Abstract
Kinases are principal components of signal transduction pathways and the focus of intense basic and drug discovery research. Irreversible inhibitors that covalently modify non-catalytic cysteines in kinase active-sites have emerged as valuable probes and approved drugs. Many protein classes, however, possess functional cysteines and therefore understanding the proteome-wide selectivity of covalent kinase inhibitors is imperative. Here, we accomplish this objective using activity-based protein profiling coupled with quantitative mass spectrometry to globally map the targets, both specific and non-specific, of covalent kinase inhibitors in human cells. Many of the specific off-targets represent non-kinase proteins that, interestingly, possess conserved, active-site cysteines. We define windows of selectivity for covalent kinase inhibitors and show that, when these windows are exceeded, rampant proteome-wide reactivity and kinase target-independent cell death conjointly occur. Our findings, taken together, provide an experimental roadmap to illuminate opportunities and surmount challenges for the development of covalent kinase inhibitors.
Protein kinases are one of the largest and most diverse enzyme classes in Nature, with more than 500 members in the human proteome1. The size of the kinome, coupled with the central roles that kinases play in cell signaling, physiology, and disease, has inspired widespread effort to create selective kinase inhibitors as basic research probes and therapeutics2,3.
Most kinase inhibitors described to date are reversible compounds that interact with the conserved ATP-binding pocket of kinases. A number of technology platforms have been developed to profile inhibitor selectivity across the kinome (and, in some cases, the broader ATP-binding proteome) in purified protein and native biological systems4-6. The implementation of these platforms, combined with structure-guided medicinal chemistry, has greatly increased the number of kinases for which selective inhibitors have been developed. Despite these advances, the vast majority of human kinases still lack selective inhibitors for investigating their functions in biological systems3.
A second, emerging class of inhibitors block kinase activity irreversibly by forming covalent bonds with nucleophilic residues, most commonly cysteine, found in the ATP-binding pockets of a substantial fraction of kinases3,7-9. Irreversible inhibitors can exhibit advantages over reversible compounds3,7-11 such as achieving more complete and sustained target engagement in living systems by being less susceptible to competition by high intracellular concentrations of ATP and requiring the physical turnover of kinase proteins to restore inhibited signaling pathways. The cysteine(s) targeted by covalent kinase inhibitors also offer a potential selectivity filter12, as these residues are not uniformly conserved across the kinome12,13. Thus far, covalent inhibitors have been developed for several protein kinases3, including oncogenic drug targets like the epidermal growth factor receptor (EGFR) and Bruton's tyrosine kinase (BTK), for which the corresponding irreversible inhibitors afatinib (or BIBW-2992)14 and ibrutinib (or PCI-32675)15 were recently approved to treat non-small cell lung cancer and chronic lymphocytic leukemia (CLL), respectively. Initial studies with fluorescent or clickable probes suggest that at least some covalent kinase inhibitors can exhibit good selectivity16-18, although this critical parameter has not, to our knowledge, been thoroughly examined across the proteome for any covalent kinase inhibitor. It has been suggested that, as long as covalent kinase inhibitors are selective within the kinome, specificity across the greater proteome may be inferred9. Cysteine, however, is the most intrinsically nucleophilic proteinaceous residue, and many protein classes rely on cysteines for function19-21. These factors suggest that covalent kinase inhibitors have the potential to cross-react, either specifically or non-specifically, with proteins outside of the kinome. Such “off-target” activity complicates the assignment of biological functions to kinases in chemical biology experiments and could lead to unanticipated toxicities in drug development programs.
Here, we use activity-based protein profiling (ABPP22) combined with quantitative mass spectrometry (MS) to perform a global and in-depth analysis of proteins targeted by covalent kinase inhibitors in human cancer cells. We find that covalent kinase inhibitors, including approved drugs, have defined, but limited concentration windows across which selective target inhibition can be achieved. Once this selectivity range is breached, substantial off-target protein reactivity and kinase target-independent cytotoxicity is observed. Our results thus indicate that medicinal chemistry efforts aimed at optimizing the selectivity of covalent kinase inhibitors should account for their reactivity across the entire human proteome in order to ensure suitable windows of selectivity for basic pharmacology and drug development initiatives. Herein we present an experimental framework for achieving this objective.
Results
Proteomic profiling of covalent kinase inhibitors
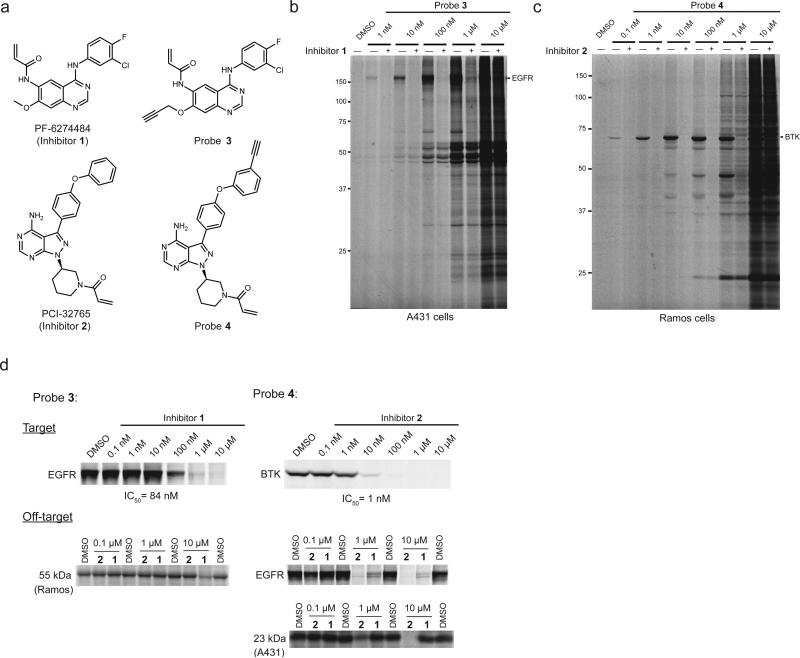
For our studies of covalent kinase inhibitors, we selected representative agents that target EGFR (PF-6274484, 1)23 and BTK (ibrutinib, 2)18 (Fig. 1a). Inhibitors 1 and 2 both use αβ-unsaturated amide electrophiles (hereafter referred to as Michael acceptors) to covalently react with active-site cysteines in the ATP-binding pockets of their target kinases. We synthesized alkynylated variants of inhibitors 1 and 2 (probes 3 and 4, respectively; Fig. 1a), taking care to install the alkyne at positions that would not perturb the intrinsic reactivity of the Michael acceptors nor interfere with target kinase interactions based on established structural-activity relationships18,23.
Figure 1.
Characterization of the proteome reactivities of covalent kinase inhibitors in human cancer cells as determined by gel-based ABPP. (a) Structures of parent inhibitors and alkyne-modified (clickable) ABPP probes. (b, c) In situ reactivity profiles for probes 3 (b) and 4 (c) in A431 and Ramos cells, respectively, with pretreatment by inhibitors (1 and 2, respectively) where indicated. Cells were treated with DMSO or inhibitor (10 μM, 1 hr) followed by probes (0.001-10 μM, 1 hr) and then analyzed by gel-based ABPP. Membrane and soluble proteomes are shown for 3-treated A431 cells and 4-treated Ramos cells, respectively. Specific protein targets with molecular masses matching EGFR and BTK are marked in A431 and Ramos cells, respectively. See Supplementary Fig. 1 for similar profiles performed in cancer cell lysates (in vitro reactivity profiles). (d) Concentration-dependent blockade of probe labeling by parent inhibitors in cancer cells. Representative specific targets and off-targets are shown for inhibitor 1/probe 3 (left gels) and inhibitor 2/probe 4 (right gels). For (b-d), gel-based ABPP experiments were performed in duplicate or triplicate with consistent results. See Supplementary Fig. 12 for full-length gels containing cropped gel data shown in these panels.
Initial ABPP experiments were performed in A431 and Ramos cells, which possess high levels of the primary inhibitor targets EGFR and BTK, respectively. We first evaluated the in vitro and in situ reactivity of probes 3 and 4 by gel-based ABPP, where cell lysates or cells were treated with probes (0.001-10 μM, 1 hr), and then reacted with an azide-rhodamine (N3-Rh) reporter tag under copper-catalyzed azide-alkyne cycloaddition (CuACC or click chemistry) conditions24 and probe-labeled proteins visualized by SDS-PAGE and in-gel fluorescence scanning25. Competitive ABPP experiments, where proteomes or cells were pre-treated with inhibitors 1 and 2 (10 μM, 30 min) facilitated detection of specific targets. In probe 3-treated A431 cells, we observed the concentration-dependent and inhibitor 1-competed labeling of an ~150 kDa membrane protein that presumably represented EGFR (Fig. 1b and Supplementary Results, Supplementary Fig. 1a). The concentration-dependent labeling of several additional proteins was also detected in A431 cells and most of these labeling events were not competed by inhibitor 1. In probe 4-treated Ramos cells, we detected the concentration-dependent and inhibitor 2-competed labeling of an ~70 kDa soluble protein that presumably represented BTK, along with a handful of additional inhibitor 2-competed labeling events visible at low concentrations of probe (≤ 100 nM) and many additional labeling events that appeared at higher concentrations of probe (1 μM or above) and were not competed by inhibitor 2 (Fig 1c and Supplementary Fig. 1a, b).
Competitive ABPP experiments performed in vitro and in situ produced IC50 values for inhibitors 1 and 2 blockade of EGFR and BTK probe-labeling, respectively, that generally matched those reported for the same18 or related23 compounds using substrate assays (Fig. 1d, Supplementary Fig. 1c, d, and Supplementary Table 1) and were also similar to estimated half-maximal labeling values for probes 3 and 4 (Supplementary Fig. 1e), indicating that structural modifications to install alkynes did not substantially alter inhibitor potency for the target kinases. Probe 3 could detect EGFR across a range of endogenous expression levels in human cancer cell lines (Supplementary Fig. 1f). Inhibitor 1 exhibited similar in vitro potencies against EGFR in all of the cancer cell lines examined, but consistently showed lower in situ IC50 values in cancer lines expressing mutant (L858R or exon 19 deletion) forms of EGFR compared to cancer lines expressing wild type (WT)-EGFR (Supplementary Fig. 1c and Supplementary Table 1). While the basis for this differential potency is not clear, it suggests that the activation status of EGFR can impact the extent of target engagement by inhibitors in cancer cells.
ABPP studies identified additional inhibitor-competed targets in A431 and Ramos cells, including EGFR as a potent target of inhibitor 2 (a known interaction18) (Fig. 1d; also see Supplementary Fig. 1g). Other inhibitor-competed proteins were selectively labeled by probe 3 or 4 (Supplementary Fig. 1h; arrows), which mirrored their respective inhibition by 1 and 2 (Fig. 1d), albeit with weaker potencies than the target kinases EGFR and BTK. Finally, it is interesting to note that certain inhibitor-competed proteins were detected in situ, but not in vitro (Supplementary Fig. 1g), underscoring the importance of performing competitive ABPP studies in living systems.
Taken together, these gel-based ABPP data indicated that clickable probes can detect “on”- and “off”-targets for covalent kinase inhibitors in cancer cells, where off-targets appear to include both specific (inhibitor-competed and/or probe-selective) and non-specific reactivity events. We next set out to identify the targets of covalent kinase inhibitors by combining ABPP with quantitative mass spectrometry (MS).
Protein targets of covalent kinase inhibitors
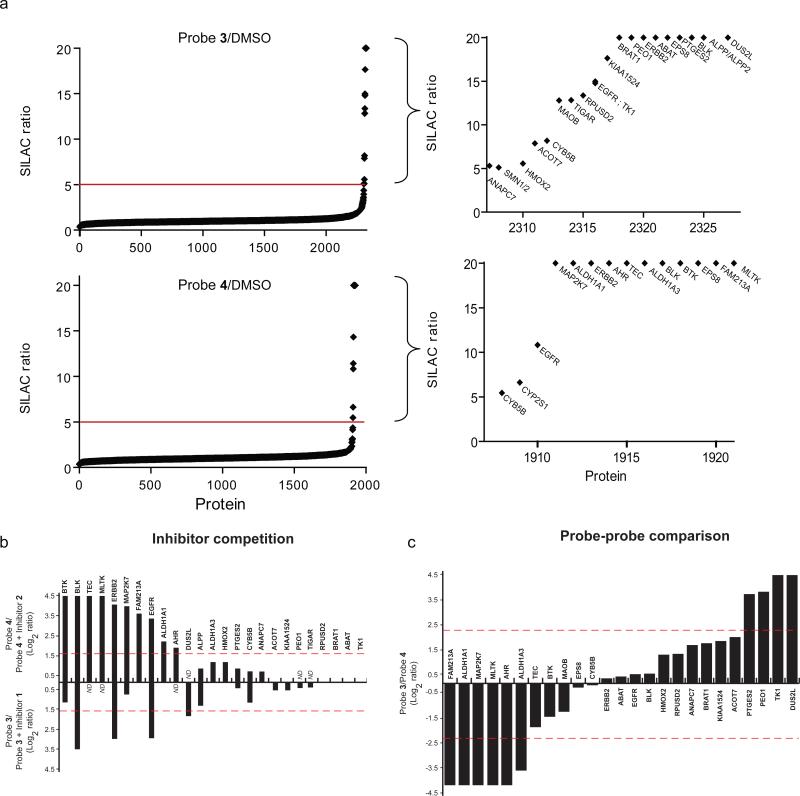
We enriched and identified proteins that reacted with probes 3 and 4 by combining ABPP with SILAC (stable isotope labeling by amino acids in cell culture26) MS analysis (see Supplementary Fig. 2 and refs. 27 and 28 for more details on the ABPP-SILAC methodology). In our initial ABPP-SILAC studies, we treated cancer cells grown in isotopically ‘light’ or ‘heavy’ media with probe (3 or 4; 1 μM, 1 hr) and DMSO, respectively, which enabled a full inventory of all probe-labeled proteins, defined as proteins that showed light:heavy ratios > 5. Combined analyses of both A431 and Ramos cells treated with probes 3 and 4 identified a total of 29 probe targets (Fig. 2a and Supplementary Tables 2 and 3), including EGFR and BTK, additional known kinase targets of the tested covalent inhibitors – ERBB218,23, BLK18,29, and TEC18 – and many other non-kinase proteins. Competitive ABPP-SILAC experiments revealed that probe labeling of a subset of these targets, including the aforementioned kinases and a handful of additional proteins, were blocked (> 3-fold) by pre-treatment with inhibitor (1 or 2, 10 μM, 1 hr) (Fig. 2b and Supplementary Tables 2 and 3). An overlapping, but distinct subset of targets showed preferential reactivity (> 5-fold) with probe 3 or 4 in ABPP-SILAC experiments that compared cells treated with equal concentrations of each probe (1 μM; 1 hr; Fig. 2c and Supplementary Tables 2 and 3).
Figure 2.
Characterization of the proteomic reactivities of covalent kinase inhibitors in cancer cells by ABPP-SILAC. (a) SILAC ratio plots for total proteins identified in experiments comparing cells treated with probe (3 or 4, 1 μM, 1 hr) versus DMSO (no-probe). Proteins with median SILAC ratios ≥ 5 (probe/DMSO) are designated as probe-labeled targets. Ratios ≥ 20 are listed as 20; results are a combination of duplicate ABPP-SILAC experiments performed in A431 and Ramos cells for each probe. (b) SILAC ratios for probe targets from competitive ABPP-SILAC studies performed in duplicate, where cancer cells were pre-treated with inhibitor (1 or 2, 10 uM, 1 hr) or DMSO, followed by treatment with probe (3 or 4, respectively; 10 μM, 1 hr). Data for inhibitor 2 + probe 4 and inhibitor 1 + probe 3 are plotted above and below the x-axis, respectively, as Log2 of the median SILAC ratios (DMSO/inhibitor). Proteins with SILAC ratios ≥ 3 are designated as competed targets. Not detected (ND). (c) SILAC ratios for probe targets from an ABPP-SILAC study performed in duplicate, where cancer cells were treated with probe 3 (1 uM, 1 hr) or probe 4 (1 μM, 1 hr). Protein targets exhibiting preferential labeling with probe 3 or 4 extend above and below the x-axis, respectively, plotted as Log2 of the median SILAC ratios (probe 3/probe 4). Proteins with SILAC ratios ≥ 5 are designated as probe-selective targets. Competition and probe-probe comparison data were obtained for 24 of the 29 probe targets identified in part a.
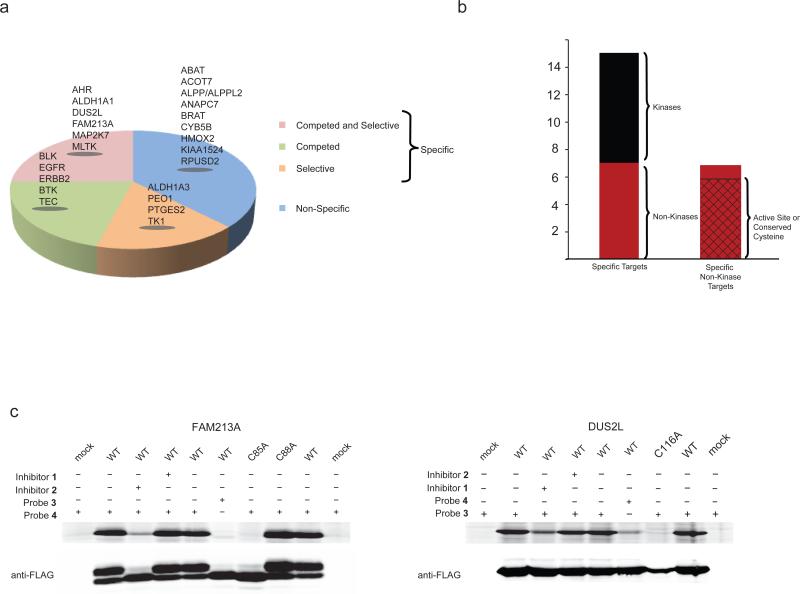
Combining data from our ABPP-SILAC experiments, we could categorize 24 of the 29 probe targets as shown in Figure 3a. We first note that the majority of the probe targets exhibited evidence of specificity in terms of being competed by parent inhibitors (“competed”; green, Fig. 3a), selectively reacting with probes 3 or 4 (“selective”; orange, Fig. 3a), or both (“competed and selective”; pink, Fig. 3a). Only a limited number of putatively non-specific targets (not-competed, not-selective; blue, Fig. 3a) were detected in cancer cells at the tested concentrations of covalent kinase inhibitors. Consistent with previous studies18,23, EGFR and ERBB2 were identified as specific targets of both inhibitor 1/probe 3 and inhibitor 2/probe 4 (Fig. 3a and Supplementary Table 2 and 3). BTK, on the other hand, was primarily competed by inhibitor 2 (Fig. 3a and Supplementary Table 2). Additional protein kinase targets also distributed into the specific categories of competed (BLK) or competed and selective (MAP2K7, TEC, MLTK) (Fig. 3a and Supplementary Table 2 and 3). Most of these kinases are established off-targets that were identified in previous in vitro kinome selectivity screens18. Of particular interest, however, was the discovery of several specific nonkinase targets of probes 3 and 4 that included enzymes (e.g., DUS2L, PTGES2, ALDH1A1), receptors (e.g., AHR), and uncharacterized proteins (e.g., FAM213A) (Fig. 3a, b and Supplementary Table 2 and 3). The non-kinase targets did not share any obvious structural or functional relatedness to kinases or to one another. Intriguingly, however, most of these proteins possess conserved active site or functional cysteines (Fig. 3b; Supplementary Table 2 and Supplementary Fig. 3), pointing to a potential unifying basis for their cross-reactivity with covalent kinase inhibitors.
Figure 3.
Analysis of protein targets of covalent kinase inhibitors. (a) Categorization of protein targets of probes 3 and 4 based upon ABPP-SILAC experiments. (b) Breakdown of specific targets of probes 3 and 4 as kinase (black) or non-kinase (red) proteins. Further analysis of the non-kinase targets demonstrates that most possess active-site or conserved cysteine residues (hatched section of right bar). (c) Characterization of representative non-kinase, specific off-targets of inhibitor 2/probe 4 (FAM213A) and inhibitor 1/probe 3 (DUS2L). WT and indicated mutants of FAM213A and DUS2L were recombinantly overexpressed in HEK 293T cells and evaluated for their probe reactivity and the sensitivity of these reaction to inhibitor competition by gel-based ABPP (upper panels). Anti-FLAG western blotting was used to confirmed protein expression levels (lower panels). Note that probe 4-modification causes an upward migration of FAM213A on SDS-PAGE. Gel-based ABPP experiments and western blots were performed in duplicate or triplicate with consistent results. See Supplementary Fig. 12 for full-length gels containing cropped gel data shown in this panel.
We next recombinantly expressed a subset of off-targets in mammalian cells and assayed their probe reactivity by gel-based ABPP. Consistent with our ABPP-SILAC studies, recombinant DUS2L and FAM213A reacted selectively with probes 3 and 4, respectively (Fig. 3c), and these reactions were competed by inhibitors 1 and 2 (Fig. 3c) with corresponding IC50 values of 1.6 and 1.0 μM (Supplementary Fig. 4a). These inhibition values were similar to those observed for endogenous proteins in cancer cells predicted by molecular mass to represent FAM213A and DUS2L (Fig. 1d). Mutation of the conserved catalytic cysteine in DUS2L to alanine (C116A) modestly reduced protein expression and completely eliminated 3-labeling (Fig. 3c). FAM213A is an uncharacterized protein that contains two conserved cysteines that form a CXXC motif as part of its predicted thioredoxin-like domain (Supplementary Fig. 3). Mutation of one of these cysteines to alanine (C85A) completely blocked labeling by probe 4 (Fig. 3c).
Most of the kinase off-targets identified by ABPP-SILAC share the active-site cysteine residue targeted by covalent kinase inhibitors in EGFR and BTK3. One exception is MLTK (or ZAK), which was identified as a specific target of probe 4. We confirmed that probe 4, but not 3 reacted with recombinant MLTK and this reaction depended on a distinct active site-proximal cysteine (C22) and was competed by inhibitor 2 (but not 1) with an IC50 value of 0.7 μM (Supplementary Fig. 4b-d).
These data, taken together, indicate that off-targets of covalent kinase inhibitors originate from structurally and functionally diverse protein families and that these reactions depend on functional cysteines in off-target active sites.
Inhibitor modifications that alter selectivity
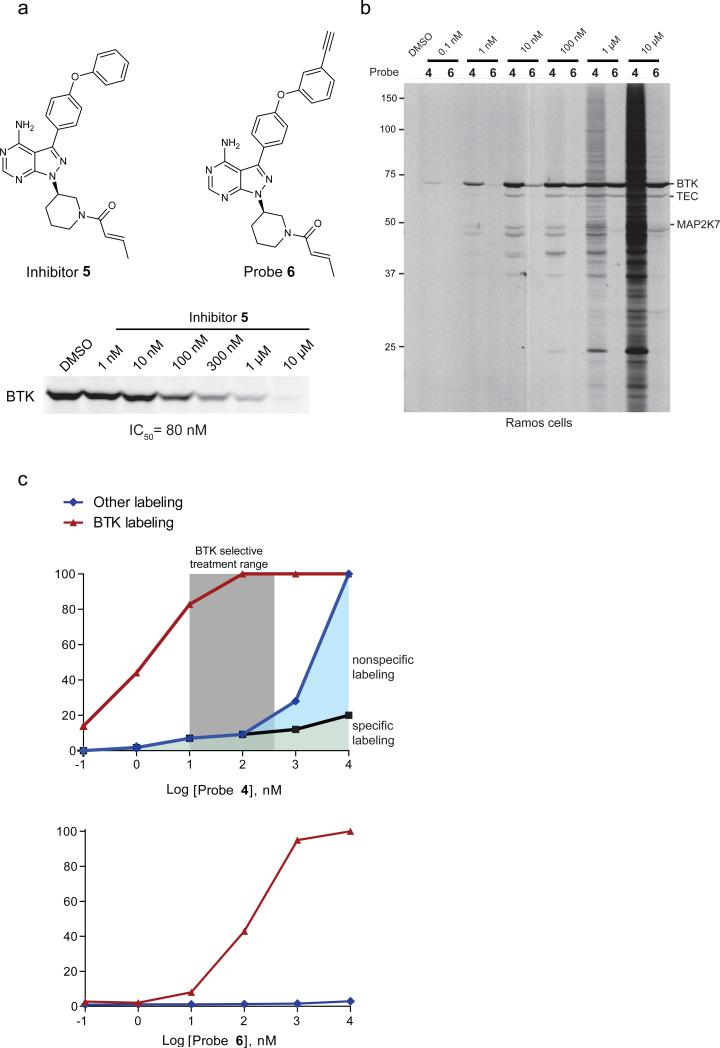
We next asked whether modifications to the Michael acceptor might alter covalent kinase inhibitor reactivity. Simple methylation of the terminal Michael acceptor of inhibitor 2/probe 4 produced an inhibitor/probe pair [inhibitor 5 (ref. 30) and probe 6, respectively; Fig. 4a] that still covalently inhibited BTK with good potency in cells (in situ IC50 = 80 nM for inhibitor 5, compared to 2 nM for 2) and exhibited dramatically lower background proteome reactivity as measured by both concentration-dependent (Fig. 4b and Supplementary Fig. 5a) and time-course (Supplementary Fig. 5b) experiments comparing probes 4 and 6. The reduced cross-reactivity observed with inhibitor 5/probe 6 included both specific (e.g., BLK, MAP2K7, MLTK, FAM213A; Supplementary Fig. 5c, d) and non-specific targets (Supplementary Fig. 5d), as determined by gel-based ABPP and ABPP-SILAC studies. Indeed, the only specific off-target still labeled well by 6 in RAMOS cells was TEC kinase (Supplementary Fig. 5d). A direct comparison of the in situ proteome-reactivity profiles of probes 4 and 6 enabled rigorous assignment of suitable concentration windows across which these probes could achieve near-complete inhibition of BTK without substantial non-specific proteome reactivity. For probe 4, this window extended from 10 to 300 nM, while for probe 6, the window spanned from 500 nM to ≥ 10 μM (Fig. 4c). These data should help to guide ongoing and future studies that aim to study the biological effects of selective BTK inactivation (see ref. 31 and Discussion below).
Figure 4.
Minimizing the off-target reactivity of covalent kinase inhibitors by modification of the Michael acceptor reactive group. (a) Structures of β-methyl agents (inhibitor 5 and probe 6) and in situ IC50 value for inhibition of BTK by inhibitor 5 as measured by competitive ABPP. (b) A comparison of in situ reactivity profiles for probes 4 and 6 in Ramos cells (soluble proteome) reveals much less proteomic reactivity for probe 6. For (a, b), gel-based ABPP experiments were performed in duplicate or triplicate with consistent results. See Supplementary Fig. 12 for full-length gels containing cropped gel data shown in these panels. (c) Comparative graphical analysis of the in situ target and off-target reactivity profiles of probes 4 and 6 in Ramos cells, as measured by gel band (BTK) or full gel lane (Other) intensities in the representative ABPP experiments shown in Figs. 1c and 4b. Gels were quantified for probe 4 and 6 analyses, respectively (note that data from Fig. 1c were used for probe 4 to determine the fraction of inhibitor 2-competed targets that constitute “specific labeling”). The red lines denote % BTK probe occupancy; the blue lines denote labeling of all other proteins, with 100% taken from lane intensity measurement at 10 μM of probe 4 (data from Fig. 4b). The black trace denotes percent of gel-lane labeling competed by pretreatment with inhibitor 2 (10 μM; data from Fig. 1c).
Inhibitor modifications that alter in situ reactivity
One common substitution to the Michael acceptor of covalent kinase inhibitors is the dimethylaminomethyl (DMAM) group, which can be found on several covalent kinase inhibitors, including afatinib3. The DMAM group is thought to improve drug solubility and, at the same time, maintain potency, possibly by providing a proximal basic amine to activate the attacking cysteine thiol32.
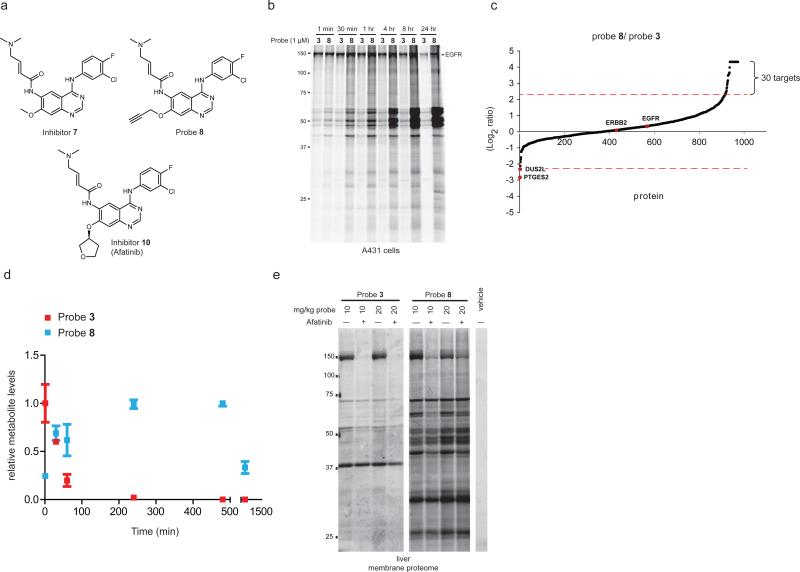
We synthesized DMAM-substituted analogues of inhibitor 1 and probe 3 (inhibitor 7 and probe 8, respectively; Fig. 5a) and found that 1 and 7 showed similar in vitro and in situ potencies for inhibiting EGFR in cancer cells, as measured by competitive ABPP (Supplementary Table 1). Probes 3 and 8 also showed similar potencies for labeling EGFR in cancer cell lysates, where 3 appeared to exhibit a somewhat greater degree of background proteome reactivity (Supplementary Fig. 6a). In striking contrast to these in vitro profiles, however, probe 8 exhibited much greater time- (Fig. 5b and Supplementary Fig. 6b) and concentration- (Supplementary Fig. 6c) dependent proteome reactivity in living cancer cells. Pre-treatment with inhibitor 7 failed to block most of the extensive proteome reactivity of probe 8 (with EGFR labeling being a notable exception; Supplementary Fig. 6d). ABPP-SILAC experiments confirmed that many more protein targets in cancer cells exhibited greater reactivity with probe 8 compared to probe 3 (Fig. 5c and Supplementary Table 3). The only exceptions were DUS2L and PTGES2, two proteins that we earlier identified as selective targets of probe 3 (Figs. 2 and 3) and found here to preferentially react with 3 over 8 (Fig. 5c). We also compared the protein labeling profiles of cells treated with 1 versus 10 μM of probe 8 and found that virtually all targets showed substantially higher labeling at 10 μM probe (EGFR and ERBB2 being notable exceptions; Supplementary Fig. 6e and Supplementary Table 3). Taken together, these findings indicate that introduction of a DMAM group onto the inhibitor 1/probe 3 scaffold dramatically increased protein reactivity in cancer cells. Examination of a DMAM analogue of probe 4 (probe 9) revealed that the DMAM modification did not confer higher cellular protein reactivity to the ibrutinib scaffold of covalent kinase inhibitors (Supplementary Fig. 7).
Figure 5.
The in situ proteomic reactivity of covalent kinase inhibitors is altered by modification of the Michael acceptor. (a) Structures of dimethylaminomethyl (DMAM)-modified agents [inhibitor 7, probe 8, and afatinib (10)]. (b) Gel-based ABPP showing markedly different in situ reactivity profiles for probes 3 and 8 (1 μM) in A431 cells treated for the indicated time. (c) SILAC ratios for probe targets from a probe-probe comparison study performed in duplicate, where cancer cells were treated with probe 3 (1 uM, 4 hr) or 8 (1 μM, 4 hr). Results are plotted as Log2 of the median SILAC ratios (probe 8/probe 3). Proteins exhibiting preferential labeling with probe 8 or 3 extend above and below the x-axis, respectively. Proteins with SILAC ratios ≥ 5 are designated as probe-selective targets. (d) Time course measurement of probe 3 (red) and 8 (green) concentrations in A431 cells. Data represent relative amounts of each compound normalized to the highest measured cellular concentration across the time course (set to an arbitrary value of 1.0) and are reported as average values + standard errors for three biological replicates. (e) Gel-based ABPP of liver membrane proteomes from mice treated with probes 3 or 8 (10 or 20 mg/kg, 1 hr). Pre-treatment with afatinib (20 mg/kg, 1 hr) blocked probe-labeling of an ~150 kDa protein interpreted to represent EGFR. For (b, d), gel-based ABPP experiments were performed in duplicate or triplicate with consistent results.
We found that probe 8 displayed much greater stability in cancer cells compared to probe 1 (Fig. 5d), providing a plausible explanation for its enhanced in situ proteomic reactivity. Inhibitor 7 also showed more prolonged accumulation in cells compared to inhibitor 1, as did afatinib (10; Fig. 5a) compared to its unsubstituted Michael acceptor analogue (11) (Supplementary Fig. 8a, b), indicating that the DMAM group conferred a general increase in the cellular half-lives of EGFR inhibitors.
Finally, we evaluated the proteome reactivity of probes 3 and 8 in vivo by treating mice with 10 or 20 mg/kg of each compound for 1 hr, after which animals were sacrificed and tissues removed for ABPP. Both probes 3 and 8 labeled an ~150 kDa protein in mouse liver and these labeling events were competed by pre-treatment with afatinib (20 mg/kg, 1 hr) (Fig. 5e). We presume that this protein represents EGFR, which is highly expressed in mouse liver (BioGPS http://biogps.org/). Probe 8 also labeled many additional proteins in mouse liver compared to probe 3 (Fig. 5e and Supplementary Fig. 8c). Similar in vivo studies were performed with the BTK probes 4 and 6, where we found that probe 4 showed higher proteomic reactivity than probe 6 in vivo (Supplementary Fig. 8d). These data indicate that the differences in proteomic reactivity displayed by covalent kinase inhibitors in cell culture are also observed in vivo. Importantly, both the EGFR and BTK probes were tested at pharmacologically relevant doses that others have used for structurally related inhibitors (e.g., afatinib and ibrutinib, respectively) to block cancer progression in mouse xenograft models33,34.
Extensive proteomic reactivity promotes cytotoxicity
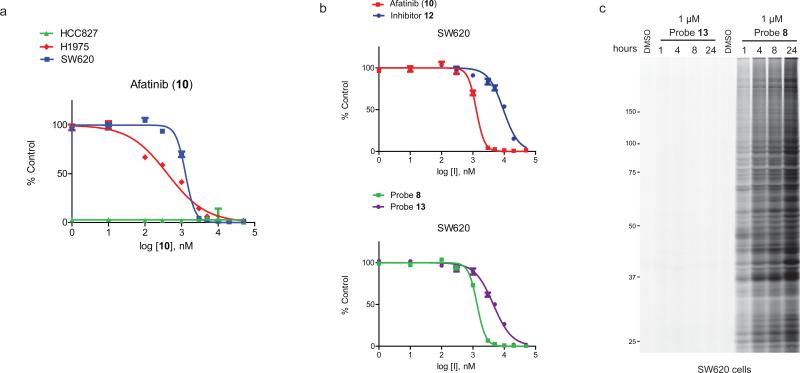
A critical question is whether off-target protein reactivity is pharmacologically relevant at concentrations of covalent inhibitors required to produce biological effects through blocking target kinases. The EGFR system provided an excellent opportunity to address this question, as afatinib has been approved for the treatment of exon 19-deletion and L858R EGFR mutant cancers14 and remains under clinical development for the treatment of T790M EGFR mutant lung tumors (http://clinicaltrials.gov/ct2/show/NCT01647711?term=t790m&rank=1). Past preclinical studies have shown that afatinib displays much greater potency for killing exon 19-deletion and L858R-EGFR cancer cells compared to T790M-EGFR cancer cells (low-nM versus high-nM, respectively)33,35. We replicated these findings using the HCC827 (exon 19-del EGFR) and H1975 (L858R/T790M EGFR) cancer lines, where afatinib (and inhibitor 7 and probe 8) showed cytotoxicity IC50 values of < 1 nM and 300-400 nM, respectively (Fig. 6a and Supplementary Fig. 9). We also tested these compounds in three EGFR-independent cancer cell lines (MDA-MB-435S, NIH3T3, SW620), where they displayed cytotoxicity values that were within two to five-fold (0.7-2.2 μM) of those observed in H1975 cells (Fig. 6a and Supplementary Fig. 9). Interestingly, the unreactive, saturated analogues of the EGFR inhibitors (12-14; Supplementary Fig. 9) showed uniformly weaker cytotoxic activity across the EGFR-independent cell lines (Fig. 6b and Supplementary Fig. 9).
Figure 6.
Cytotoxicity and proteomic reactivity of DMAM-modified EGFR inhibitors. (a) Afatinib shows much greater cytotoxic activity in HCC827 cancer cells compared to H1975 or SW620 cancer cells. Data represent average values ± standard errors for triplicate measurements from two independent experiments (six total replicates). (b) Afatinib and probe 8 show greater cytotoxic activity in the EGFR-independent cell line SW620 than their unreactive saturated analogues 12 and 13, respectively (see Supplementary Fig. 9 for saturated analogue structures and additional cytotoxicity data). I, inhibitor or probe tested. Data represent average values ± standard errors for triplicate measurements from two independent experiments (six total replicates). (c) Probe 8, but not the saturated analogue 13 (in situ treatment with 1 μM of each probe), shows substantial time-dependent proteomic reactivity in SW620 cells as evaluated by gel-based ABPP. Gel-based ABPP experiments were performed in duplicate with consistent results.
We next correlated the cytotoxicity profiles of EGFR inhibitors with their proteome reactivity as measured by gel-based ABPP. In the low-nM range required to kill HCC827 cells, probe 8 showed high selectivity for EGFR (Supplementary Fig. 6c). In contrast, at high nM-low μM concentrations (approximating the IC50 values for killing H1975 and EGFR-independent cancer cells), probe 8 showed extensive concentration-(Supplementary Fig. 6c) and time-dependent (Fig. 6c, Supplementary Fig. 6b, and Supplementary Fig. 10) reactivity with the proteomes of all cell lines examined. Negligible time-dependent proteomic reactivity was observed with saturated analogue probe 13 (Fig. 6c and Supplementary Fig. 10). ABPP-SILAC experiments performed with 3 μM of 8 [corresponding to the IC90 value for killing of H1975 cells (Fig. 6a)] confirmed the remarkably broad proteomic reactivity of this probe, which canvased > 80 and 400 proteins after 1 and 8 h probe treatments, respectively (Supplementary Fig. 11 and Supplementary Table 3). Pre-treatment with afatinib (3 μM) revealed that the 8-labeled proteins corresponded to a limited number of specific off-targets (e.g., afatinib-competed) and many additional non-specific (afatinib-insensitive) off-targets (Supplementary Fig. 11 and Supplementary Table 3).
These studies, taken together, indicate that covalent EGFR inhibitors bearing a DMAM modification cause cell death in EGFR-independent cancer cells through a mechanism that depends on the reactivity of the inhibitors. Additionally, the potency of this effect approaches the potency at which these compounds kill T790M-EGFR cancer cells (e.g., the IC50 for killing EGFR-independent cancer cells equals the IC70-80 for killing H1975 cells; Fig. 6a and Supplementary Fig. 9).
Discussion
Understanding target engagement and off-target interactions is important for all small-molecule probes36. These parameters are, somewhat ironically, much easier to measure for irreversible inhibitors than reversibly acting agents, as the very feature that has historically concerned scientists about irreversible probes, namely their covalent binding to proteins37, also provides a means to detect, enrich, and identify interacting proteins in a proteome-wide manner directly in living systems. We27,38-40, and others17,41,42, have used bioorthogonal probes to assess the proteome-wide specificity of irreversible inhibitors in cell and animal models. Nonetheless, these methods have only been applied to a handful of probes and the basis for and pharmacological relevance of off-target activity (specific or non-specific) has gone mostly uninvestigated. Recognizing a growing interest in the development of irreversible inhibitors in both academia and industry3,7-11, including the recent approval of the first covalent kinase inhibitors afatinib and ibrutinib for treatment of non-small cell lung cancer14 and CLL15, respectively, we felt that establishing a more general and rigorous strategy for characterizing the targets of irreversible probes was warranted. Here, we have presented a chemoproteomic platform that meets this goal and applied it to determine the protein reactivity profiles of irreversible kinase inhibitors in both cultured cancer cells and in vivo.
Several important insights were gained from our analysis. First, for most of the covalent kinase inhibitors examined, the off-targets were principally specific in that they showed evidence of either competition by excess parent inhibitor or selective labeling by a subset of the probes under investigation. Specific off-targets were not, however, restricted to kinases, and included proteins from diverse mechanistic classes, including enzymes, receptors, and proteins of uncharacterized function. A unifying feature among these proteins was the presence of active site or functional cysteines, which we confirmed in several instances were required for inhibitor reactivity. These data indicate that proteins bearing functional cysteine residues represent a common source for off-target activity of covalent kinase inhibitors.
An exceptional probe was the DMAM agent 8, which exhibited extensive, time-dependent proteome reactivity in cancer cells. This outcome could be explained, at least in part, by the high and persistent cellular concentrations of probe 8, which converted a selective labeling profile for EGFR observed shortly after treatment to widespread proteome-wide reactivity at later time points (Fig. 5b). That the concentrations at which DMAM-modified EGFR inhibitors started to exhibit substantial proteomic reactivity (≥ 0.1 μM) correlated with the emergence of cytotoxic activity in EGFR-independent cancer cells (IC50 values of 0.7-2.2 μM) leads us to conclude that off-target protein modification is a principal driver of the EGFR-independent cell death effects of these compounds. Also consistent with this interpretation is the uniformly greater cytotoxicity displayed by probe 8, inhibitor 2, and afatinib compared to their saturated analogues. We suspect that the cytotoxic effects of DMAM-modified EGFR inhibitors are caused by reactivity with many proteins in cells, rather than being due to the specific inactivation of a single protein for multiple reasons: 1) the tested DMAM-modified EGFR inhibitors killed all three EGFR-independent cancer cell lines with similar potencies; and 2) a very large number of time-dependent protein labeling events were observed (> 400) at inhibitor concentrations that killed these cancer cells.
While the DMAM-modified inhibitors killed exon 19-del-EGFR cancer cells at concentrations where selective EGFR labeling was observed, the concentration of these compounds required to kill T790M-EGFR cancer cells traversed into the range where extensive off-target reactivity and EGFR-independent cytotoxicity was observed (Fig. 6a, b and Supplementary Fig. 6, 9, and 10). Our results thus highlight potential challenges for current efforts to expand the clinical utility of afatinib beyond exon 19-deletion and L858R-EGFR tumors to include T790M-EGFR tumors. We should note that the mechanistic basis for the reduced activity of DMAM-modified inhibitors against T790MEGFR cancer cells remains unclear, with some33,43, but not all studies44 suggesting reduced potency for this mutant form of the kinase. Regardless, the effect appears to be intrinsic to the T790M-EGFR protein, since introduction of this mutant kinase into cancer cells is sufficient to reduce their sensitivity to afatinib35.
The reduced proteomic reactivity of probe 6, which still maintained good activity against its target kinase BTK, indicates that it may be possible, for exceptionally potent covalent inhibitors, to trade some potency in exchange for improved selectivity. This could be particularly important for tool compounds, where a defined, but restricted window of selectivity, as we observed for inhibitor 2/probe 4, may complicate biological experiments. Indeed, we have found many papers in the literature where 2 (ibrutinib) was applied to cells at ≥ 5 μM concentrations and its effects interpreted to occur through BTK45-48. Based on our chemoproteomic profiles, it would be difficult to draw such conclusions with confidence given the large number of proteins that react with probe 4 in this concentration range. We should also note, however, that reductions in covalent reactivity may not correlate with improvements in selectivity for reversible binding to off-targets. As we have shown for EGFR inhibitors, unreactive, saturated analogues of Michael acceptor-containing kinase inhibitors can serve as useful control probes for discerning pharmacological effects that are mediated by covalent versus non-covalent mechanisms. Finally, from a technical perspective, our finding that modifications to the Michael acceptor can dramatically alter proteome reactivity indicates that efforts to create chemoproteomic probes that authentically report on inhibitor selectivity should avoid attaching reporter tags directly to the inhibitor's reactive group (as has been done previously for 218).
Projecting forward, we believe that the chemoproteomic platform described herein should be applicable to any irreversible inhibitor, assuming that it can be modified by an alkyne without substantial alterations in structure or activity. We elected to characterize advanced kinase inhibitors in this study, but chemoproteomics can also facilitate the discovery of kinases that are targeted by covalent inhibitors in phenotypic screens41. As a greater number of covalent kinase inhibitors are examined using chemoproteomics, it will be interesting to determine whether a finite set of specific off-targets is identified. Considering the limited overlap in specific off-targets observed for 1 and 2 in this study, combined with the large and diverse number of proteins that require cysteine residues for function19-21, we suspect that many additional cysteine-dependent proteins in the human proteome may prove sensitive to covalent kinase inhibitors. Finally, we are emboldened by the discovery that irreversible kinase inhibitors targeting cysteine can have, in general, only a limited number of off-target interactions in cells and these off-targets appear to be confined to proteins that themselves possess active-site or functional cysteines. That the off-targets also exhibit a clear structure-activity relationship indicates achieving selectivity for covalent kinase inhibitors, or for that matter, any compound that acts via cysteine adduction, should have a medicinal chemistry solution, as long as these efforts are integrated with chemoproteomics as a guiding platform.
Online Methods
Cell culture
Cells were grown at 37 °C under a humidified 5% CO2 atmosphere, in a culture medium consisting of high-glucose DMEM (Caisson Labs) for A431, HEK293T, NIH-3T3 and HeLa cells or RPMI (Caisson Labs) for Ramos, HCC827, and A549 cells. The SW620 and MDA-MB-435S cells were grown in L-15 media in a humidified atmosphere with no CO2 added. All media was supplemented with 10% FBS (Gemini), penicillin (50 IU/mL), streptomycin (50 μg/mL), and glutamine (2 mM) (Cellgro). For SILAC experiments, the culture medium was replaced with either SILAC DMEM or SILAC RPMI (Thermo) as appropriate, and the medium was supplemented with 10% dialyzed FBS, penicillin, streptomycin, and glutamine. For the isotopically heavy cell samples, 100 μg/mL of both [13C6,15N4]L-arginine-HCl and [13C6,15N2]L-lysine-HCl (Sigma-Aldrich) was also added to the culture medium. For the isotopically light cell samples, the culture medium was supplemented with 100 μg/mL of both L-arginine-HCl and L-lysine-HCl. Cells were passaged at least six times in isotope-containing medium before being used for ABPP-SILAC experiments.
In situ cell treatment, gel-based ABPP
Adherent cell lines were grown to ~ 90% confluence in 10 mL of growth media in 10 cm plates. The growth media was aspirated off, and the cells were washed 2X with DPBS (~ 10 mL), followed by the addition of fresh media (10 mL). The inhibitor or probe (10 μL of 1000X DMSO stock) was then added, the plate was swirled to disperse the compound and then incubated at 37 °C for 1 hr. For IC50 measurements, a 1 hr inhibitor pre-incubation was followed by a 1 hr probe chase. Following incubation, the media was removed and the cells were washed 3X with 10 mL DPBS. The cells were then harvested and stored as pellets at − 80 °C. For suspension cell lines (Ramos cells), the cells were grown to ~ 3 × 106 cells/mL in large culture flasks and then were aliquoted (10 mL) into 10 cm plates for treatment. The cells were then treated and stored as described above for the adherent cell lines.
In situ cell treatment, ABPP-SILAC
For adherent cell lines, the same procedure as above was followed, except that 10 cm plates were used and the treatment volume was increased to 10 mL. A total of 4 plates were treated for each biological replicate. For Ramos cells, the cells were grown as described above and then were aliquoted (15 mL) into 100 mL tissue culture flasks for treatment. The cells were then treated and harvested as described above.
Sample processing for in situ-labeled samples, gel-based ABPP
Cell pellets were thawed on ice and cold DPBS (200 μL) plus protease inhibitor cocktail (Roche) was added to each sample. The cells were lysed using a probe sonicator and the lysate was fractionated by centrifugation at 100,000 × g for 45 min. The supernatant (soluble proteome) was transferred to a separate microfuge tube and the pellet (membrane proteome) was resuspended in 200 μL cold DPBS (plus protease inhibitors) using a probe sonicator. Protein concentrations for each sample were determined using the DC protein assay (BioRad) and then nomalized to 1 mg/mL in a volume of 43 μL. Click chemistry was performed on each sample using final concentrations of 25 μM rhodamine azide, 1 mM Tris(2-carboxyethyl)phosphine (TCEP, Sigma Aldrich), 100 μM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, Sigma Aldrich) and 1 mM CuSO4 in a final volume of 50 μL. The samples were incubated at room temperature for 1 hr, followed by addition of 20 μL 4X SDS loading buffer. The samples were then loaded (30 μL) and resolved on a 10% SDS-PAGE gel and imaged using a Hitachi FMBIO-II flatbed fluorescence scanner. Fluorescence images are shown in grayscale.
In vitro cell lysate labeling, gel-based ABPP
Soluble and membrane proteome samples from untreated cells were obtained by lysis and ultracentrifuge fractionation as described above and were normalized to a concentration of 1 mg/mL in a volume of 49 μL. Samples were then treated with inhibitor or probe (1 uL 50X stock in DMSO), mixed by agitation, and incubated at 37 °C for 1 hr. For IC50 measurements, a 1 hr inhibitor treatment was followed by a 1 hr probe treatment. Click chemistry was then set up (reagents and concentrations same as described above) in a total volume of 57 μL and the samples were incubated at room temperature for 1 hr. The samples were treated with 20 μL 4X SDS-PAGE loading buffer and then loaded (30 μL) and resolved on a 10% SDS-PAGE gel. The gel was then imaged as described above.
Determination of IC50 values by competitive gel-based ABPP
Determination of IC50 values from gel-based competitive ABPP experiments was performed as previously described49.
Recombinant expression and labeling of proteins in HEK293T cells
The cDNA constructs for DUS2L, MAP2K7, and FAM213A were obtained from Origene in the pCMV6-entry vector, with expression of C-terminal FLAG tags. The cDNA construct for ALDH1A1 was obtained from Thermo Fisher and cloned into the pFLAG-CMV2 vector, with expression of an N-terminal FLAG tag. For MLTK, a HEK293T cell line with stable overexpression of an MLTK construct containing an N-terminal FLAG tag was used. Overexpression of the other proteins in HEK293T cells was obtained by transient transfection using the FuGENE HD transfection system according to the manufacturers protocol. Forty-eight hours after transfection, in situ gel-based ABPP was then performed as detailed above. Western blotting was accomplished as previously described50, using an anti-FLAG primary antibody, IRdye 680 (Licor Biosciences) secondary antibody and Licor Odyssey CLx near-infrared imager.
Sample processing for analysis by LC-MS/MS
Sample preparation for LC-MS/MS was performed as previously described51, with minor modifications. In brief, light and heavy cell pellets were processed as described above for in-situ labeled gel-based ABPP samples to obtain the membrane and soluble proteomes of both light and heavy samples. All samples were normalized to 2 mg/mL in 430 μL of DPBS and click chemistry was performed with final concentrations of 200 μM biotin-azide (Chempep Inc.), 1 mM TCEP, 100 μM TBTA, and 1 mM CuSO4 in a final volume of 500 μL. The reaction was allowed to proceed at room temperature for 1 hr, and the excess reagents were then removed by CHCl3/MeOH precipitation with 2 mL MeOH, 1.5 mL H2O, and 0.5 mL CHCl3. Following isolation, the heavy and light proteome pellets were then combined into 600 μL of MeOH and the sample was precipitated again by addition of 600 μL of H2O and 150 μL of CHCl3. The pellet obtained was air dried and then resuspended in 500 μL of aqueous 6M urea, 25 mM NH4HCO3. The peptides were reduced (DTT) and alkylated at cysteine (iodoacetamide) and then diluted to 6 mL with DPBS and enriched over avidin agarose (Sigma; 100 μL slurry) for 4 hrs at room temperature. The avidin beads were washed and an overnight on-bead trypsin digestion was performed as previously described51. The final trypsin digest sample (300 μL) was acidified with 16 μL of formic acid and pressure loaded onto a biphasic (strong cation exchange/reverse phase) capillary column for analysis by two-dimensional LC-MS/MS.
Mass spectrometry and data analysis
Mass spectrometry was performed using a Thermo Orbitrap Velos mass spectrometer, using a previously described protocol51,52. Peptides were eluted using a five-step multidimensional LC-MS (MudPIT53) protocol (using 0%, 25%, 50%, 80% and 100% salt bumps of 500 mM aqueous ammonium acetate, followed by an increasing gradient of aqueous acetonitrile and 0.1% formic acid in each step), and data were collected in data-dependent acquisition mode (400–1800 mass to charge ratio (m/z) and 30 data-dependent fragmentation (MS2) scans) with dynamic exclusion enabled (repeat count of 1, exclusion duration of 20 s). Prolucid searches allowed for variable oxidation of methionine (+15.9949 m/z), static modification of cysteine residues (+57.0215 m/z; iodoacetamide alkylation) and one end specificity for trypsin. Each data set was independently searched with light and heavy parameter files; for the light search, all other amino acids were left at default masses; for the heavy search, static modifications on lysine (+8.0142 m/z) and arginine (+10.0082 m/z) were specified. The precursor-ion mass tolerance was set to 50 ppm and the fragment-ion mass tolerance was the default assignment of 600 ppm. The data were searched using a human reverse-concatenated nonredundant (gene-centric) FASTA database that was assembled from the Uniprot database (www.uniprot.org). The resulting matched MS2 spectra were assembled into protein identifications, then filtered using DTASelect (version 2.0.47), and only half-tryptic or fully-tryptic peptides were accepted for identification, and only fully-tryptic peptides were considered for quantification. Peptide false positive rates for all datasets were < 0.3%. Redundant peptide identifications common between multiple proteins were allowed, but the database was restricted to a single consensus splice variant. SILAC ratios were quantified using in-house software as described (CIMAGE19). Briefly, extracted MS1 ion chromatograms (± 10 ppm) from both 'light' and 'heavy' target peptide masses (m/z) are generated using a retention time window (± 10 min) centered on the time when the peptide ion was selected for MS/MS fragmentation, and subsequently identified. Next, the ratio of the peak areas under the light and heavy signals (signal-to-noise ratio > 2.5) are calculated. Computational filters used to ensure that the correct peak-pair is used for quantification include a co-elution correlation score filter (R2 ≥ 0.8), removing target peptides with bad co-elution profile, and an 'envelope correlation score' filter (R2 > 0.8) that eliminates target peptides whose predicted pattern of the isotopic envelope distribution does not match the experimentally observed high-resolution MS1 spectrum. Also, peptides detected as singletons, where only the heavy or light isotopically labeled peptide was detected and sequenced, but which passed all other filtering parameters, were given a standard ratio of 20, which is the maximum SILAC ratio reported here.
Metabolite analysis
A431 cells were seeded at 2 × 106 cells/60 mm dish and cultured in complete DMEM medium (4 mL) for 24 h. The media was removed and the cells were washed once with 3 mL of DPBS. Fresh media (3 mL) was added to the plates followed by treatment with probe 3 or probe 6 (1000X stock in DMSO, 3 μL). Plates were incubated at 37 °C until time point, after which the media was aspirated and the plates were washed 5 x 5 mL of DPBS and the cells were harvested and the cell pellet collected. H2O-soluble cellular metabolites were extracted using a MeOH/H2O extraction protocol, essentially as previously described3. In brief, cell pellets were resuspended in 200 μL of an 80:20 mixture of MeOH/H2O and an internal deuterated standard, 1mM of deuterated serine-d3 (Cambridge Isotope Laboratories), was added to the extraction solution for absolute quantification and sample normalization. The mixture was sonicated for 2 x 8 pulses followed by a 5 min centrifugation at 14,000g. The supernatant was collected and stored at −80 °C until LCMS analysis.
Samples of metabolites from both probe treatments (3 replicates/probe) were run sequentially. H2O-soluble cellular metabolites were separated by reverse phase chromatography (Phenomenex, C18) using elution conditions as described previously1. MS analysis was performed on an Agilent G6410B tandem mass spectrometer using ESI source setting as described previously1. The mass spectrometer was run in MRM mode, monitoring the transition of m/z from 454.1 to 343.1 for probe 6 , 397.1 to 343.1 for probe 3, and 109.1 to 63.2 for serine-d3. Metabolites were quantified by measuring the area under the peak in comparison to an external calibration curve. Ion abundances were normalized against serine-d3.
Cytotoxicity Assays
All cell lines were plated at 5,000 cells/well in 96-well plates in100 μL growth media (except SW620, which was plated at 10,000 cells/well). The cells were then treated with compounds or DMSO (1% DMSO final) and incubated at 37 °C for 72 hrs. Cell density was then measured using the Sulforhodamine B assay54. In brief, the cells were fixed by addition of 100 μL of cold 10% aqueous trichloroacetic acid and incubation for 1 hr at 4 °C. The TCA solution was decanted and the plates were washed gently with H2O (3X) and incubated at room temperature until dry. The plates were then stained by addition of 100 μL of 0.057% (wt/vol) Sulforhodamine B solution and incubation at room temperature for 30 min. The staining solution was decanted and the plates were washed gently with 1% acetic acid (3X) and then allowed to dry at room temperature. The plates were then treated with 200 μL 10 mM Tris (pH 10.5) solution and incubated at room temperature for 30 minutes followed by OD measurements at 535 nM using a Tecan Infinite F500 instrument.
In vivo studies
C57BL/6 mice were injected (i.p.) with compounds using a vehicle composed of 17:1:1:1 (v/v/v/v) solution of saline/EtOH/DMSO/Cremophor EL (10 μL g−1). Mice were treated with inhibitors (20 mg/kg) or vehicle for 1 hr, followed by treatment with probes (10 or 20 mg/kg) for 1 hr. Mice were then anesthetized with isoflurane and euthanized by cervical dislocation. Tissues were harvested and washed with cold DPBS (3X) followed by homogenization using a NextAdvance bullet blender and Zirconium Oxide beads. Samples were then centrifuged at 1,500 × g for 5 minutes and supernatant was removed and sonicated using a probe sonicator to ensure complete cell lysis. The samples were centrifuged at 100,000 × g for 45 min to provide the membrane proteome as a pellet and cytosolic proteome as supernatant. The pellet was washed with cold DPBS and resuspended by sonication. Protein concentrations were normalized to 2 mg/mL and analyzed by gel-based ABPP as described above. All mouse studies were performed following protocols that received approval from the TSRIIACUC office.
Supplementary Material
Acknowledgments
We thank Gabriel Simon for assistance with proteomic data analysis. This work was supported by the National Institutes of Health (CA087660), Pfizer, and the Skaggs Institute for Chemical Biology.
Footnotes
Author contributions. B.R.L., L.R.W., M.M.D., and B.F.C. designed the experiments; B.R.L., L.R.W., and M.M.D. performed the experiments and analyzed data; C.J. assisted with experiments; C.W. and J.J.H. assisted with data analysis; J.D., A.M.G., E.C.H., T.O.J., J.C.K., S.N., L.R.R., M.E.S., B.W., L.O.W., and M.M.H. provided intellectual input for probe design, experimental strategy, and interpretation of proteomic data. M.M.H. and B.F.C. provided inter-institutional leadership and coordination; B.R.L, L.R.W, M.M.H, and B.F.C. wrote the manuscript.
Competing Financial Interests. The authors declare competing financial interests. J.D., A.M.G., E.C.H., T.O.J., J.C.K., S.N., L.R.R., M.E.S., B.W., L.O.W., and M.M.H. are employees of Pfizer. The research was partially funded by Pfizer.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, et al. Developing irreversible inhibitors of the protein kinase cysteinome. Chem Biol. 2013;20:146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenblum JS, Nomanbhoy TK, Kozarich JW. Functional interrogation of kinases and other nucleotide-binding proteins. FEBS Lett. 2013;587:1870–1877. doi: 10.1016/j.febslet.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Krishnamurty R, Maly DJ. Chemical genomic and proteomic methods for determining kinase inhibitor selectivity. Comb Chem High Throughput Screen. 2007;10:652–666. doi: 10.2174/138620707782507368. [DOI] [PubMed] [Google Scholar]
- 6.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 7.Singh J, Petter RC, Kluge AF. Targeted covalent drugs of the kinase family. Curr Opin Chem Biol. 2010;14:475–480. doi: 10.1016/j.cbpa.2010.06.168. [DOI] [PubMed] [Google Scholar]
- 8.Carmi C, Mor M, Petronini PG, Alfieri RR. Clinical perspectives for irreversible tyrosine kinase inhibitors in cancer. Biochem Pharmacol. 2012;84:1388–1399. doi: 10.1016/j.bcp.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 9.Barf T, Kaptein A. Irreversible protein kinase inhibitors: balancing the benefits and risks. J Med Chem. 2012;55:6243–6262. doi: 10.1021/jm3003203. [DOI] [PubMed] [Google Scholar]
- 10.Johnson DS, Weerapana E, Cravatt BF. Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med Chem. 2010;2:949–964. doi: 10.4155/fmc.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potashman MH, Duggan ME. Covalent modifiers: an orthogonal approach to drug design. J Med Chem. 2009;52:1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 12.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leproult E, Barluenga S, Moras D, Wurtz JM, Winssinger N. Cysteine mapping in conformationally distinct kinase nucleotide binding sites: application to the design of selective covalent inhibitors. J Med Chem. 2011;54:1347–1355. doi: 10.1021/jm101396q. [DOI] [PubMed] [Google Scholar]
- 14.Nelson V, Ziehr J, Agulnik M, Johnson M. Afatinib: emerging next-generation tyrosine kinase inhibitor for NSCLC. Onco Targets Ther. 2013;6:135–143. doi: 10.2147/OTT.S23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron F, Sanford M. Ibrutinib: first global approval. Drugs. 2014;74:263–271. doi: 10.1007/s40265-014-0178-8. [DOI] [PubMed] [Google Scholar]
- 16.Blair JA, et al. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Biol. 2007;3:229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 17.Cohen MS, Hadjivassiliou H, Taunton J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat Chem Biol. 2007;3:156–160. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honigberg LA, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pace NJ, Weerapana E. Diverse functional roles of reactive cysteines. ACS Chem Biol. 2013;8:283–296. doi: 10.1021/cb3005269. [DOI] [PubMed] [Google Scholar]
- 21.Shin NY, Liu Q, Stamer SL, Liebler DC. Protein targets of reactive electrophiles in human liver microsomes. Chem Res Toxicol. 2007;20:859–867. doi: 10.1021/tx700031r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cravatt BF, Wright AT, Kozarich JW. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu Rev Biochem. 2008 doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 23.Smaill JB, et al. Tyrosine kinase inhibitors. 17. Irreversible inhibitors of the epidermal growth factor receptor: 4-(phenylamino)quinazoline- and 4-(phenylamino)pyrido[3,2-d]pyrimidine-6-acrylamides bearing additional solubilizing functions. J Med Chem. 2000;43:1380–1397. doi: 10.1021/jm990482t. [DOI] [PubMed] [Google Scholar]
- 24.Rostovtsev VV, Green JG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Amer. Chem. Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 26.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 27.Bachovchin DA, et al. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proc Natl Acad Sci U S A. 2011;108:6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adibekian A, et al. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat Chem Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hur W, et al. Clinical stage EGFR inhibitors irreversibly alkylate Bmx kinase. Bioorg Med Chem Lett. 2008;18:5916–5919. doi: 10.1016/j.bmcl.2008.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan Z, et al. Discovery of selective irreversible inhibitors for Bruton's tyrosine kinase. ChemMedChem. 2007;2:58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 31.Woyach JA, et al. Bruton's tyrosine kinase (BTK) function is important to the development and expansion of chronic lymphocytic leukemia (CLL). Blood. 2014;123:1207–1213. doi: 10.1182/blood-2013-07-515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsou HR, et al. 6-Substituted-4-(3-bromophenylamino)quinazolines as putative irreversible inhibitors of the epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor (HER-2) tyrosine kinases with enhanced antitumor activity. J Med Chem. 2001;44:2719–2734. doi: 10.1021/jm0005555. [DOI] [PubMed] [Google Scholar]
- 33.Li D, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponader S, et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119:1182–1189. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y, et al. The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor. Mol Cancer Ther. 2012;11:784–791. doi: 10.1158/1535-7163.MCT-11-0750. [DOI] [PubMed] [Google Scholar]
- 36.Simon GM, Niphakis MJ, Cravatt BF. Determining target engagement in living systems. Nat Chem Biol. 2013;9:200–205. doi: 10.1038/nchembio.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama S, et al. A zone classification system for risk assessment of idiosyncratic drug toxicity using daily dose and covalent binding. Drug Metab Dispos. 2009;37:1970–1977. doi: 10.1124/dmd.109.027797. [DOI] [PubMed] [Google Scholar]
- 38.Ahn K, et al. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol. 2009;16:411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang JW, Cognetta AB, 3rd, Niphakis MJ, Cravatt BF. Proteome-Wide Reactivity Profiling Identifies Diverse Carbamate Chemotypes Tuned for Serine Hydrolase Inhibition. ACS Chem Biol. 2013;8:1590–1599. doi: 10.1021/cb400261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsuboi K, et al. Potent and selective inhibitors of glutathione S-transferase omega 1 that impair cancer drug resistance. J Am Chem Soc. 2011;133:16605–16616. doi: 10.1021/ja2066972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishino M, et al. Hypothemicin, a fungal natural product, identifies therapeutic targets in Trypanosoma brucei. eLife. 2013;2:e00712. doi: 10.7554/eLife.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang PY, et al. Activity-based proteome profiling of potential cellular targets of Orlistat--an FDA-approved drug with anti-tumor activities. J Am Chem Soc. 2010;132:656–666. doi: 10.1021/ja907716f. [DOI] [PubMed] [Google Scholar]
- 43.Solca F, et al. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. Journ Pharmacol Exp Ther. 2012;343:342–350. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz PA, et al. Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. Proc Natl Acad Sci U S A. 2014;111:173–178. doi: 10.1073/pnas.1313733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herman SE, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117:6287–6296. doi: 10.1182/blood-2011-01-328484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dasmahapatra G, et al. The Bruton tyrosine kinase (BTK) inhibitor PCI-32765 synergistically increases proteasome inhibitor activity in diffuse large-B cell lymphoma (DLBCL) and mantle cell lymphoma (MCL) cells sensitive or resistant to bortezomib. Br J Haematol. 2013;161:43–56. doi: 10.1111/bjh.12206. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 47.Rushworth SA, et al. BTK inhibitor ibrutinib is cytotoxic to myeloma and potently enhances bortezomib and lenalidomide activities through NF-kappaB. Cell Signal. 2013;25:106–112. doi: 10.1016/j.cellsig.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Tai YT, et al. Bruton tyrosine kinase inhibition is a novel therapeutic strategy targeting tumor in the bone marrow microenvironment in multiple myeloma. Blood. 2012;120:1877–1887. doi: 10.1182/blood-2011-12-396853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu KL, et al. DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat Chem Biol. 2012;8:999–1007. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weerapana E, Simon GM, Cravatt BF. Disparate proteome reactivity profiles of carbon electrophiles. Nat Chem Biol. 2008;4:405–407. doi: 10.1038/nchembio.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods. 2013;10:259–264. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 54.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.