Abstract
The Interferon-gamma Enzyme-Linked ImmunoSpot (IFN-γ ELISpot) assay has been developed and used as an end-point assay in clinical trials for infectious diseases and cancer to detect the magnitude of antigen-specific immune responses. The ability to compare data generated by different laboratories across organizations is pivotal to understand the relative potency of different therapeutic and vaccine strategies. We developed an external proficiency program for the IFN-γ ELISpot assay that evaluates the laboratory performance based on five parameters: timeliness for data reporting; ability to handle cellular samples; detection of background (non-specific) responses; accuracy to consensus of the results; and precision of the measurements. Points are awarded for each criterion, and the sum of the points is used to determine a numeric and adjectival performance rating. Importantly, the evaluation of the accuracy to the consensus mean for the detection of antigen-specific responses using laboratory-specific procedures informs each laboratory and its sponsor on the degree of concordance of its results with those obtained by other laboratories. This study will ultimately provide the scientific community with information on how to organize and implement an external proficiency program to evaluate longitudinally the performance of the participating laboratories and, therefore, fulfill the requirements of the GCLP guidelines for laboratories performing end-point IFN-γ ELISpot assay for clinical trials.
1. Introduction
The enzyme-linked Immunospot (ELISpot) assay was described more than 21 years ago for the detection of antigen-specific immune cells at the single cell level (Czerkinsky et al., 1984). The utility of the IFN-γ ELISpot assay in detecting antigen-specific T-cells was initially demonstrated in models of autoimmune and infectious diseases (Link et al., 1991; Mahanty et al., 1992; Olsson et al., 1990). It has been reported that many components of the ELISpot assay can contribute to variability of the results obtained by laboratories utilizing different assay procedures (Cox et al., 2005). A follow-up to this initial study, provided more details on the possible variables that influences the results obtained with the IFN-γ ELISpot assay (Janetzki et al., 2007). Further efforts have been devoted to perform formal validation of the IFN-γ ELISpot assay to be used as end-point assay in vaccine clinical trials (Russell et al., 2003) and to provide the field with specific information on the aspect for the validation of this assay (Janetzki and Britten, 2011; Janetzki et al., 2005). Over all these studies have led to the optimization of the assay through the introduction of specifically designed antibodies, 96-well plates, substrate kits, and other modifications has broadened the potential uses for the IFN-γ ELISpot assay. Today, it is being used for a wide range of applications including the following: monitoring responses in cancer patients undergoing immunotherapeutic treatment (Leffers et al., 2009; Palmer et al., 2009; Schuetz et al., 2009), and monitoring specific immune response patterns in patients with infectious (reviewed by Walker and Slifka (Walker and Slifka, 2010)), neoplastic (Kabingu et al., 2009; Leffers et al., 2009), or autoimmune diseases (Zanone et al., 2010). Additionally, it has been an important tool in the identification of immunodominance and escape mutations in HIV-1 infection (Goonetilleke et al., 2009; Streeck et al., 2008) as well as in the development of specific AIDS vaccine strategies (Goepfert et al., 2005; 2007; Graham et al., 2010; Russell et al., 2003; Spearman et al., 2009).
Overall, for the past two decades the IFN-γ ELISpot assay has been a highly sensitive, yet reproducible and simple platform to detect and quantify antigen-specific T-cell responses. Because of these properties and its applications to monitoring the immune responses using cryopreserved cells in multi-national clinical trials (Mashishi and Gray, 2002; Russell et al., 2003), this assay has become the benchmark for analysis of T cell responses according to Good Clinical Laboratory Practice (GCLP) guidance (Ezzelle et al., 2008).
One of the requirements of the GCLP guidelines is that laboratories performing validated end-point clinical assays must participate in an external proficiency (EP) program (Sarzotti-Kelsoe et al., 2009). To support the HIV vaccine trials efforts, the National Institute of Allergy and Infectious Diseases, Division of AIDS (NIAID DAIDS) contracted SeraCare Bioservices, Inc. to develop an IFN-γ ELISpot PT program starting in 1998. During the time SeraCare Bioservices had this contract, eight EP rounds were completed with sites receiving summary statistics for each EP, but no grading of performance evaluation.
In 2011, Duke University was awarded the NIAID DAIDS External Quality Assurance Program Oversight Laboratory (EQAPOL). One aim of the EQAPOL program was to continue the IFN-γ ELISpot EP program with the implementation of grading criteria to assess assay performance when different assay protocols are utilized to evaluate T cell responses in a common set of samples using the same antigens. Since inception of the EQAPOL ELISpot program, we have completed five EP rounds and implemented grading criteria starting in the fourth external proficiency round (EP4). The ELISpot program was instituted with a primary goal to evaluate the performance of different laboratory-specific assay protocols and determine their level of concordance in detecting and quantifying T cell responses. In addition, EQAPOL developed a reference ELISpot assay that can be performed by the participating laboratory as a common testing platform to identify problems related to reagents and/or In-house testing procedures. We utilized this assay to troubleshoot the performance of each laboratory and to provide constructive feedbacks to sites with suboptimal performance.
In this study, we report our effort to organize an external proficiency program that allow the participant laboratories to compare their results using the in-house validated assay procedures and to receive a score that can help to evaluate their performance over time. In addition, we provided the participating laboratories with a common assay platform that could be used to resolve differences in the results. We believe that this EP approach can be utilized by other network and provide useful information to the institutions sponsoring the end-point laboratory programs.
2. Methods
2.1 Cellular samples
Peripheral Blood Mononuclear Cells (PBMCs) utilized to perform the IFN-γ ELISpot assay were collected, processed, and stored according to the procedures described elsewhere in this special issue (Garcia, et al., in this issue). The cellular samples utilized in the SeraCare BioServices Inc. and EQAPOL ELISpot programs were collected utilizing similar procedures. All the samples were collected according to protocols approved by the Duke University Medical Center Institutional Review Board.
2.2 Peptide pools
The peptide pools utilized for this EP program were purchased from JPT Peptide Technologies (Berlin, Germany). The peptides were >90% purity as defined by the certificate of analysis and provided in small lyophilized aliquots that were reconstituted with DMSO and dH20 (1:1) just prior to shipment of the reagents to the participating site laboratories. The first pool was constituted by the 23 peptides that represent the CMV, EBV, and Influenza MHC class I restricted epitopes previously described (CEF pool) (Currier et al., 2002). The second pool represented the sequence of the CMV pp65 protein and was constituted by 15 amino acid peptides overlapping by 11 amino acids.
2.3 EQAPOL ELISpot assay
The EQAPOL ELISpot Oversight Laboratory (EOL) performs the ELISpot assay according to the method described below. Cryopreserved PBMCs were thawed, washed and rested overnight before use in the IFN-γ ELISpot assay. The recovery and viability after thawing and after overnight rest were evaluated using the Guava cell counter (Millipore: Billerica, Massachusetts). The IFN-γ ELISpot assay was performed using Mabtech's pre-coated plates (Mabtech; Mariemont, Ohio); Mabtech biotinylated anti-IFN-γ 7-B6-1 monoclonal antibody was used as detecting Ab; Streptavidin-HRP complex (Mabtech; Mariemont, Ohio) and Nova Red substrate (Vector Laboratories; Burlingame, CA) were used to reveal the presence of spots. Spots formed by IFN-γ-secreting cells were counted with an automated ImmunoSpot plate reader (Cellular Technologies; Cleveland Ohio), and results are presented as spot-forming cells (SFC) per 106 PBMCs. For all samples, the cells were evaluated for their functionality using the CEF and CMVpp65 peptide pools at the concentration of 1μg/ml per peptide.
2.4 EQAPOL Oversight Laboratory ELISpot kit and assay
For each proficiency panel, we provided the participant laboratories with cryopreserved PBMCs and peptide pools to be utilized in their standard assays as a kit. Sites were instructed to use the PBMCs and peptides in their assay according to their own protocol. In addition, we provided the reagents and the standard operating procedure documents to perform the reference assay as described in the previous section (Section 2.3). The EQAPOL kit was provided to the laboratories for troubleshooting purposes only as discussed in this manuscript.
2.5 Centralized reading of the ELISpot plates
We required each site to send the plates utilized for each EP, post-completion, to the EOL for centralized reading. These readings were not utilized for the purpose of generating grading criteria. We implemented this procedure to verify the In-house reading and determine if there were issues related to the In-house ELISpot reader. The centralized reading was performed with the automated ImmunoSpot plate reader (Cellular Technologies, Cleveland Ohio). The maintenance of the instrument and the reading procedure were performed under oversight of the Quality Assurance Unit according to established standard operating procedures (see Todd et al. in this issue)
2.6 Shipping of reagents to participating laboratories
For each EP, we provided the sites with nine vials of cryopreserved PBMCs from the three selected donors randomized as Sample 1,2 or 3 and utilized a liquid nitrogen dryshipper as reported elsewhere in this issue (Garcia, et al.). The donor samples utilized for each EP included a non-responder, low responder, and high responder. We also provided three vials of peptide pools (Control, CMV, and CEF) labeled as Reagent A, B, or C reconstituted in DMSO using the same shipper. In order for the sites to test the samples using the EQAPOL reference assay, we provided the sites with reagents described in the ELISpot assay section that included: two capture-antibody coated plates; one vial of the secondary antibody; one vial of the streptavidin-HRP; and one Nova Red Substrate kit. These reagents were supplied using a wet ice shipment. All reagents were tested by the EOL in bridging studies to determine their performance and provide the sites with the correct information on the amount/dilution to be used (data not shown).
2.7 Participating laboratories
Eleven laboratories participated in the EQAPOL ELISpot EP program. Six of these laboratories were located in the US and one in each of the following countries: United Kingdom, Germany, Switzerland, Tanzania, and Thailand.
2.8 Analysis of the results
During the first two years of the program, EQAPOL developed a web-based system that allows the laboratory to automatically upload all the data and information required by the statistical group to analyze the results and for the laboratories to communicate with the program manager and the principal investigator (for further description of EQAPOL web-based application see Todd et al, in this issue). Each site completed an online questionnaire that collects the information relative to any aspect of laboratory practice and assay procedures that have been previously reported as relevant source of variation in assay performance (Cox et al., 2005; Janetzki et al., 2007). The requested information included the operational status of the laboratory with regard to reagent utilized for the assay, the type of instruments utilized for the assay and acquisition of the data (i.e., plate washer and ELISpot reader), and other GCLP related information (see Supplementary Figure 1). The statistical analysis of the results performed to implement the scoring system is reported by Rountree and collaborators (Rountree et al., 2014).
2.9 Site Performance Evaluation System
We developed a scoring system to evaluate the performance of each laboratory with input from the contract sponsor (NIAID/DAIDS), the ELISpot Advisory Committee and the EQAPOL Scientific Advisory Board. The scoring system is based on five grading criteria identified by the EQAPOL ELISpot team: 1) timeliness for data reporting; 2) PBMC sample handling; 3) background; 4) accuracy to the consensus; 5) precision. These criteria are summarized in Table 1. We requested each participating laboratory to perform the assay according to their established protocols (in-house assay) and with the provided ELISpot kit (EQAPOL assay) using 200,00 PBMC per wells. The instructions required to test each sample in 9 replicate wells using a common plate layout. Only the results of the in-house assay and not the EQAPOL assay were used for final grading purposes. The descriptions of each criterion and data for each evaluation were included in each report provided to the participating laboratories. Criteria were weighted to allow for a maximum of 100 points to be earned in a given EP. We utilized the scores to establish ranges that can define the performance as excellent, good, fair, and poor (Table 2). All EQAPOL programs have currently adopted these standard ranges. The goal of establishing these grading criteria and performance rating are to aid sponsors in identifying areas of strength and weakness in site proficiency with respect to ELISpot assays.
Table 1.
EQAPOL ELISpot Grading Criteria
| Criteria | Description | Target | Points |
|---|---|---|---|
| Timeliness | On time EP valid data and questionnaire upload | Per due date set by EQAPOL | • 10 points total |
| PBMC Handling | Viability: Pass/Fail per each donor sample | Donor must be >80% after overnight rest (day 1) | •9 points total •1.5 points available per donor for viability •1.5 points available per donor for recovery |
| Recovery: Pass/Fail per each donor sample | Donor must be between 70-120% after overnight rest (day 1) | ||
| Background | • Pass/Fail per each donor sample | Average for each donor must be <10 | •9 total •3 points available per donor |
| Accuracy to the consensus | • Assessed for each donor sample and for each stimulation condition (CMV and CEF) | Site's average for 9 wells must not be significantly different from consensus average | •54 points total •9 point deduction for each donor by stim condition significantly different from consensus average |
| Precision | • Assessed for each donor sample for each stimulation condition (CMV and CEF) | The dispersion must be less than 95% CI boundary | •18 points total •3 points deducted for each donor/stimulation outside of range |
Table 2.
EQAPOL Performance Ranges
| Overall Performance Score | Performance |
|---|---|
| 91-100 | Excellent |
| 75-90 | Good |
| 66-74 | Fair |
| 0-65 | Poor |
2.10 Timeliness grading criteria
Sites were given four weeks from kit receipt to complete the assay and return the data reporting Excel file and post-assay questionnaire. Failure to upload valid data and survey responses by the due date resulted in a site's loss of all proficiency points for timeliness (10 points).
2.11 PBMC handling grading criteria
Sites were instructed to thaw and count PBMCs according to their own in-house methods. Viability and recovery were reported at the time of thaw (Day 0) and after an overnight rest (D1). PBMC samples were pre-screened prior to the send-out to ensure that were within the acceptability criteria for viability and recovery. For assessment, we adopted the criteria from the Immunology Quality Assessment (IQA) program of acceptable PBMC viability and recovery after overnight resting (Weinberg et al., 2010), unless the mean for all sites was outside of these boundaries. In this regard, we considered that viability after overnight rest should be greater than 80%, and any donor viability less than 80% resulted in a 1.5 point deduction. In addition, the recovery after overnight rest should be between 70 to 120%, and any donor recovery outside of the boundary resulted in a 1.5 point deduction.
2.12 Background grading criteria
Although we recognize that each assay can be validated to accept a certain degree of background, defined as presence of spots in absence of any cellular stimulation, we implemented an overall evaluation of the background observed by the sites and reported to the program. For proficiency grading, we established that the average background (PBMC in the presence of culture medium + DMSO only) SFC per 2×105 PBMC for the 9 wells per donor had to be less than 10. Only site reported In-house assay data were assessed for grading. For the purposes of grading assessment, background data are rounded up so the result is an integer (e.g., 8.2 is rounded up to 9).
2.13 Accuracy to the consensus average grading criteria
Accuracy is commonly defined as the closeness of an estimate to either a true (known) value or an accepted reference standard. However, for assays that evaluate individual antigen-specific cellular responses such as the ELISpot assay, there is not a true value. Therefore, we used the consensus average as the accepted reference. As a consequence, accuracy to the consensus average was assessed for each donor by stimulation (CEF and CMV) based on a comparison of the site's average of the nine wells to the consensus average for all sites reporting data. All antigen-specific data were background-subtracted; the average background for that donor was subtracted from all CEF and CMV SFC values. Accuracy to consensus average was assessed using a mixed effects model based on a Poisson distribution (SAS 9.2 Proc GLIMMIX) using the consensus average of all participating sites with acceptable data (e.g., exclude if all data is missing or near zero for a condition that should have a response, an indication of potential technical errors). This model estimates the mean (the population average using data from all sites) as the intercept, which is a fixed effect, and each site as a random effect that deviates from the intercept. Thus each site is compared to the consensus average of all sites.
A False Discovery Rate correction for the significance level was used to determine which sites are significantly different from the consensus average. When a site is significantly different for a donor/stimulation condition, a nine point deduction is made (max of 54 point reduction). For proficiency grading, only In-house data was utilized. A further description of accuracy to consensus measures is reported by Rountree et al. in this issue.
2.14 Precision grading criteria
Precision describes the amount of variability between repeated measures under unchanged conditions (i.e. the same operator, same instrument, etc.). These are the type of data collected in the EQAPOL ELISpot Program. Thus, precision was based on an acceptable level of variability (dispersion = variance/mean) within each site's 9 wells for each of the 6 donor by stimulation conditions. For proficiency grading, a dispersion upper limit was calculated using background-subtracted data. First, the 90th percentile for all 6 donor by stimulation conditions was calculated and used as a cutoff for extreme values. Using these remaining 90% of the data the mean and standard deviation was calculated and a 95% CI was calculated and the upper bound is the dispersion limit. Once again, we did not use data for which the presence of noticeable errors was observed, such as high background in unstimulated condition or clear plating errors for samples/reagents. Three points were deducted for each donor/stimulation outside of range. For proficiency grading, only In-house data were utilized.
2.15 SeraCare Proficiency Panel
Between 1998 and 2008, an external EP was implemented by SeraCare. The cellular samples were collected using procedures similar to those described for the EQAPOL ELISpot program. Storage and distribution of the reagents was also performed using procedure similar to those described for the EQAPOL ELISpot program. The EPs were performed by sending four cellular samples to represent negative, low, medium, and high responses to the CEF and CMVpp65 antigens previously described. The responses to each stimulation condition were tested in nine replicates.
We analyzed the data obtained by the last two EPs using the In-house assay by each laboratory to verify the performance overtime of four laboratories that have also enrolled in the EQAPOL ELISpot program.
2.16 Results Remediation Process
The secondary goal of our program was to provide the sites with feedback that could help them in improving their performance. In this context, the EQAPOL ELISpot team contacted the sites that received a rating of Fair or Poor to review performance and identify ways to improve proficiency.
3. Results
3.1 Selection of the cellular samples
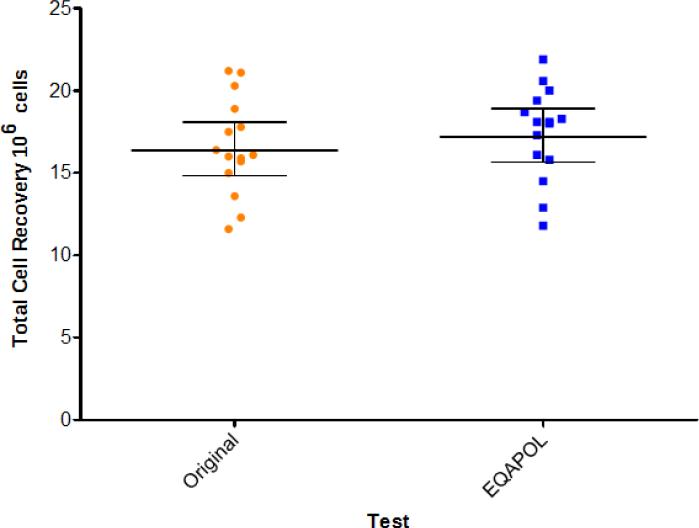
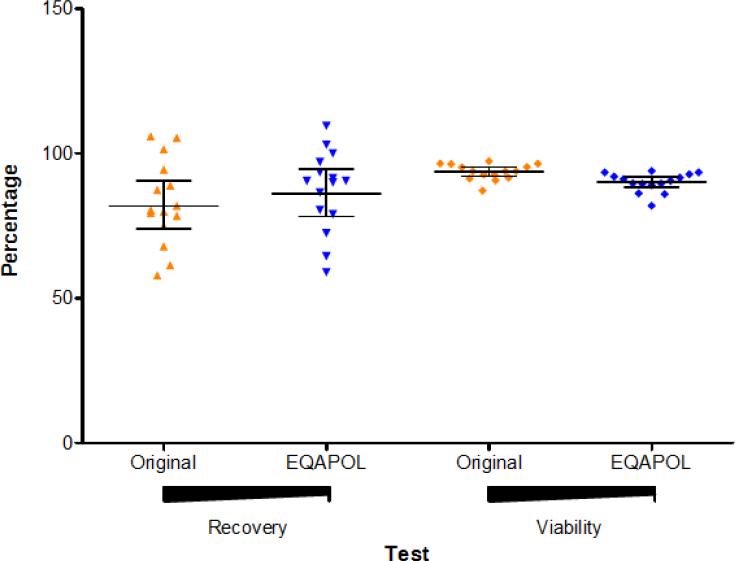
Fifteen donor samples were initially selected among over 100 donor samples collected between June 2008 and June 2010 by the Duke PBMC repository program funded by NIAID. These donors represented different levels of antigen-specific responses as determined by the initial testing performed within four weeks from sample collections (data not shown). At the time the EQAPOL ELISpot program started, the 15 samples were tested to re-evaluate their viability and function. We refer to the first testing as “original” and the current testing as “EQAPOL”. The total recovery (Figure 1) and the percentage of recovery and viability (Figure 2) for each sample after thawing and overnight recovery were compared to those observed initially within the first four weeks from sample collection. The analysis of total and percentage of recovery revealed that total and percentage of recovery did not decline with time from collection. The percentage of recovery was above the 70% that is usually considered as gold standard by the IQA program (Weinberg et al., 2010). Overtime, the average percentage of viable cells declined from 93.7% to 90.1%. Though this is a significant decline (p=0.0004 using a Wilcoxon Sign Ranked Test), it had a modest impact because the % viability was always more than 16% above the 66% required for samples to qualify for testing in most of the IFN-γ ELISpot procedures (Gill et al., 2010; Goepfert et al., 2005; Russell et al., 2003).
Figure 1. Total Cell Recovery.
The absolute number of cells in 106 is reported as observed at the time of initial testing (original, orange circle) and at the time of sample screening for the EQAPOL program (EQAPOL, blue square). Each dot represents a sample. The lines represent the geometric mean and the 95% Confidence Interval.
Figure 2. Percentage of Cell Recovery and Viability.
The percentage of cell recovery (left and triangles) and viability (right and diamonds) is shown for the samples as observed at time of sample collection (original and orange) and at time of sample screening for the EQAPOL (EQAPOL and blue). Each dot represents a sample. The lines represent the geometric means and 95% Confidence Interval.
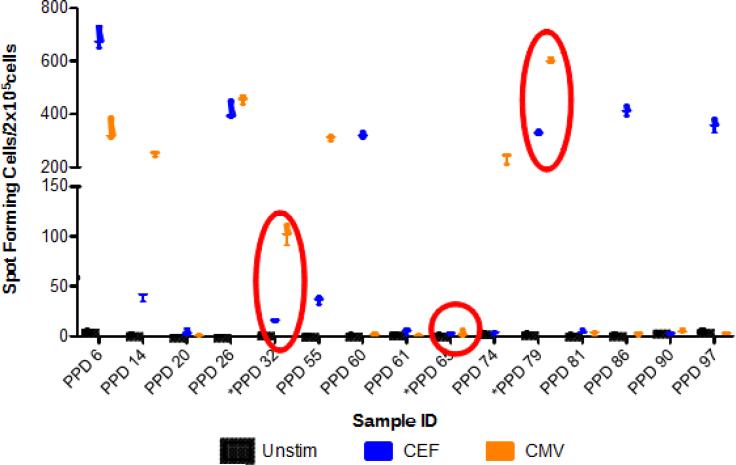
Based on the latest EQAPOL testing, we identified three samples with a functional profile that include absent, low (10-199 SFC/2×105cells), medium (200-400 SFC/2×105cells), and high (>400 SFC/2×105cells) reactivity in response to CEF or CMV stimulation. The three samples are PPD65 (non-responder), PPD32 (low) and PPD79 (medium CEF, high CMV) (Figure 3). PPD32 and PPD79 were used for EPs 1-5. PPD65 was used in EP1, but then changed to PPD90 for EPs 2-5 due to sample availability.
Figure 3. EQAPOL Functional Sample Screening Test.
The Spot Forming Cells per 2×105 cells are reported. The circles identify the samples selected for the EQAPOL program.
3.2 Grading the performance of the participating laboratories
We utilized the first 3 EPs to evaluate validity of the procedures utilized to deliver reagents, to receive the data, to perform the analysis, and to determine the variability of the results among the sites. Of note, the analysis of the variability of the results was utilized to finalize the assessment criteria of assay variability and precision.
The grading of the participating laboratories started with the EP4 at the time this manuscript was finalized, the laboratories received two reports indicating their performance for the fourth and fifth EP. The summary of the grading for each parameter in both EPs testing is reported in Table 3.
Table 3.
Summary of EP4 and EP5 grading.
| EQAPOL ELISPOT EP4 & EP5 | Site 001 | Site 003 | Site 005 | Site 007 | Site 009 | Site 010 | Site 012 | Site 013 | Site 014 | Site 015 | Site 103 | EP | EPs Average | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Criteria | Max Points | EP# | Points Earned | Average | |||||||||||
| Timeliness | 10 | 4 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 8.64 |
| 5 | 10 | 10 | 10 | 0 | 0 ^ | 0 | 10 | 10 | 10 | 10 | 10 | 7.27 | |||
| PBMC Handling | 9 | 4 | 7.5 | 9 | 9 | 6 | 4.5 | 9 | 9 | 9 | 4.5 | 7.5 | 9 | 7.64 | 7.57 |
| 5 | 3 | 9 | 9 | 4.5 | 7.5 | 7.5 | 9 | 6 | 9 | 9 | 9 | 7.5 | |||
| Background | 9 | 4 | 9 | 6 | 9 | 9 | 9 | 9 | 9 | 0 | 9 | 9 | 9 | 7.91 | 7.64 |
| 5 | 9 | 3 | 9 | 9 | 9 | 9 | 9 | 0 | 9 | 6 | 9 | 7.36 | |||
| Accuracy | 54 | 4 | 45 | 36 | 54 | 0 | 45 | 45 | 54 | 9 | 18 | 54 | 54 | 37.64 | 40.91 |
| 5 | 45 | 54 | 54 | 54 | 45 | 54 | 54 | 0 | 18 | 54 | 54 | 44.18 | |||
| Precision | 18 | 4 | 18 | 15 | 18 | 3 | 18 | 12 | 18 | 9 | 15 | 18 | 18 | 14.73 | 14.45 |
| 5 | 18 | 15 | 18 | 18 | 15 | 18 | 18 | 3 | 6 | 12 | 15 | 14.18 | |||
| Total Performance Score | 100 | 4 | 89.5 | 76 | 100 | 28 | 86.5 | 85 | 100 | 37 | 56.5 | 98.5 | 100 | 77.9 | 79.20 |
| 100 | 5 | 85 | 91 | 100 | 85.5 | 76.5 | 88.5 | 100 | 19 | 52 | 91 | 97 | 80.5 | ||
| Performance Rating | 4 | Good | Good | Excellent | Poor | Good | Good | Excellent | Poor | Poor | Excellent | Excellent | |||
| 5 | Good | Excellent | Excellent | Good | Good | Good | Excellent | Poor | Poor | Excellent | Excellent | ||||
The EOL received the samples in the same blinded manner as the other sites, and its score is reported as laboratory 103 (L103).
3.3 Evaluation of Timeliness
We determined that among the parameters used to evaluate the sites, timeliness would reflect the requirements that most of the sites that work under GCLP guidelines have to fulfill when it is time to report data to sponsors and principal investigators after testing samples collected for clinical trials. Therefore, we proposed that on time completion of proficiency panel assays, data upload and survey completion would be essential to the success of any program and to being a proficient lab working in a multi-center consortium. We asked that all sites complete an EP panel and provide valid data via our online application by a specified due date, which was set four weeks after kit receipt. Excel templates/survey results that were uploaded into our system were automatically imported into our database. Data was automatically validated by the system to be sure it was in the correct format. The results reported in Table 3 demonstrate that laboratories performed in an excellent way (all score 10) for EP4, but three failed to report in time for EP5. This is the biggest difference in score between the two EPs.
3.4 Evaluation of PBMC handling
It has been pointed out in previous studies investigating laboratory performance in testing cryopreserved cells for the presence of antigen-specific responses (Bull et al., 2007; Cox et al., 2005) that proper handling of cryopreserved PBMCs at the time of assay performance is critical to ELISpot assay success. Therefore, we consider monitoring this parameter an important step in the assay and pivotal in understanding the performance of each site. Each site received three vials of PMBCs per donor and was instructed to thaw two vials per donor and pool them for both the In-house and EQAPOL reference assay. The third vial was reserved in case of an error.Based on the post-assay questionnaire, all sites rested their cells overnight prior to stimulation. The sites reported viability and recovery on the day of thaw (Day 0), as well as on the day after the thaw following the overnight rest of the cells (Day 1). For grading, sites were assessed as a pass or fail for each donor sample for both viability and recovery using Day 1 data. As reported in Table 3, we noticed that three sites (001, 007, and 010) performed better in EP4 than in EP5. Conversely, three sites (009, 014, and 015) improved their performance in EP5 compared to EP4. Overall, no significant difference was observed in the average PBMC handling score between EP4 and EP5.
In the post-assay questionnaire, sites were asked to report the method used for assessing viability and recovery. For EP4 and EP5, Guava counters were used by five of the 11 sites (45.5%) with the remaining sites using alternative methods (e.g., ViCell, Trypan Blue/hemacytometer, Sysmex XP100, CellDyn). We did not find any evidence for scoring differences based on the method used for cell quantification. 3.5 Evaluation of Background Activity. As reported in Table 3, two sites (003 and 013) had consistent problems with assay background in both EPs. One site (015) reported high background for one donor in EP5, which was not present in EP4. Nevertheless, the average background score across the two EPs was consistent.
3.6 Evaluation of Accuracy to the consensus average
The analysis of the accuracy to the consensus average of the results revealed that seven sites scored at least 75% of the points (40.5 points) for this category and could be considered at least “Good” for their accuracy to the consensus in EP4. Of the four sites that performed sub-optimally in EP4 (003, 007, 013, and 014), two (003 and 007) improved their performance to an “Excellent” in EP5. This suggests that internal review of the data (Site 003) and remediation with the EQAPOL team (Site 007) were successful in identifying and addressing the problems revealed by the analysis of accuracy to the consensus. Overall, there was an improvement from EP4 to EP5, and only one site that had already received a low score in EP4 performed even less proficiently.
3.7 Evaluation of Precision
The analysis of the results to evaluate the precision revealed that eight laboratories were able to achieve a score that was greater than 75% of the maximum points for this category (13.5 points) in both EP4 and EP5 (Table 3). Two laboratories (007 and 010) improved their performance in EP5, whereas two laboratories (014 and 015) performed with a precision lower than 75% of the maximum score. Overall, precision was found to be a problematic aspect for a third of the laboratories participating in this program.
3.8 Overall Evaluation of the Sites
Of the original eleven participating sites, one was never included (016) because the results were never considered valid due to irreproducible technical problems (not shown). The site has also been working on the In-house assay to re-validate its platform and did not participate in the EP4 and EP5. Seven out of the ten participating sites (excluding the EOL, Site 103) scored greater than 74%, and their performance was “Good” (4) or “Excellent” (3). Site 007 improved from 28% (“Poor”) to 85.5% (“Good”), and Site 003 also improved from 76% (“Good”) to 91% (“Excellent”). The overall average of the scores was 77.9 and 80.5 for EP4 and EP5, respectively.
3.9 Evaluation of the Results obtained using the EQAPOL reference assay
The performance scores for the EQAPOL program utilize the results that sites obtain using their own in-house assay. For the program, however, sites are also required to run an EQAPOL reference assay, as well as return all assay plates for centralized reading at the EOL. The results from the EQAPOL reference assay read by the site (IH-EQA), EQAPOL assay read by the EOL (EOL-EQA) and In-house assay read by the EOL (EOL-IH) are analyzed for accuracy to the consensus and precision using the same scoring procedure as the In-house assay. The scores are not reported to the sites, but rather are utilized to pinpoint potential problems with the In-house assay, technical performance of an ELISpot assay or the reader. The scores for these three assays/readings are presented in Table 4 for EP5 only. With EP5, sites 003, 007, and 014 showed discrepant results with the In-house assay and EQAPOL reference assay for accuracy to the consensus; the results indicate that with sites 014 and 007 their score in EP5 is likely attributed to a problem with the In-house assay. It is unclear why Site 003 performed poorly with the EQAPOL assay. The problem noted with Site 014's In-house assay was also evident in the difference in precision scores (Table 4).
Table 4.
Comparison of Accuracy and Precision with In-House EQAPOL assay and In-house EOL readers for EP5.
| Accuracy to the Consensus | Reader-Assay | Site 001 | Site 003 | Site 005 | Site 007 | Site 009 | Site 010 | Site 012 | Site 013 | Site 014 | Site 015 | Site 103 |
| IH-IH | 54 | 54 | 54 | 54 | 45 | 45 | 54 | 0 | 18 | 54 | 54 | |
| EOL-IH | 54 | 54 | 54 | 36 | 54 | 54 | 54 | 0 | 9 | 36 | 36 | |
| IH-EQA | 54 | 18 | 54 | 18 | 54 | 54 | 54 | 0 | 54 | 54 | 54 | |
| EOL-EQA | 45 | 18 | 54 | 54 | 54 | 54 | 54 | 0 | 54 | 54 | 54 | |
| Precision | IH-IH | 18 | 15 | 18 | 15 | 18 | 18 | 18 | 3 | 6 | 12 | 15 |
| EOL-IH | 18 | 15 | 18 | 15 | 18 | 18 | 18 | 18 | 9 | 15 | 15 | |
| IH-EQA | 15 | 18 | 12 | 18 | 18 | 15 | 15 | 0 | 15 | 15 | 15 | |
| EOL-EQA | 15 | 18 | 18 | 18 | 18 | 18 | 18 | 18* | 15 | 15 | 15 |
The table report the scores for the accuracy and precision based on the combination of reading site (Reader) and assay procedure (Assay). The site specific reader and assay procedure are reported as IH; the standardized EQAPOL assay is reported as EQA; the centralized reader operated by the EQAPOL Oversight Laboratory (EOL) is reported as EOL. The numbers represent the points awarded to the site under each combination.
3.10 Effect of the remediation process
The results of EP4 indicated that three of the 10 participating sites encountered problems with their in-house assays. The EQAPOL ELISpot team organized conference calls with each site to discuss the results and discuss the possible causes and solutions for each problem. For site 007, it was determined that samples were plated incorrectly and this caused a problem with accuracy to the consensus. In addition, the magnitude of the responses was lower compare to that detected by the other site. This was found related to a bad batch of culture medium. Upon identification of the problems and solution to prevent them from occurring in the future EPs, site 007 score improved from 28 to 85.5.
For Site 013, we observed a high background of unknown cause. As follow-up of the conference call, EQAPOL provided the site with a new set of PBMC samples, culture media, fetal bovine serum (FBS), and antibiotics to test different combinations of the in-house and centralized components of the culture medium and reduce the level of background activity. The beta-testing of the reagents revealed that a problem could have been related to a batch of FBS during the performance of EP4. However, the same problems were observed during the performance of EP5, and the site's score decreased from 37 to 19.
Lastly, for site 014, we observed a lower recovery of viable cells and decreased level of responses to the antigens. The first problem could be related to components of the culture medium utilized by the laboratory. However, we observed that the performance of the laboratory using the EQAPOL assay was within the range of the other laboratories (not shown). This suggests that the laboratory may have problem with the In-house assay. The score for this laboratory decreased from 56.2 in EP4 to 52 in EP5. However, based on the differences in the accuracy and precision for the EQAPOL assay compared to the In-house assay (Table 4), the site’ s problem appears to lie with the In-house assay only.
These analyses pointed out the importance to perform remediation of the results and how this may lead to correctly troubleshooting the problems of the testing site. Nevertheless, it is possible further actions may be required for some of the sites to include a remediation site visit by the EQAPOL team.
3.11 Longitudinal Analysis of Mean SFC and Dispersion of EP1-5
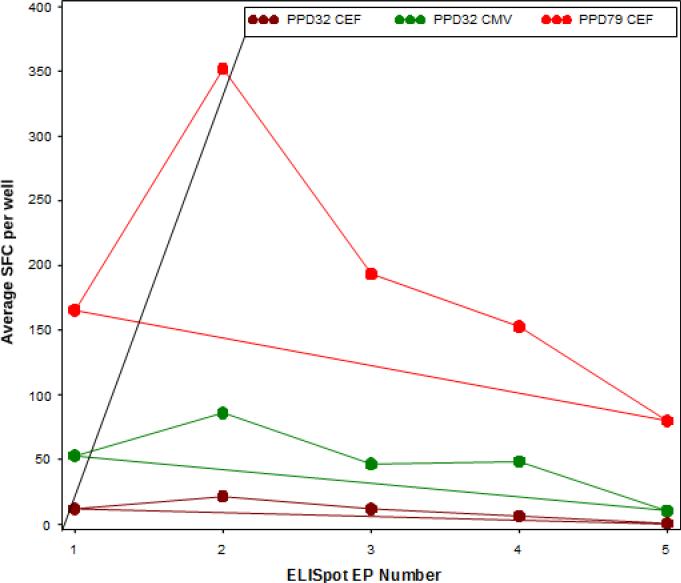
Linear mixed effects models were used to test for linear trends over time in EQAPOL ELISpot EPs 1 through 5. As previously mentioned, two donors (PPD32 and PPD79) were included in all reported EPs, although they were blinded so that the sites did not know. These models were run in SAS 9.2 with Proc GLIMMIX with the assumption of Poisson distributed data. We did not use data from laboratories reporting plating error or high background. For this reason, the data were not background subtracted for these analyses. The testing was performed using the results from testing of donors with antigen-specific responses (PPD32 and PPD79 with CMV and CEF). We tested for trends of the average of SFC (the outcome) for each laboratory using a model that had laboratory-specific random effects for the intercept and EP number. Significant EP number effects would indicate an increase or decrease in the laboratory's average SFC over time. Only the results from one site indicated a significant (p < 0.001) decreasing reactivity over time for PPD32 CEF and CMV as well as PPD79 CEF responses and are shown as example in Figure 4.
Figure 4. Longitudinal Mean Spots Forming Cell (SFC).
The lines represent the average SFC per 2×105 reported over time by one of the sites for each EP. The SFC for sample PPD32 in response to CEF and CMV stimulation are reported in brown and green, respectively. The SFC for sample PPD79 in response to CEF stimulation are reported in red. The significant decreasing trends over time (EPs 1-5), all from Site 014, were calculated using mixed effects models.
To test for trends in the dispersion (variability), we used the same criteria listed above for data inclusion, background subtraction, and data set. We tested for trends in dispersion the outcome for each laboratory using a model that had laboratory specific random effects for the intercept and EP number. Significant EP number effects would indicate an increase or decrease in the laboratory's dispersion over time. Site 003 had a significant decrease over time for PPD79 CEF because of a very high dispersion in EP1. If EP1 is removed, there is no significant effect for Site 003. Otherwise, there were no significant effects for the laboratory specific dispersion. However, the same site with decreasing mean SFC was identified for having an increase in dispersion at EP3 and EP5 for PPD79 CEF and CMV. However, this increase was not significant because it was not linear (Figure 5).
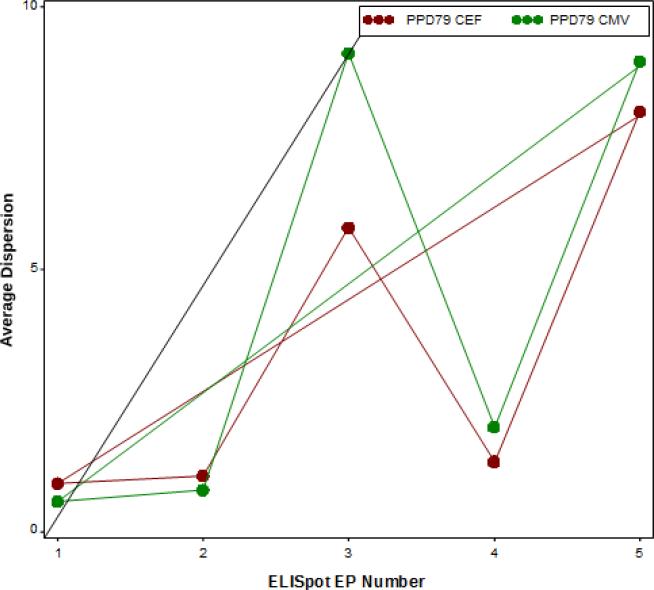
Figure 5. Longitudinal Dispersion.
The lines represent the dispersion observed over time for one site for each EP. The dispersion for the PPD79 response to CEF and CMV stimulation are reported in brown and green, respectively. The fluctuation of dispersion over time (EPs 1-5), all from the same site, calculated using mixed effects models. This is not a linear trend and thus not significant.
3.12 Longitudinal Performance of Laboratories for Accuracy to the Consensus and Precision
We utilized the data collected in context of the SeraCare BioServices EP7 and 8 and EQAPOL EP4 and EP5 to evaluate the applicability of the scoring system for accuracy to the consensus average and precision to the previous PT program and to assess the longitudinal performance of four laboratories in each program. The four laboratories performed their assays under GCLP guidelines and all served as reference laboratory for different vaccine evaluation programs. As reported in Table 5, the individual score for the four laboratories was always greater than 84 during the SeraCare EP7 and EP8 and greater than 79 during the EQAPOL EP4 and EP5. The average score of the four laboratories was consistently greater than 91 and graded as excellent. These results indicated that GCLP compliant laboratories performed consistently well across programs and over time.
Table 5.
Longitudinal comparison of site's performance based on accuracy to the consensus and precision.
| Site Number SC/EQAPOL§ | SC* EP7 | SC EP8 | EQAPOL EP4 | EQAPOL EP5 | Average Score |
|---|---|---|---|---|---|
| 005/003 | 93 | 93 | 79 | 97 | 90.5 |
| 006/103 | 84 | 93 | 100 | 97 | 93.5 |
| 008/005 | 100 | 100 | 100 | 100 | 100 |
| 010/001 | 91 | 98 | 91 | 91 | 92.75 |
| Average Score | 92 | 96 | 92.5 | 96.25 |
SC = SeraCare External Proficiency.
We report the sites that participated in both programs and worked under GCLP guidelines.
The site numbers are based on the original SeraCare and EQAPOL nomenclature.
4. Discussion
In the past, the field has devoted time and efforts in assessing the concordance of the results obtained using different Interferon-γ ELISpot assay protocols(Cox et al., 2005) and in harmonizing the results among different laboratories (Gill et al., 2010; Janetzki et al., 2007; Janetzki and Britten, 2011). Once the assay procedures have become more consistent, there has been the need to perform longitudinal evaluation of the results obtained by the individual laboratories by implementing an external proficiency program. Thus, the main goal of the EQAPOL program was to develop an external proficiency program with set criteria to grade the most critical aspects of ELISpot assay performance. The evaluation of laboratory performance is essential if data generated from testing of clinical samples by different laboratories is to be compared to determine the relative potency of candidate therapeutics or vaccines tested by different organizations. We designed and completed three external proficiency (EP1-3) testing rounds to generate data used to develop grading criteria for proficiency. We graded the sites on the following criteria: timeliness of data submission, PBMC handling, background, accuracy to the consensus and precision. Points were awarded for each criterion, and the sum of the points was used to determine a performance rating (e.g., Excellent to Poor). The choice of the criteria for grading the performance of the laboratory was based on the consideration that the ultimate goal of each laboratory is to perform in compliance to GCLP guidelines while testing clinical samples. In this perspective, the criteria were selected to appropriately evaluate the ability of a laboratory to work under predefined deadlines for assay execution and data reporting (timeliness), for PBMC handling, and assay performance (background, accuracy, and precision). Therefore, this assessment was implemented for the subsequent two EPs (EP4 and EP5), and will continue on a biannual basis going forward.
Through the development of grading criteria we were able to assess performance of each laboratory in key components that reveal strength and weaknesses in the laboratory's procedures. These grading criteria can be used: 1) by the field to determine the level of reproducibility among assays and therefore the ability to provide comparable results; 2) by the laboratory to improve performance; 2) by the sponsor to understand the performance of the endpoint laboratory compared to others in the same field.
An aspect of assay proficiency that is not formally including in our grading criteria is a site's inter-assay performance. However, because we have included two of the sample donor samples in all EPs, we have been able to conduct the analysis present in Section 3.11 to examine if a site has any trends with respect to accuracy and precision across multiple EPs. While the initial analysis shows most sites have excellent inter-assay reproducibility, more data will need to be generated before we could consider adding this as a formal grading criterion. Because each EP is scored individually, adding this as a criterion would mandate a change in our system such that multiple EPs would contribute to a cumulative score. In addition, site-specific reports include analysis comparing the in-house and EQAPOL assays, and sites are provided with comments if there are discrepancies between the two assays. These analyses have been used for site remediation purposes as reported in the Results section.
One of the main initial efforts of the EQAPOL ELISpot program was to define “Gold Standards” for the ELISpot assay. These “Gold Standards” could be implemented to evaluate the performance of different laboratories. The definition of a “Gold Standard” is not merely a statistical issue, but also a scientific one where once the science is decided on the biological relevance of the observations, then, the statistics will produce a range of likely values and those outside that range are deemed suspect. The definition of these ranges is complicated by the wide variability in the magnitude of the cellular responses that can be generated by the human immune system against each antigen. To address this problem, the EQAPOL decided to use mixed effects models for each donor by stimulation, to compare the site average to the consensus average, and to adjust the significance level based on the number of sites in the program. The approach was used to define the grading criteria for the performance of each laboratory based on the accuracy, and precision of the results. These parameters were applied to evaluate the performance of four laboratories that used validated assays and worked under Good Laboratory Clinical Program (GCLP) guidelines. The retrospective and prospective results obtained from the SeraCare program (EP7 and EP8) and for the EQAPOL program (EP4 and EP5), respectively, were analyzed according the established criteria. The results indicated that our grading criteria are applicable to define the performance of each laboratory based on the concordance of the results generated in the testing of samples collected during clinical studies.
In addition, we considered that a secondary valuable aspect of our mandate could reside in our ability to provide each site with the opportunity to discuss their results on follow-up calls and to compare the performance of their assay to that of an external standardized assay platform. Therefore, we implemented the remediation process and developed an assay that can be performed by the participating laboratory as a common testing platform and utilized to identify problems related to reagents and/or in-house testing procedures. We utilized this assay to troubleshoot the performance of each laboratory and to provide constructive feedback to sites with suboptimal performance. As reported in the results section, the parallel evaluation of the results obtained using the in-house and EQAPOL assay lead us to correctly identify areas of improvement for two laboratories and enhance their performance upon the implement of the remediation process.
Overall, the EQAPOL program has been able to finalize grading criteria and detailed data reporting systems that can provide valuable feedback to the participating sites and their sponsors to determine how comparable their results might be. Moreover, we also provide the opportunity for each site to identify possible weaknesses in their performance and the way to resolve them.
Supplementary Material
Acknowledgments
We are grateful for the guidence and leadership of Jim Lane (NIAID). We would like to acknowledge the contributions of Mark de Souza (Cooper Human Systems Llc), who was intially a member of the EQAPOL ELISpot Advisory Committee. The authors are grateful to Linda Walker, Sara Brown, Holly Alley and Jennifer Baker (EQAPOL Repository), and Chris Todd (EQAPOL CQAU). Most importantly, the authors thank the sites for their participation in the EQAPOL ELISpot program: Robert Bailer (Vaccine Research Center), Dan Barouch (Beth Israel Deaconess Medical Center), Sybil D'Costa (ImmunoSite Technologies), Amitabh Gaur (Custom Technology Team, Becton Dickinson), Peter Hayes (IAVI Core Immune Monitoring Laboratory), Leonard Maboko (Mbeya Medical Research Center), Julie McElrath (HVTN), Giuseppe Pantaleo (Centre Hospitalier Universitaire Vaudois), Silvia Ratto-Kim (US Military HIV Research Program), Alexandra Schuetz (AFRIMS), and Hagen von Briesen (Fraunhofer IBMT).
This project was funded in whole, or in part, by the Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract No. HHSN272201000045C entitled ”External Quality Assurance Program Oversight Laboratory (EQAPOL).”
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bull M, Lee D, Stucky J, Chiu Y-L, Rubin A, Horton H, McElrath MJ. Defining blood processing parameters for optimal detection of cryopreserved antigen-specific responses for HIV vaccine trials. J Immunol Methods. 2007;322:57–69. doi: 10.1016/j.jim.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JH, Ferrari G, Kalams SA, Lopaczynski W, Oden N, D'souza MP, Elispot Collaborative Study Group Results of an ELISPOT proficiency panel conducted in 11 laboratories participating in international human immunodeficiency virus type 1 vaccine trials. AIDS Res Hum Retroviruses. 2005;21:68–81. doi: 10.1089/aid.2005.21.68. [DOI] [PubMed] [Google Scholar]
- Currier JR, Kuta EG, Turk E, Earhart LB, Loomis-Price L, Janetzki S, Ferrari G, Birx DL, Cox JH. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J Immunol Methods. 2002;260:157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- Czerkinsky CC, Tarkowski A, Nilsson LÅ, Ouchterlony Ö, Nygren H, Gretzer C. Reverse enzyme-linked immunospot assay (RELISPOT) for the detection of cells secreting immunoreactive substances. J Immunol Methods. 1984;72:489–496. doi: 10.1016/0022-1759(84)90017-6. [DOI] [PubMed] [Google Scholar]
- Ezzelle J, Rodriguez-Chavez IR, Darden JM, Stirewalt M, Kunwar N, Hitchcock R, Walter T, D'Souza MP. Guidelines on good clinical laboratory practice: bridging operations between research and clinical research laboratories. Journal of pharmaceutical and biomedical analysis. 2008;46:18–29. doi: 10.1016/j.jpba.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DK, Huang Y, Levine GL, Sambor A, Carter DK, Sato A, Kopycinski J, Hayes P, Hahn B, Birungi J, Tarragona-Fiol T, Wan H, Randles M, Cooper AR, Ssemaganda A, Clark L, Kaleebu P, Self SG, Koup R, Wood B, McElrath MJ, Cox JH, Hural J, Gilmour J. Equivalence of ELISpot assays demonstrated between major HIV network laboratories. PLoS ONE. 2010;5:e14330. doi: 10.1371/journal.pone.0014330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert PA, Horton H, McElrath MJ, Gurunathan S, Ferrari G, Tomaras GD, Montefiori DC, Allen M, Chiu Y-L, Spearman P, Fuchs JD, Koblin BA, Blattner WA, Frey S, Keefer MC, Baden LR, Corey L, NIAID HIV Vaccine Trials Network High-dose recombinant Canarypox vaccine expressing HIV-1 protein, in seronegative human subjects. J Infect Dis. 2005;192:1249–1259. doi: 10.1086/432915. [DOI] [PubMed] [Google Scholar]
- Goepfert PA, Tomaras GD, Horton H, Montefiori D, Ferrari G, Deers M, Voss G, Koutsoukos M, Pedneault L, Vandepapeliere P, McElrath MJ, Spearman P, Fuchs JD, Koblin BA, Blattner WA, Frey S, Baden LR, Harro C, Evans T, NIAID HIV Vaccine Trials Network Durable HIV-1 antibody and T-cell responses elicited by an adjuvanted multi-protein recombinant vaccine in uninfected human volunteers. Vaccine. 2007;25:510–518. doi: 10.1016/j.vaccine.2006.07.050. [DOI] [PubMed] [Google Scholar]
- Goonetilleke N, Liu MKP, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, CHAVI Clinical Core B. Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BS, McElrath MJ, Keefer MC, Rybczyk K, Berger D, Weinhold KJ, Ottinger J, Ferarri G, Montefiori DC, Stablein D, Smith C, Ginsberg R, Eldridge J, Duerr A, Fast P, Haynes BF, AIDS Vaccine Evaluation Group Immunization with cocktail of HIV-derived peptides in montanide ISA-51 is immunogenic, but causes sterile abscesses and unacceptable reactogenicity. PLoS ONE. 2010;5:e11995. doi: 10.1371/journal.pone.0011995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetzki S, Britten CM. Handbook of ELISPOT, Methods in Molecular Biology. Humana Press; Totowa, NJ: 2011. The Impact of Harmonization on ELISPOT Assay Performance; pp. 25–36. [DOI] [PubMed] [Google Scholar]
- Janetzki S, Cox JH, Oden N, Ferrari G. Standardization and validation issues of the ELISPOT assay. Methods Mol Biol. 2005;302:51–86. doi: 10.1385/1-59259-903-6:051. [DOI] [PubMed] [Google Scholar]
- Janetzki S, Panageas KS, Ben-Porat L, Boyer J, Britten CM, Clay TM, Kalos M, Maecker HT, Romero P, Yuan J, Martin Kast W, Hoos A, the Elispot Proficiency Panel of the CVC Immune Assay Working Group Results and harmonization guidelines from two large-scale international Elispot proficiency panels conducted by the Cancer Vaccine Consortium (CVC/SVI). Cancer Immunol Immunother. 2007;57:303–315. doi: 10.1007/s00262-007-0380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabingu E, Oseroff AR, Wilding GE, Gollnick SO. Enhanced systemic immune reactivity to a Basal cell carcinoma associated antigen following photodynamic therapy. Clin Cancer Res. 2009;15:4460–4466. doi: 10.1158/1078-0432.CCR-09-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers N, Lambeck AJA, Gooden MJM, Hoogeboom B-N, Wolf R, Hamming IE, Hepkema BG, Willemse PHB, Molmans BHW, Hollema H, Drijfhout JW, Sluiter WJ, Valentijn ARPM, Fathers LM, Oostendorp J, van der Zee AGJ, Melief CJ, van der Burg SH, Daemen T, Nijman HW. Immunization with a P53 synthetic long peptide vaccine induces P53-specific immune responses in ovarian cancer patients, a phase II trial. Int J Cancer. 2009;125:2104–2113. doi: 10.1002/ijc.24597. [DOI] [PubMed] [Google Scholar]
- Link H, Olsson O, Sun J, Wang WZ, Andersson G, Ekre HP, Brenner T, Abramsky O, Olsson T. Acetylcholine receptor-reactive T and B cells in myasthenia gravis and controls. J Clin Invest. 1991;87:2191–2196. doi: 10.1172/JCI115253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S, Abrams JS, King CL, Limaye AP, Nutman TB. Parallel regulation of IL-4 and IL-5 in human helminth infections. J Immunol. 1992;148:3567–3571. [PubMed] [Google Scholar]
- Mashishi T, Gray CM. The ELISPOT Assay: An Easily Transferable Method for Measuring Cellular Responses and Identifying T Cell Epitopes. Clinical Chemistry and Laboratory Medicine. 2002;40:903–910. doi: 10.1515/CCLM.2002.159. [DOI] [PubMed] [Google Scholar]
- Olsson T, Zhi WW, Höjeberg B, Kostulas V, Jiang YP, Anderson G, Ekre HP, Link H. Autoreactive T lymphocytes in multiple sclerosis determined by antigen-induced secretion of interferon-gamma. J Clin Invest. 1990;86:981–985. doi: 10.1172/JCI114800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DH, Midgley RS, Mirza N, Torr EE, Ahmed F, Steele JC, Steven NM, Kerr DJ, Young LS, Adams DH. A phase II study of adoptive immunotherapy using dendritic cells pulsed with tumor lysate in patients with hepatocellular carcinoma. Hepatology. 2009;49:124–132. doi: 10.1002/hep.22626. [DOI] [PubMed] [Google Scholar]
- Rountree W, Vandergrift N, Bainbridge J, Sanchez AM, Denny TN. Statistical methods for the assessment of EQAPOL proficiency testing: ELISpot, Luminex, and Flow Cytometry. J Immunol Methods. 2014 doi: 10.1016/j.jim.2014.01.007. (ahead of publication: http://dx.doi.org/10.1016/j.jim.2014.01.007) [DOI] [PMC free article] [PubMed]
- Russell ND, Hudgens MG, Ha R, Havenar-Daughton C, McElrath MJ. Moving to human immunodeficiency virus type 1 vaccine efficacy trials: defining T cell responses as potential correlates of immunity. J Infect Dis. 2003;187:226–242. doi: 10.1086/367702. [DOI] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M, Cox J, Cleland N, Denny T, Hural J, Needham L, Ozaki D, Rodriguez-Chavez IR, Stevens G, Stiles T, Tarragona-Fiol T, Simkins A. Evaluation and Recommendations on Good Clinical Laboratory Practice Guidelines for Phase I–III Clinical Trials. PLoS Med. 2009;6:e1000067. doi: 10.1371/journal.pmed.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz F, Ehlert K, Ge Y, Schneeweiss A, Rom J, Inzkirweli N, Sohn C, Schirrmacher V, Beckhove P. Treatment of advanced metastasized breast cancer with bone marrow-derived tumour-reactive memory T cells: a pilot clinical study. Cancer Immunol Immunother. 2009;58:887–900. doi: 10.1007/s00262-008-0605-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman P, Kalams S, Elizaga M, Metch B, Chiu Y-L, Allen M, Weinhold KJ, Ferrari G, Parker SD, McElrath MJ, Frey SE, Fuchs JD, Keefer MC, Lubeck MD, Egan M, Braun R, Eldridge JH, Haynes BF, Corey L, NIAID HIV Vaccine Trials Network Safety and immunogenicity of a CTL multiepitope peptide vaccine for HIV with or without GM-CSF in a phase I trial. Vaccine. 2009;27:243–249. doi: 10.1016/j.vaccine.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck H, Brumme ZL, Anastario M, Cohen KW, Jolin JS, Meier A, Brumme CJ, Rosenberg ES, Alter G, Allen TM, Walker BD, Altfeld M. Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS Med. 2008;5:e100. doi: 10.1371/journal.pmed.0050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JM, Slifka MK. Longevity of T-Cell Memory following Acute Viral Infection. In: Zanetti M, Schoenberger SP, editors. Memory T Cells. Springer; New York: 2010. pp. 96–107. [DOI] [PubMed] [Google Scholar]
- Weinberg A, Song LY, Wilkening CL, Fenton T, Hural J, Louzao R, Ferrari G, Etter PE, Berrong M, Canniff JD, Carter D, Defawe OD, Garcia A, Garrelts TL, Gelman R, Lambrecht LK, Pahwa S, Pilakka-Kanthikeel S, Shugarts DL, Tustin NB. Optimization of storage and shipment of cryopreserved peripheral blood mononuclear cells from HIV-infected and uninfected individuals for ELISPOT assays. J Immunol Methods. 2010;363:42–50. doi: 10.1016/j.jim.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanone M, Favaro E, Quadri R, Miceli I, Giaretta F, Romagnoli R, David E, Perin P, Salizzoni M, Camussi G. Association of cytomegalovirus infections with recurrence of humoral and cellular autoimmunity to islet autoantigens and of type 1 diabetes in a pancreas transplanted patient. Transpl Int. 2010;23:333–337. doi: 10.1111/j.1432-2277.2009.00994.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.