defined constraints and topologies that must be matched precisely for successful inhibitor design.
Online Methods
Protein-protein structures were downloaded from the PDB (August, 2013) using the advanced search function. Structures with > 4 Å resolution or with > 90% similarity were excluded. The resultant structures were than analyzed with LoopFinder, a program written in C++. For all PDB files, the headers were first removed. For NMR structures, only the first structure in the file was considered. For multi-chain biological assemblies, PDB files were split into new files containing each possible binary interface.
Interface residues were identified as any amino acid containing at least one heavy atom within 6.5 Å of another heavy atom on the partner chain. Loops were then identified using the following criteria. 1) Loops contain 4-8 consecutive amino acids, a length suitable for incorporation into a synthetically feasible cyclic peptide. 2) The Cα-Cα distance of the loop termini is limited to a maximum of 6.2 Å for loops of 6–8 amino acids (4.67 Å for 4-amino-acid loops; 5.83 Å for 5-amino-acid loops), to exclude extended secondary structure elements such as alpha helices and beta strands and to ensure that the N- and C-termini of the loop are in relative positions amenable to cyclization. 3) Loops must contain at least 80% interface residues (at least one heavy atom within 6.5 Å of another heavy atom on the partner chain).
Interface loops were then subjected to a computational alanine scan using PyRosetta v2.012 and Rosetta 3.0. The PyRosetta alanine scanning script originally developed by the Gray lab (http://graylab.jhu.edu/pyrosetta/downloads/scripts/demos/D090_Ala_scan.py) was modified to implement a modified score function and to be run in a parallel manner on Tufts’s research cluster. The score function was parameterized to match previously reported general computational alanine scanning protocols,14 but without environment-dependent hydrogen-bonding terms:
ddG_scorefxn = create_score_function_ws_patch('standard','score12')
ddG_scorefxn.set_weight(fa_atr, 0.44)
ddG_scorefxn.set_weight(fa_rep, 0.07)
ddG_scorefxn.set_weight(fa_sol, 0.32)
ddG_scorefxn.set_weight(hbond_bb_sc, 0.49)
The results of the alanine scans were then combined with the loop data from LoopFinder using a Python script. Sequence data for each loop was combined with the computational alanine scan results to derive a set of hot loops. ΔΔG values that exceeded 4.5 kcal/mol were reduced to 4.5 prior to determining ΔΔGloop,avg., ΔΔGloop,sum, or ΔΔGinterface based on previous limits set for computation alanine scan values and to avoid biasing the data set in favor of loops with a single hot spot.14,44 These hot spots represented 8.2% of the hot spots found in interface loops, and were further enriched in the set of hot loops (14.8%).
A web-based interface for LoopFinder is currently under construction. In the meantime, LoopFinder is freely available for use. Requests for binary files or code can be sent to joshua.kritzertufts.edu.
Supplementary Material
Acknowledgments
J.G. was supported in part by NIH/NIGMS IRACDA grant K12GM074869. T.R.S. was supported in part by Dept. of Education GAANN grant P200A090303. This work was supported in part by NIH DP20D007303 to J.A.K. The authors thank the Tufts Technology Services for research cluster access and Prof. Rebecca Scheck for helpful conversations.
Footnotes
Author Contributions
B.S. wrote the LoopFinder code. J.G., B.S. and J.A.K. troubleshooted, debugged, and parameterized Loopfinder and Rosetta-based computational alanine scanning. J.G., T.S., M.E. and J.A.K. analyzed and contextualized data. J.G., T.S., and J.A.K. produced figures and tables and wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 2.Marsault E, Peterson ML. Macrocycles Are Great Cycles: Applications, Opportunities, and Challenges of Synthetic Macrocycles in Drug Discovery. J. Med. Chem. 2011;54:1961–2004. doi: 10.1021/jm1012374. [DOI] [PubMed] [Google Scholar]
- 3.Bock JE, Gavenonis J, Kritzer JA. Getting in Shape: Controlling Peptide Bioactivity and Bioavailability Using Conformational Constraints. ACS Chem. Biol. 2012;8:488–499. doi: 10.1021/cb300515u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vagner J, Qu H, Hruby VJ. Peptidomimetics, a synthetic tool of drug discovery. Curr. Opin. Chem. Biol. 2008;12:292–296. doi: 10.1016/j.cbpa.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowick JS. Exploring β-Sheet Structure and Interactions with Chemical Model Systems. Acc. Chem. Res. 2008;41:1319–1330. doi: 10.1021/ar800064f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of [alpha]-helix-mediated protein-protein interactions using designed molecules. Nat Chem. 2013;5:161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- 7.Guharoy M, Chakrabarti P. Secondary structure based analysis and classification of biological interfaces: identification of binding motifs in protein–protein interactions. Bioinformatics. 2007;23:1909–1918. doi: 10.1093/bioinformatics/btm274. [DOI] [PubMed] [Google Scholar]
- 8.Guney E, Tuncbag N, Keskin O, Gursoy A. HotSprint: database of computational hot spots in protein interfaces. Nucleic Acids Res. 2008;36:D662–D666. doi: 10.1093/nar/gkm813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koes D, et al. Enabling Large-Scale Design, Synthesis and Validation of Small Molecule Protein-Protein Antagonists. PLoS ONE. 2012;7:e32839. doi: 10.1371/journal.pone.0032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jochim AL, Arora PS. Systematic Analysis of Helical Protein Interfaces Reveals Targets for Synthetic Inhibitors. ACS Chem. Biol. 2010;5:919–923. doi: 10.1021/cb1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergey CM, Watkins AM, Arora PS. HippDB: A database of readily targeted helical protein-protein interactions. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.London N, Raveh B, Movshovitz-Attias D, Schueler-Furman O. Can self-inhibitory peptides be derived from the interfaces of globular protein–protein interactions? Proteins Struct. Funct. Bioinforma. 2010;78:3140–3149. doi: 10.1002/prot.22785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chrencik JE, et al. Structure and Thermodynamic Characterization of the EphB4/Ephrin-B2 Antagonist Peptide Complex Reveals the Determinants for Receptor Specificity. Structure. 2006;14:321–330. doi: 10.1016/j.str.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein–protein complexes. Proc. Natl. Acad. Sci. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kortemme T, Kim D, Baker D. Computational alanine scanning of protein-protein interfaces. Sci. STKE Signal Transduct. Knowl. Environ. 2004;2004 doi: 10.1126/stke.2192004pl2. pl2. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhury S, Lyskov S, Gray JJ. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics. 2010;26:689–691. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ. Spatial chemical conservation of hot spot interactions in protein-protein complexes. BMC Biol. 2007;5:43. doi: 10.1186/1741-7007-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golovin A, Henrick K. MSDmotif: exploring protein sites and motifs. BMC Bioinformatics. 2008;9:312. doi: 10.1186/1471-2105-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 20.Janin J, Miller S, Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 21.Tsai C-J, Lin SL, Wolfson HJ, Nussinov R. Studies of protein-protein interfaces: A statistical analysis of the hydrophobic effect. Protein Sci. 1997;6:53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham BC, Wells JA. Rational design of receptor-specific variants of human growth hormone. Proc. Natl. Acad. Sci. 1991;88:3407–3411. doi: 10.1073/pnas.88.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham BC, Wells JA. Comparison of a Structural and a Functional Epitope. J. Mol. Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 24.Sundström M, et al. Crystal Structure of an Antagonist Mutant of Human Growth Hormone, G120R, in Complex with Its Receptor at 2.9 Å Resolution. J. Biol. Chem. 1996;271:32197–32203. doi: 10.1074/jbc.271.50.32197. [DOI] [PubMed] [Google Scholar]
- 25.Hong DS, et al. A Phase I First-in-Human Trial of Bardoxolone Methyl in Patients with Advanced Solid Tumors and Lymphomas. Clin. Cancer Res. 2012;18:3396–3406. doi: 10.1158/1078-0432.CCR-11-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo S-C, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1 : Nrf2 interface provides mechanistic insight into Nrf2 signaling. Embo J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Inoyama D, Kong A-NT, Beamer LJ, Hu L. Kinetic Analyses of Keap1–Nrf2 Interaction and Determination of the Minimal Nrf2 Peptide Sequence Required for Keap1 Binding Using Surface Plasmon Resonance. Chem. Biol. Drug Des. 2011;78:1014–1021. doi: 10.1111/j.1747-0285.2011.01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock R, Schaap M, Pfister H, Wells G. Peptide inhibitors of the Keap1-Nrf2 protein-protein interaction with improved binding and cellular activity. Org. Biomol. Chem. 2013;11:3553–3557. doi: 10.1039/c3ob40249e. [DOI] [PubMed] [Google Scholar]
- 29.Horer S, Reinert D, Ostmann K, Hoevels Y, Nar H. Crystal-contact engineering to obtain a crystal form of the Kelch domain of human Keap1 suitable for ligand-soaking experiments. Acta Crystallogr. Sect. F. 2013;69:592–596. doi: 10.1107/S174430911301124X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Q, et al. Targeting the p27 E3 ligase SCFSkp2 results in p27-and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao B, et al. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCFSkp2 ubipuitin ligase. Mol. Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Sitry D, et al. Three Different Binding Sites of Cks1 Are Required for p27-Ubiquitin Ligation. J. Biol. Chem. 2002;277:42233–42240. doi: 10.1074/jbc.M205254200. [DOI] [PubMed] [Google Scholar]
- 33.Huang K, Vassilev LT. High-throughput screening for inhibitors of the Cks1-Skp2 interaction. Methods Enzymol. 2005;399:717–728. doi: 10.1016/S0076-6879(05)99047-2. [DOI] [PubMed] [Google Scholar]
- 34.Ungermannova D, et al. High-Throughput Screening AlphaScreen Assay for Identification of Small-Molecule Inhibitors of Ubiquitin E3 Ligase SCFSkp2-Cks1. J. Biomol. Screen. 2013;18:910–920. doi: 10.1177/1087057113485789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu L, et al. Specific Small Molecule Inhibitors of Skp2-Mediated p27 Degradation. Chem. Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, et al. Structural insight into the regulation of MOF in the male-specific lethal complex and the non-specific lethal complex. Cell Res. 2012;22:1078–1081. doi: 10.1038/cr.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadlec J, et al. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol. 2011;18:142–149. doi: 10.1038/nsmb.1960. [DOI] [PubMed] [Google Scholar]
- 38.Clackson T, Wells J. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 40.Gohlke H, Kiel C, Case DA. Insights into Protein–Protein Binding by Binding Free Energy Calculation and Free Energy Decomposition for the Ras–Raf and Ras–RalGDS Complexes. J. Mol. Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 41.Moreira IS, Fernandes PA, Ramos MJ. Hot spots—A review of the protein–protein interface determinant amino-acid residues. Proteins Struct. Funct. Bioinforma. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 42.DeLano WL. The PyMol Molecular Graphic System. DeLano Scientific LLC; at < www.delanoscientific.com>. [Google Scholar]
- 43.Wisniewska M, et al. Structural Determinants of the ADAM Inhibition by TIMP-3: Crystal Structure of the TACE-N-TIMP-3 Complex. J. Mol. Biol. 2008;381:1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- 44.Fersht AR, et al. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.