Abstract
To investigate the role of Hippo pathway signaling during vertebrate development transgenic zebrafish lines were generated and validated to dynamically monitor and manipulate Yap/Taz-Tead activity. Spatial and temporal analysis of Yap/Taz-Tead activity suggested the importance of Hippo signaling during cardiac precursor migration and other developmental processes. When the transcriptional co-activators, Yap and Taz were restricted from interacting with DNA-binding Tead transcription factors through expression of a dominant negative transgene, cardiac precursors failed to migrate completely to the midline resulting in strong cardia bifida. Yap/Taz-Tead activity reporters also allowed us to investigate upstream and downstream factors known to regulate Hippo signaling output in Drosophila. While Crumbs mutations in Drosophila eye disc epithelia increase nuclear translocation and activity of Yorkie (the fly homolog of Yap/Taz), zebrafish crb2a mutants lacked nuclear Yap positive cells and down-regulated Yap/Taz-Tead activity reporters in the eye epithelia, despite the loss of apical-basal cell polarity in those cells. However, as an example of evolutionary conservation, the Tondu-domain containing protein Vestigial-like 4b (Vgll4b) was found to down-regulate endogenous Yap/Taz-Tead activity in the retinal pigment epithelium, similar to Drosophila Tgi in imaginal discs. In conclusion, the Yap/Taz-Tead activity reporters revealed the dynamics of Yap/Taz-Tead signaling and novel insights into Hippo pathway regulation for vertebrates. These studies highlight the utility of this transgenic tool-suite for ongoing analysis into the mechanisms of Hippo pathway regulation and the consequences of signaling output.
Keywords: Hippo signaling, fluorescent reporter, heart development, eye development, in vivo imaging
1. Introduction
The Hippo signaling pathway is tightly controlled and critical during development and is often deregulated in disease. This evolutionary conserved signaling network influences tissue growth by regulating cell proliferation, apoptosis, and cell fate decisions. The core components of the pathway include the Serine Threonine Kinases 3 and 4 (Stk3/4; Hippo in Drosophila); two other serine threonine kinases called Large Tumor Suppressor 1 and 2 (Lats1/2; Warts in Drosophila); two scaffolding proteins Sav1 (Salvador in Drosophila) and Mps One Binder Kinase Activator-like 1A and 1B (Mobkl1a/1b; Mob as Tumor Suppressor in Drosophila). These proteins form a complex whose activity regulates the phosphorylation state, stability, and localization of the downstream transcriptional co-activators Yes-associated protein and WW Domain-containing transcriptional regulator 1 (Yap and Wwtr1 or more commonly, Taz; Yorkie in Drosophila). When the core kinases are inactive, Yap/Taz are free to translocate to the nucleus to activate transcription via interaction with members of the Tea Domain Family of transcription factors (Tead1–4; Scalloped in Drosophila) (Halder and Johnson, 2011; Yu and Guan, 2013). Regulatory factors both upstream and downstream of the core kinase complex are not well understood. However, some recently discovered components have been investigated in invertebrates and cell culture, but their role and significance has not been fully evaluated within vertebrates. Examples include Crumbs as an upstream component for controlling Hippo signaling activity, and the role of Vestigial-like 4 (Vgll4) as a co-repressor of Tead transcription factors.
Crumbs homologs (Crb1–3) are transmembrane proteins that contribute to a protein complex associated with apical cell-cell junctions of epithelial tissues and regulate various aspects of polarity. The Crumbs polarity complex was recently linked to the Hippo pathway in Drosophila through the effectors Kibra, Expanded, and Merlin. Crumbs manipulation in Dropsophila revealed tissue and developmental timing specificity on Hippo signaling output. Specifically, it was reported that either Crumbs overexpression or deletion in eye and wing imaginal discs resulted in mis-localized Expanded, increased nuclear Yorkie, and tissue overgrowth (Chen et al., 2010; Ling et al., 2010). In mammalian cell culture, several protein-binding assays showed that Yap and Taz interacted with the Crumbs polarity complex. When crb3 was knocked down, phosphorylation of the cytoplasmic retention domain for Yap was reduced and there was a concomitant increase in nuclear Yap (Varelas et al., 2010). Together, the authors concluded that the Crumbs complex can sequester Yap/Taz at apical junctions in cultures of high-density, therefore preventing Yap/Taz-mediated proliferation. These observations in flies and cell culture provide strong rationale for investigation into the role of Crumbs and other upstream components on Hippo signaling in vertebrate animals.
The Tondu-domain containing protein Tgi was recently characterized as a downstream regulator of Hippo signaling in Drosophila (Koontz et al., 2013). Tgi interacts with Yorkie and competes for Scalloped binding, suggesting a model where Tgi acts as a co-factor to enhance Scalloped-mediated default repression. Interestingly, the mammalian ortholog of Tgi, Vestigial-like 4 (Vgll4) did not interact with Yap, but was found to bind Tead2 and block transcription within in vitro assays. Consistent with its role as a co-repressor of Scalloped/Tead-type transcription factors, overexpression of Vgll4 in mouse transgenic livers that also over-expressed Yap, reduced the Yap-mediated overgrowth phenotype. It will be important to confirm the role of Vgll4 as a co-repressor in other contexts in vivo and investigate whether this occurs through endogenous Yap/Taz-Tead signaling.
We are interested in how the Hippo network and other polarized signaling pathways function during development. To augment our understanding of Hippo signaling, particularly across different tissues in vivo, we have generated a tool-suite for monitoring and manipulating the Hippo-Yap/Taz-Tead signaling network in zebrafish. In particular, we generated fluorescent Hippo-Yap/Taz-Tead responsive transgenic lines based on the previously characterized 4xGTIIC enhancer (Mahoney et al., 2005). This synthetic transcriptional enhancer contains four copies of the GTIIC sequence of the SV40 proximal promoter. The GTIIC element, as well as the multimerized variant, was found to bind Tead proteins, which subsequently interact with Yap or Taz to strongly activate transcription (Davidson et al., 1988; Mahoney et al., 2005; Sawada et al., 2005). We have verified the 4xGTIIC transgenic lines as Hippo-Yap/Taz-Tead reporters by using gain and loss of function experiments, along with the analysis of endogenous Yap localization. We next used these lines to investigate the significance of Hippo signaling during early heart morphogenesis and to test the role of Crb2a and Vgll4b as potential endogenous, upstream and downstream regulators of vertebrate Hippo signaling.
2. Results
2.1. Establishment of a Hippo-Yap/Taz-Tead responsive transgenic reporter lines
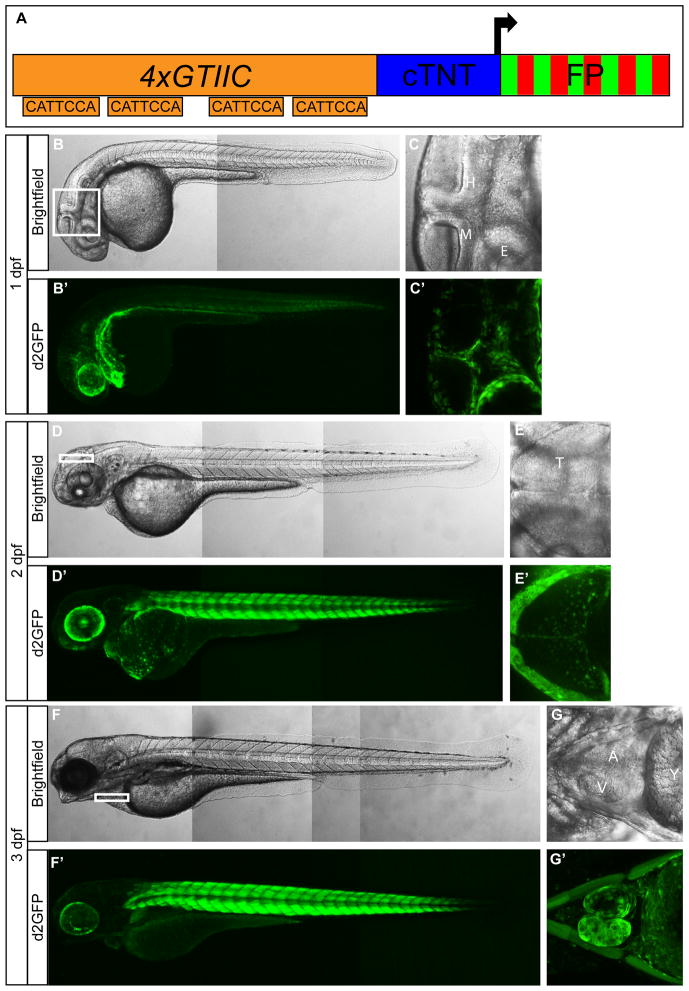
The 4xGTIIC promoter contains 4 multimerized SV40 proximal promoter GTIIC sequences, which are consensus Tead binding sites (Fig. 1A) and was previously reported to be responsive to Yap/Taz-Tead activity (Davidson et al., 1988; Mahoney et al., 2005). The 4xGTIIC and other Tead and Scalloped multimerized binding site promoters have been shown to be responsive to Hippo pathway manipulation in other models and contexts (Dupont et al., 2011; Ota and Sasaki, 2008; Zhang et al., 2008). Therefore, we generated stably transgenic zebrafish that contain the 4xGTIIC promoter driving expression of d2GFP, eGFP, or mCherry. Multiple founders were generated, isolated, and characterized to ensure consistency in the pattern of expression. For each transgenic construct, offspring from one founder was used to establish stable expressing lines. High expression in the developing larvae was noted in the epidermis, cardiac progenitor cells, presumptive sinus venosus, undifferentiated endoderm, otic and lens vesicles, retinal pigmented epithelium (RPE), cranial mesenchymal cells, multiple cell types in the heart, and within striated muscle of the trunk (Fig. 1). Several of the 4xGTIIC positive cell types have been shown to express Yap/Taz-Tead target genes ctgfa anc cyr61 in zebrafish (Fernando et al., 2010). At 1 day post fertilization (dpf) the 4xGTIIC reporter is highly active in migratory cells located at the midbrain/hindbrain boundary and in the craniofacial region (Fig. 1C′, Fig. 2, Fig. 3A′ and B′). Co-expression analysis of these migratory cells revealed active transgene expression in foxc1b-positive mesenchymal cells, but not sox10-positive neural crest cells (Fig. 2). Expression profile comparisons between the d2GFP, eGFP, and mCherry transgenes revealed significant overlap in the areas of high expression (Supplemental Fig 1). In order to verify that the transgenic reporter was active in tissues with elevated nuclear Yap expression, we performed immunofluorescence to assess endogenous Yap protein localization. Co-staining the 4xGTIIC:(d2GFP/eGFP) with a Yap antibody showed strong correlation between nuclear located Yap and GFP expression (Fig. 3A‴, B‴, C‴, and D‴), suggesting our transgenic lines faithfully report endogenous Yap-Tead activity. For further verification we manipulated levels of Yap and Taz in specific tissues within the 4xGTIIC reporter lines.
Figure 1. 4xGTIIC:d2GFP expression during zebrafish development.
(A) Schematic of the Yap/Taz-Tead reporter construct. (B′,D′,F′) Low magnification images show 4xGTIIC:d2GFP expression in whole embryos/larvae at 1 dpf (B′), 2 dpf (D′) and 3 dpf (F′). (C′, E′, G′) Higher magnification images show d2GFP expression in non-neural cells located at the (C′) midbrain-hindbrain boundary and around the eye at 1 dpf, within the tectum at 2 dpf (E′), and in the heart at 3 dpf (G′). Brightfield images (B,C,D,E,F,G) are placed above each fluorescent image. cTNT=chicken troponin T minimal promoter; FP= fluorescent protein; M=midbrain; H=hindbrain; E=eye; A=Atrium; V=Ventricle; Y=Yolk. B, B′, C, C′, D, D′, F, F′ = lateral view; E, E′ = dorsal view; G, G′ = ventral view.
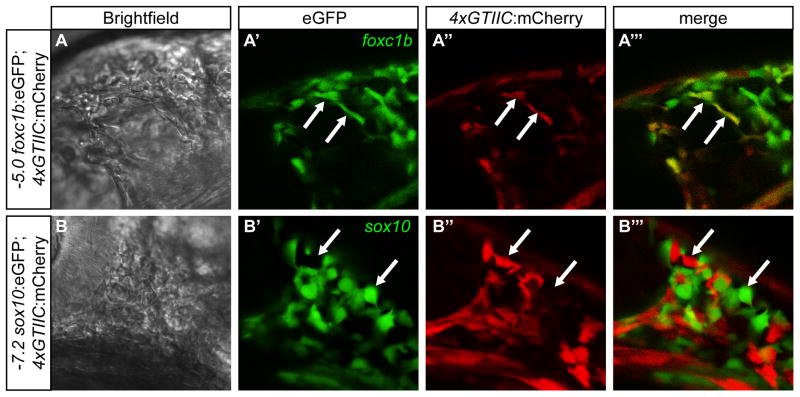
Figure 2. 4xGTIIC:mCherry co-localizes with -5.0 foxc1b:eGFP but not -7.2sox10:eGFP positive cells.
(A–A‴) Co-expression of 4xGTIIC:mCherry and -5.0 foxc1b:eGFP in superficial migratory cranial mesenchymal cells located above the midbrain/hindbrain boundary. The arrows indicate co-localzation between the two transgenes. (B–B‴) Non-overlapping expression of 4xGTIIC:mCherry and -7.2sox10:eGFP in superficial migratory neural crest cells located between the eye and midbrain/hindbrain boundary. The arrows indicate cells that only express one of the transgenes. Embryos are at the 22 somite stage.
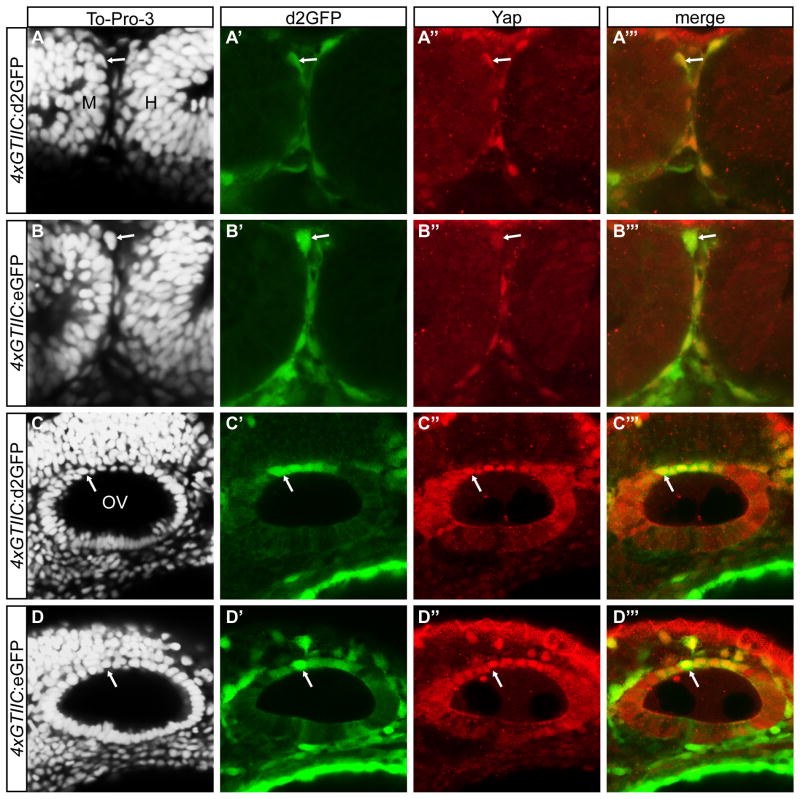
Figure 3. Nuclear Yap protein co-localizes with 4xGTIIC:d2GFP/eGFP positive cells.
(A–A‴, B–B‴) d2GFP/eGFP positive cells located at the midbrain-hindbrain boundary contain nuclear Yap (red) as detected by immunofluorescence. (C–C‴, D–D‴) Cells located in the dorsal otic vesicle express d2GFP/eGFP and nuclear Yap (red). M=midbrain; H=hindbrain; OV=otic vesicle.
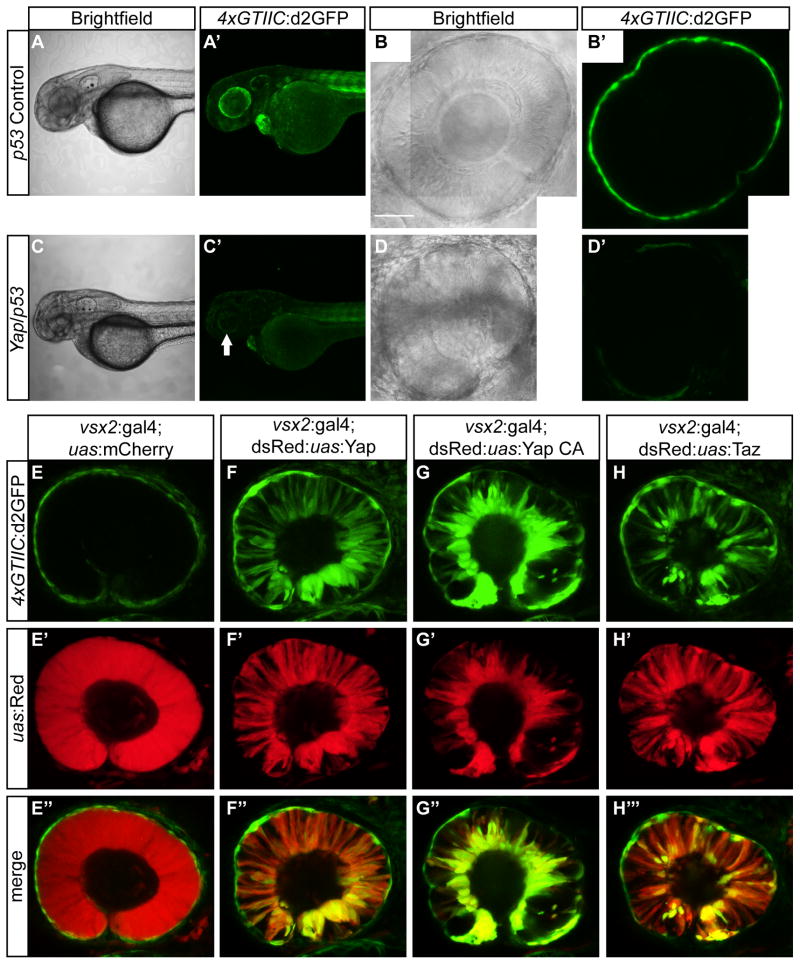
Yap knockdown was accomplished by microinjecting a splice morpholino (MO) that targets yap exon 2/intron 2 border (Skouloudaki et al., 2009). Embryos that were injected with yap MO showed decreased 4xGTIIC:d2GFP expression throughout the entire embryo at 2 dpf (Fig. 4C′). Particularly strong d2GFP reductions were noted in the RPE of the eyes (Fig. 4D′). To test the responsiveness of the reporter in tissues of low d2GFP expression, we overexpressed Yap, YapCA, or Taz in neural retinal progenitors. Overexpression of each factor resulted in enhanced and ectopic expression of d2GFP in this tissue (Fig. 4F, G, and H).
Figure 4. 4xGTIIC:d2GFP expression is decreased in response to Yap knockdown and enhanced with Yap or Taz overexpression.
(A′,C′) d2GFP expression levels are decreased in yap/p53 MO injected embryos (C′) compared to the p53 control embryos (A′) at 2 dpf. (B′,D′) yap/p53 MO embryos have a smaller eye and decreased RPE expression (D′) compared to p53 MO control embryos. (E–E″) Overexpression of mCherry does not cause enhanced or ectopic d2GFP expression. (F–F″) Overexpression of Yap, (G–G″) Yap constitutive active (CA) transgene (yapS87A) or (H–H″) Taz results in enhanced and ectopic d2GFP expression in retinal progenitor cells (embryos are 30 hpf). Scale bar=50 μm in B.
2.2. 4xGTIIC reporter activity in migratory cells and morphogenic tissues
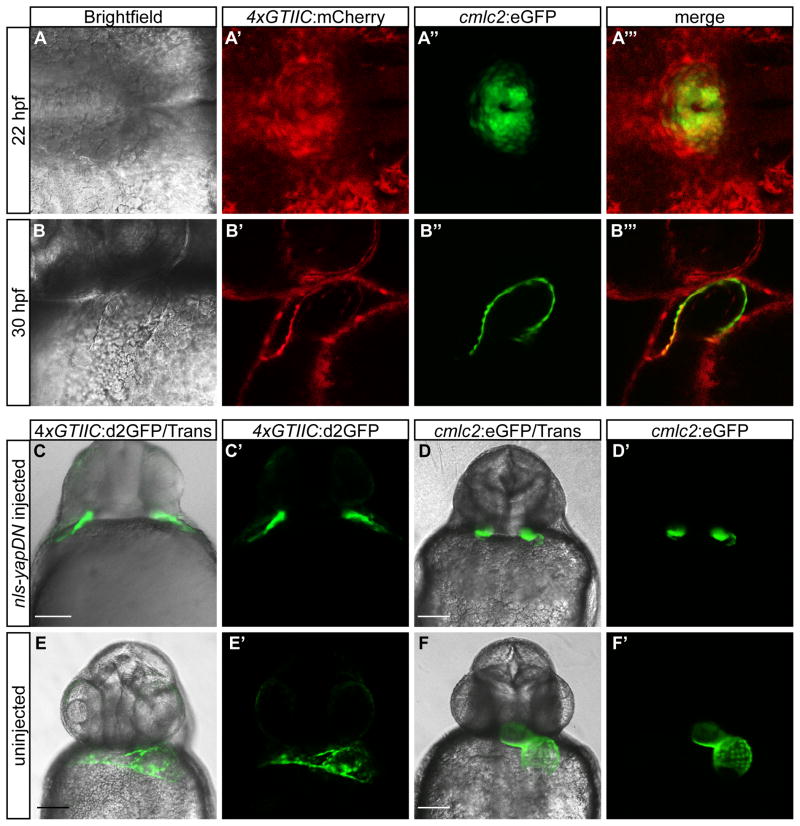
Throughout development exquisite regulation of cell migration and cell shape changes facilitate proper tissue morphology. In the 4xGTIIC transgenic lines, high expression is observed in cells and tissues undergoing these dynamic processes. For example, high Yap/Taz-Tead activity was observed in mesenchymal cells migrating away from the lateral mesoderm (Fig. 2; Supplemental Movie 1), in vascular progenitor cells forming the sinus venous (Fig. 1; Supplemental Movie 2), and in presumptive cardiac precursors migrating to the midline during heart tube formation (Fig 5A′, Supplemental Movie 2). To confirm the identity of cells migrating to the midline where the heart tube forms, we crossed the 4xGTIIC:mCherry line to a fish transgenic for cmlc2:eGFP. The cmlc2 promoter marks cardiac progenitors, as well as differentiated cardiomyocytes. Co-localization between the Yap/Taz-Tead reporter and cmlc2:eGFP expression was noted at 22 hours post fertilization (hpf) (Fig. 5A‴), just prior to heart formation, as well as at 30 hpf when the tubular heart has formed (Fig. 5B‴). The adjacent endoderm also appeared to be positive, but marker co-localization is needed to confirm this possibility. Based on the high expression within migrating cardiac progenitors prior to heart tube formation we predicted a functional role for nuclear Yap activity. Additional rationale for this hypothesis comes from the known requirement of S1P signaling for midline migration of cardiac progenitors during heart development (Kupperman et al., 2000; Osborne et al., 2008). Recent studies have demonstrated S1P as a regulator of Hippo signaling that results in nuclear Yap localization (Miller et al., 2012; Yu et al., 2012). To downregulate nuclear Yap/Taz-Tead transcriptional activation we injected mRNA into zebrafish embryos that encoded a dominant negative form of Yap (NLS-YapDN). Expression of NLS-YapDN in 4xGTIIC:d2GFP or cmlc2:eGFP positive embryos caused cardia bifida, but did not completely eliminate the differentiation of beating cardiac myocytes (Fig 5C, C′, D, and D′, Supplemental Movie 3). Similar phenotypes were observed with the overexpression of dominant negative Taz (data not shown). Indeed, either dominant negative protein should inhibit both Yap and Taz activity. We did not observe cardia bifida in Yap morphants. These results suggest a potential link between S1P and Hippo signaling during zebrafish cardic progenitor cell migration and heart development, and highlight the utility of the Yap/Taz-Tead reporters to analyze Hippo signaling during dynamic processes of development.
Figure 5. Yap/Taz-Tead nuclear activity is high in cardiac precursors and cardiomyocytes, and is important in cardiac precursor migration.
(A–B‴) Expression of (A′,B′) 4xGTIIC:mCherry and (A″,B″) cmlc2:eGFP in the developing heart field at (A–A‴) 22 hpf and (B–B‴) 30 hpf. (C–D′) Cardia bifida in (C,C′) 4xGTIIC:d2GFP and (D,D′) cmlc2:eGFP positive embryos injected with NLS-YapDN mRNA (28 hpf). (E–F′) Heart expression in uninjected (E,E′) 4xGTIIC:d2GFP and (F,F′) cmlc2:eGFP positive embryos (28 hpf). Scale bar = 100 μm in C, D, E, and F.
2.3. 4xGTIIC:d2GFP expression is downregulated in crb2a mutants
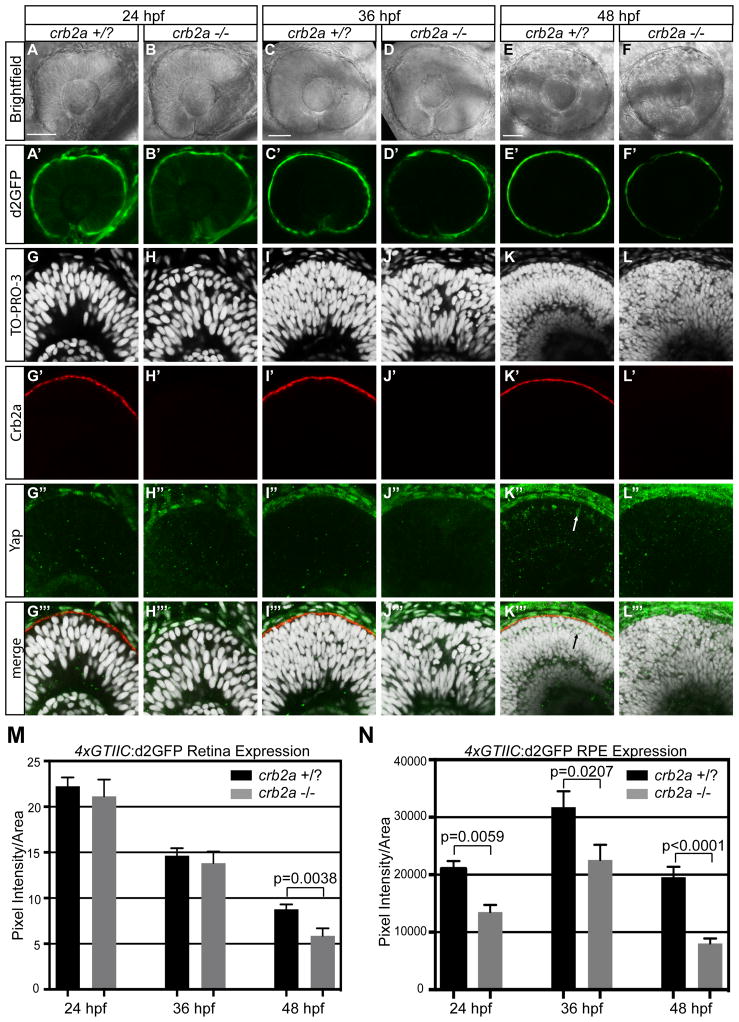
Evidence for the involvement of Crumbs proteins in the regulation of Hippo signaling has been primarily limited to Drosophila and cell culture (Chen et al., 2010; Ling et al., 2010; Varelas et al., 2010). In those studies, the requirement and role of Crumbs showed temporal and cell type specificity. To probe the role of Crumbs on Hippo signaling during vertebrate ocular development, we analyzed crb2a mutants that carried the 4xGTIIC:d2GFP transgene. Pixel intensity of d2GFP fluorescence was measured in the neural retina and RPE at 24, 36, and 48 hpf (Fig. 6M, N). Previous analysis of the crb2a mutants has shown that at 24 hpf neuroepithelial integrity is intact, but apical-basal polarity quickly becomes disrupted, resulting in severe patterning and lamination defects (Omori and Malicki, 2006). Analysis of the RPE cells at 48 hpf revealed a similar progression of disorder. RPE cells in crb2a mutants are smaller in size (mean=182.1 μm2) and appear disorganized compared to control cells (mean=227.5 μm2) (p=0.0004) (Supplemental Fig. 2 C–E). In neural retina of fish with the 4xGTIIC:d2GFP transgene, no difference in pixel intensity was measured between crb2a mutants and their wild-type siblings at 24 and 36 hpf (Fig. 6A′, B′, C′, D′, and M). At 48 hpf, however, there was a small but significant decrease in 4xGTIIC:d2GFP expression (p=0.0038) (Fig. 6E′, F′, and M). Within the RPE there was significant downregulation in reporter expression at all time-points: 24 hpf (p=0.0059); 36 hpf (p=0.0207), and 48 hpf (p<0.0001) (Fig. 6A′, B′, C′, D′, E′, F′, and N). These results are surprising based on observations in Drosophila and cell culture in which deletion of Crumbs homologs caused increased nuclear Yap localization and upregulated Scalloped/Tead transcriptional activity. To further probe the effects of crb2a mutations on Yap localization in zebrafish retinal cells, Yap immunofluorescence was performed. At 24 and 36 hpf, no change was noted in Yap localization between crb2a mutants and wild-type siblings (Fig. 6G–J″). At 48 hpf crb2a mutants lacked Yap positive cells within the neural retina, yet polarity was disrupted as indicated by nuclear lamination and ectopic actin foci (Fig. 6G′–L‴; Supplemental Fig. 2).
Figure 6. Loss of crb2a results in a decrease of 4xGTIIC:d2GFP expression in the neural retina at 48 hpf and at 24, 36, 48 hpf in the RPE.
(A–F′) Neural retina and RPE 4xGTIIC:d2GFP expression in crb2a +/? and crb2a −/− embryos. (G–L‴) Immunofluorescence of Yap (green) and Crb2a (red) in crb2a +/? and crb2a −/− embryos. Arrow indicates Yap positive cells in the crb2a +/? neural retina that do not appear in crb2a −/−. (M,N) Quantification of d2GFP pixel intensity in the (M) neural retina and (N) RPE of crb2a +/?/crb2a −/− embryos at 24 (n=46/n=13), 36 (n=32/n=28), and 48 (n=26/n=23) hpf. Error bars represent S.E.M. p=unpaired t-test with equal S.D. Scale bar=50μm in A,C, and E.
2.4. Vgll4b represses Yap activity in the RPE
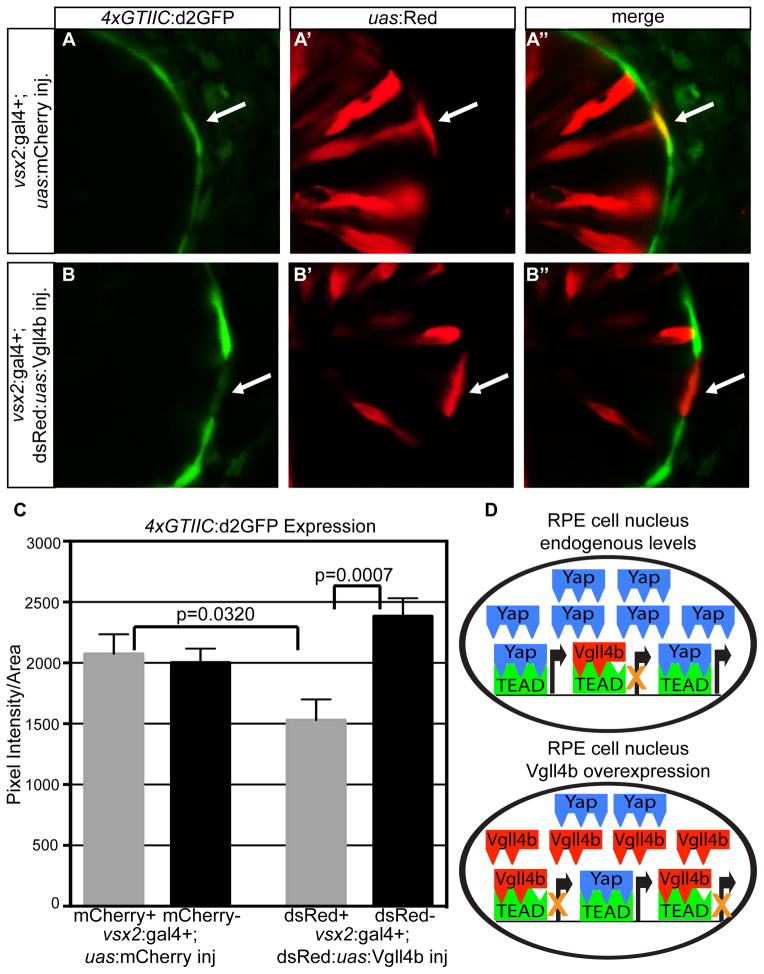
The overgrowth phenotype seen in mouse livers by the overexpression of Yap S127A can be recused by the co-expression of Vgll4 (Koontz et al., 2013). Due to the evidence of conserved function of Vgll4 with Dropsophila Tgi, we investigated whether Vgll4 can act as a co-repressor of endogenous Yap. Vgll4b, the zebrafish homologue of Vgll4, is expressed within the RPE, which is a tissue showing both strong endogenous nuclear Yap and high levels of Yap/Taz-Tead reporter activity. In cells that overexpressed Vgll4b there was a significant decrease in 4xGTIIC:d2GFP expression compared to the neighboring cells without Vgll4b overexpression (p=0.0007) (Fig. 7). There was also a significant decrease in the Yap/Taz-Tead reporter when compared to the UAS:mCherry injected controls (p=0.0320) (Fig. 7). These results indicate that Vgll4b can act to co-repress endogenous levels of Yap/Taz-Tead signaling in the developing RPE, potentially through competition with Yap/Taz binding of Teads (Fig 7D).
Figure 7. Vgll4b represses nuclear Yap/Taz-TEAD activity.
(A–B″) 4xGTIIC:d2GFP positive RPE cells expressing (A′) mCherry or (B′) Vgll4b. Arrows depict the mCherry and Vgll4b positive cells. (C) Quantification of d2GFP pixel intensity in mCherry + (n=19), mCherry – (n=38), dsRed/Vgll4b + (n=28), and dsRed/Vgll4b – (n=58) cell clones. More than 12 embryos were used for each condition. Error bars represent S.E.M. p=unpaired t-test with equal S.D. (D) Diagram of the relationship between Yap-Tead and Vgll4b-Tead in RPE cells expressing endogenous or overexpressed levels of Vgll4b.
3. DISCUSSION
Ongoing investigations of the components and function of the Hippo signaling network continues to reveal complexity and evolutionary conservation. In both invertebrates and vertebrates there is tissue and temporal specificity for the roles and regulation of the Hippo pathway during development and in disease. In this study we generated tools to monitor and manipulate Hippo-Yap/Taz-Tead signaling in zebrafish. Central among these resources is the Yap/Taz-Tead responsive transgenic reporter lines, which can be used to examine the cell type specificity and dynamics of the Hippo pathway during development. The 4xGTIIC transgenic lines incorporated the 4xGTIIC promoter, previously characterized as Tead-binding and transcriptionally responsive in cell culture (Mahoney et al., 2005). We have shown the 4xGTIIC promoter to be a faithful Yap/Taz-Tead pathway reporter through knockdown and overexpression of the downstream transcriptional co-activators Yap and Taz. Validation of these lines facilitated investigation of Hippo signaling during heart development and the relationship of Crb2a and Vgll4b on the Hippo pathway in vivo.
Temporal analysis of the expression pattern in 4xGTIIC:(d2)GFP/mCherry transgenic zebrafish implicated regulation of Hippo signaling in cardiac progenitor migration and heart formation. Consistent with high activity of the Yap/Taz-Tead reporter, disruption of Yap/Taz activity caused a failure of midline migration for cardiac progenitor cells resulting in significant cardia bifida. Although the cardiac progenitors failed to migrate properly after knockdown of Yap/Taz the rudimentary cardiomyocytes still expressed 4xGTIIC:d2GFP. A possible explanation for this maintained expression could be the result of NLS-YapDN mRNA stability. The strong effects of the overexpression of NLS-YapDN may happen early before mRNA or protein degradation occurs, allowing for the presumptive cardiac myocytes to turn on expression of the transgene after the precursors fail to migrate and start to form the rudimentary cluster of beating cardiomyocytes. Maintained expression may also occur because the levels of dominant negative protein cannot compete with up-regulation of Yap/Taz-Tead activity in differentiating cardiomyocytes. There are several previously characterized zebrafish mutants that show cardiac precursor migration defects, which lead to cardia bifida. Two mutants affect the sphingosine-1-phosphate (S1P) pathway (Kupperman et al., 2000; Osborne et al., 2008). Interestingly, two cell culture studies have linked S1P signaling to Yap/Taz nuclear activity but through slightly different mechanisms. S1P was shown to act through either a Lats1/2 dependent (Yu et al., 2012) or independent (Miller et al., 2012) mechanism. Conversely, there may be cell non-autonomous effects of Yap/Taz knockdown that result in failure of proper cardiac progenitor migration. For example Fibronectin secreted by the yolk syncytial layer can affect cadiomyocyte progenitor migration, an effect mediated by the S1P receptor (Matsui et al., 2007; Sakaguchi et al., 2006; Trinh and Stainier, 2004). Interestingly, fibronectin1 has been demonstrated as a target gene of Yap/Taz-Tead activity (Zhao et al., 2008). These results offer rationale for continued research into the role of Hippo signaling during heart development and a potential link between the S1P pathway mutants and their cardiac progenitor migration defects (Kupperman et al., 2000; Osborne et al., 2008). Continued analysis of the 4xGTIIC reporter will also aid in determining the importance of Hippo pathway regulation in other cells during tissue morphogenesis.
Previous studies on loss of the apical-basal polarity protein Crumbs in Drosophila and mammalian cell culture revealed an increase in nuclear Yorkie and Yap/Taz, respectively. In Drosophila, loss of Crumbs resulted in Yorkie dependent increased cell proliferation due to Expanded mislocalization (Chen et al., 2010; Ling et al., 2010). In cell culture, crb3 knock-down also caused increased nuclear Yap/Taz, but the resultant epithelial-to-mesenchymal transition was due to Yap/Taz-Smad interactions (Varelas et al., 2010). Other factors that regulate apical-basal cell polarity in Drosophila can also down-regulate Hippo signaling and drive nuclear Yorkie when deleted (Genevet and Tapon, 2011). That crb2a mutants showed a progressive loss of apical-basal cell polarity allowed us to address two different cellular relationships. By monitoring 4xGTIIC:d2GFP activity and endogenous Yap localization in crb2a mutants we could investigate the association between Crb2a and Yap/Taz-Tead activity separately from the relationship between cell polarity and Yap/Taz-Tead activity. Therefore, we predicted that loss of crb2a and polarity in the zebrafish retina would show increased Yap/Taz-Tead reporter activity. However, we found that despite strong loss of cell polarity, there was a modest, but significant decrease in Yap nuclear localization and activity. These results are consistent with recent analysis done in mice with crb1/2 mutant retinas. Two studies that assessed Hippo pathway signaling changes found either no change or a slight decrease of Hippo target genes and Yap protein levels (Alves et al., 2013; Pellissier et al., 2013). One possibility for this difference may lay in the fact that vertebrate Yap is subjected to ubiquitin-mediated degradation via a β-TRCP1 recognition motif termed a phosphodegron (Zhao et al., 2010), as well as through cytoplasmic retention. Potentially, loss of Crb2a and consequently the disruption of apical junctions where Yap can be sequestered may result in degradation of the transcriptional co-activator in tissues such as the retina. For Drosophila Yorkie, the phosphodegron does not exist. Alternatively, a lack of up-regulation in Yap activity in the neural retina may be due to redundancy/compensation by other Crb isoforms. Further investigation will be needed to assess the relationship across multiple cell types. Loss of Crb2a in the RPE resulted in an earlier and stronger decrease in 4xGTIIC:d2GFP expression. This may be due to the higher endogenous Yap/Taz-Tead activity in the RPE as compared to the neural retina. Alternatively, decreased Yap activity may reflect a general block in RPE cell differentiation or survival. When crb2a mutant zebrafish are allowed to pigment there is an obvious patchy pigmentation defect, which has not been well characterized.
Tgi and its mammalian orthologue Vgll4 have recently been shown to act as transcriptional co-repressors for Scalloped and Tead, respectively (Koontz et al., 2013). In that study Vgll4 was used to repress overgrowth phenotypes caused by an overexpression of Yap and a conditional knockout of nf2 in the liver. We sought to further test the concept of Vgll4 as a co-repressor and analyze its function on endogenous Yap/Taz-Tead activity. The overexpression of Vgll4b in RPE cells did not produce a noticeable phenotype, but it did down-regulate the expression of the 4xGTIIC:d2GFP reporter. These results are consistent with the previously reported function of Vgll4 and suggest that Vgll4b can act as a transcriptional co-repressor of Yap/Taz-Tead activity in the zebrafish RPE. Vestigial-like proteins have been reported to interact with Tead at two of the three same interfaces as Yap (Pobbati and Hong, 2013). Even though Yap interacts on one more interface than Vgll4b, Yap is less likely to interact with Tead in the presence of excess Vgll4b. Furthermore, the measurable changes on reporter expression without obvious phenotype consequences, suggests the 4xGTIIC:d2GFP reporter is sensitive enough to detect subtle changes in Hippo pathway regulation. The sensitivity of the 4xGTIIC reporter could therefore be leveraged for drug screens or used to identify tissues in which Hippo signaling is active, but where manipulations to Hippo components do not cause immediate or obvious defects. Indeed, targeting the Hippo pathway for pharmacologic modifiers may prove to be important in cancer treatment (Johnson and Halder, 2014). Overall, we have generated a number of transgenic tools to monitor and manipulate the Hippo-Yap/Taz-Tead signaling network in vivo and in a vertebrate. Utilization of these tools should facilitate ongoing insights into the nuances of Hippo-Yap/Taz-Tead signaling as well as potential cross-regulation between multiple other signaling pathways.
4. Experimental Procedures
4.1. Generation of Plasmids and Transgenic lines
We used the pGL3-4xGTIIC-49 plasmid (Mahoney et al., 2005), kindly provided by Dr. Iain Farrance (University of Maryland, Baltimore, MD), to generate the 4xGTIIC based transgenic zebrafish lines. Specifically, PCR was used to construct a Gateway® (Invitrogen™) entry clone containing the 4xGTIIC multimerized TEAD-binding sequence followed by the chicken troponin T (cTNT) minimal promoter or the minimal Adeno-associated virus major late promoter (AAVmlp) (Fig. 1A). Final plasmids were constructed using Gateway® recombination and the Tol2 Kit (Kwan et al., 2007) to place the 4xGTIIC promoter upstream of eGFP, d2GFP (destabilized GFP; Clonetech), or mCherry. In addition, RT-PCR was performed to generate Gateway® entry plasmids of full length cDNA sequences for zebrafish yap, taz, and vgll4b, as well as truncated forms of yap and taz. A constitutively active form of Yap (YapS87A or YapCA) was made using QuickChange® (Stratagene™) site directed mutagenesis to alter the serine 87 codon to an alanine. Yap and Taz dominant negative (YapDN, TazDN) entry clones, based on a previously described Yap dominant negative protein (Cao et al., 2008), were generated to contain an ectopic nuclear localization sequence (NLS), but lack the transactivation domain. Transposase mRNA was injected with the fully assembled Tol2 constructs to generate each transgenic line (Kawakami, 2005). All final plasmids are listed in Supplemental Table 1. Protein overexpression using each plasmid was accomplished by using the Gal4/upstream–activator sequence (UAS) system (Scheer and Campos-Ortega, 1999). The Tg(vsx2:Gal4-VP16) transgenic line expresses at the optic cup stage in presumptive neural retina and RPE cells and is maintained in the neuroepithelia, but down-regulated in the RPE as these layers become distinct (Clark et al., 2011). The vsx2:gal4 expression mimics the expression pattern of similar vsx2/chx10 promoter transgenes in other vertebrates and was used to drive expression in progenitors for both the neural retina and retinal pigmented epithelium (RPE) (Rowan and Cepko, 2005; Rowan et al., 2004). A bi-directional UAS promoter that simultaneously drives the expression of dsRed and proteins of interest was used for overexpression experiments (Paquet et al., 2009).
4.2. Transgenic and Mutant Lines
Tg(4xGTIIC:d2GFP) (This Study)
Tg(4xGTIIC:eGFP) (This Study)
Tg(4xGTIIC:mCherry) (This Study)
Tg(dsRed.T4:14xUAS:Yap) (This Study)
Tg(dsRed.T4:14xUAS:Taz) (This Study)
Tg(vsx2:Gal4vp16)mw39 (Clark et al., 2011)
Tg(-5.0 foxc1b:eGFP)mw44 (This Study)
Tg(-7.2 sox10:eGFP) zf77 (Hoffman et al., 2007)
Tg(UAS:mKO2;cmlc2:eGFP)mw45 (This Study)
Tg(UAS:mCherry)mw60 (This Study)
crb2am289 (Omori and Malicki, 2006)
4.3. Immunofluorescence
Standard methodology was used for whole mount immunofluorescence (Clark et al., 2011). The antibodies used included a 1:200 dilution of Yap rabbit polyclonal (Cat#4912; Cell signaling) and a 1:20 dilution of Crb2a/Zs4 antigen mouse monoclonal supernatant from the University of Oregon Monoclonal Antibody Facility (Hsu and Jensen, 2010). Deletion of Yap or Crb2a resulted in complete loss of immunoreactivity, demonstrating the specificity of these antibodies (data not shown) (Hsu and Jensen, 2010). Secondary antibodies included Alexa 488 anti-rabbit (Invitrogen), Alexa-547 anti-mouse (Invitrogen), and Alexa 547 anti-rabbit (Invitrogen). Nuclei were labeled with TO-PRO®-3 (Molecular Probes). Fluorescent images were collected as previously described (Clark et al., 2011).
4.4. Morpholinos
The following Morpholinos were synthesized by GeneTools, LLC: yap1 SP MO1, 5′-AGCAACATTTAACAACTCACTTTAGG-3′ (Skouloudaki et al., 2009). tp53 MO, 5′-GCGCCATTGCTTTGCAAGAATTG-3′ (Robu et al., 2007). Splice disruption was characterized by RT-PCR on whole embryos at 23 hpf.
4.5. Pixel Intensity Analysis
All pixel intensity analyses were performed using Metamorph (Molecular Devices, Inc.) software. We assessed neural retina d2GFP pixel intensity by defining the entire retina and excluding the lens and RPE, for Region of Interest analyses. The ratios of total pixel count to total neural retina area were used to compare crb2a +/? and crb2a −/− 4xGTIIC:d2GFP expression. RPE analysis was performed in a similar manner. Pixel intensity analysis for the Vgll4b overexpression experiment was done by injecting either UAS:mCherry or dsRed.T4:14xUAS:Vgll4b plasmid into vsx2:gal4/4xGTIIC:d2GFP transgene-positive, 1–4 cell stage embryos. d2GFP pixel intensity was determined for Vgll4b positive cells by outlining dsRed positive RPE cell clusters. dsRed negative cells proximal to Vgll4b (dsRed)-positive clusters were analyzed as controls. These control cells were required to be immediately adjacent to the dsRed-positive clusters and of equivalent size. Similar analyses were done for UAS:mCherry positive and negative cells.
4.6. RPE Cell Size Analysis
RPE cell borders were distinguished by using phalloidin to stain for actin. Cell borders were traced and areas determined using Metamorph (Molecular Devices, Inc.) software.
Supplementary Material
The arrow shown in the movie signifies an area where a mesenchymal cell will start migrating away from the lateral mesoderm and subsequently up-regulate 4xGTIIC:d2GFP expression. Acquisition interval equals 20 minutes/frame.
The arrows shown in the movie indicate the coalescing of 4xGTIIC:d2GFP positive cardiac progenitor cells to form the tubular heart. Acquisition interval equals 20 minutes/frame.
4xGTIIC:d2GFP and cmlc2:eGFP heart expression in wild-type and nls-yapDN injected embryos. Acquisition interval equals 0.7 second/frame.
(A–C) The expression pattern of each transgene is similar at 2 dpf. Each transgene image was captured using the same optical settings for comparison of the relative levels of expression between each transgenic line. The d2GFP transgenic line expresses in the same tissues as both the eGFP and mCherry embryos but the levels of expression are lower due to the more rapid turnover.
(A′A‴) crb2a +/? embryos maintain polarity at 48 hpf. Arrows represent intact junctions in the neural retina marked by phalloidin staining. (B–B‴) crb2a −/− embryos lose polarity at 48 hpf. Arrows represent ectopic junctions marked by phalloidin staining. (C–D) crb2a −/− embryos have RPE cells that appear abnormal in shape and pigment distribution compared to crb2a +/? Asterisk marks an area in the RPE layer that lacks pigmentation. (E–F) En face phalloidin staining at the level of the RPE. Note the disorganization in crb2a−/− mutants. Asterisks mark examples of apical neuroepithelia endfeet visible underlying some RPE cells (G) Quantification of RPE cell size in eyes of crb2a +/? (n=38 cells, <10 embryos) and crb2a −/− (n=42 cells, <10 embryos). Error bars represent S.E.M. p=unpaired t-test with equal S.D.
Supplemental Table 1. List of transgenic lines/plasmids generated and used for this study.
Highlights.
Transgenic fish were generated to report and manipulate Hippo signaling.
Loss of Yap/Taz activity resulted in cardiac progenitor migration defects.
Loss of Crb2a in retinal neuroepithelia did not up-regulate Yap/Taz-Tead activity.
Overexpression of Vgll4b down-regulated endogenous Yap/Taz-Tead activity.
Acknowledgments
We thank Anitha Ponnuswami and Jon Bostrom for assistance with molecular biology; Pat Cliff, Bill Hudzinski, and Brandon Milkulski for zebrafish husbandy; Jon Skarie for generating the -5.0foxC1b:EGFP plasmid; Iain Farrance for the pGL3-4xGTIIC-49 plasmid; Dominic Paquet for the bi-directional UAS plasmid; and all members of the Link Lab for helpful discussions. This project was supported by NIH grants T32EY014536 (JBM), R01EY014167 (BAL), a pilot project grant from the Medical College of Wisconsin Cancer Center - Advancing a Healthier Wisconsin (BAL), as well as an NEI Core Facilities Grant P30EY001931 to the vision research community of the Medical College of Wisconsin.
Footnotes
Author Contributions
JM and BL designed, carried out experiments, and co-wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alves CH, Bossers K, Vos RM, Essing AH, Swagemakers S, van der Spek PJ, Verhaagen J, Wijnholds J. Microarray and morphological analysis of early postnatal CRB2 mutant retinas on a pure C57BL/6J genetic background. PLoS One. 2013;8:e82532. doi: 10.1371/journal.pone.0082532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–34. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–5. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Winter M, Cohen AR, Link BA. Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev Dyn. 2011;240:2452–65. doi: 10.1002/dvdy.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I, Xiao JH, Rosales R, Staub A, Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988;54:931–42. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Fernando CA, Conrad PA, Bartels CF, Marques T, To M, Balow SA, Nakamura Y, Warman ML. Temporal and spatial expression of CCN genes in zebrafish. Dev Dyn. 2010;239:1755–67. doi: 10.1002/dvdy.22279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–24. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zoolog B Mol Dev Evol. 2007;308:679–91. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Jensen AM. Multiple domains in the Crumbs Homolog 2a (Crb2a) protein are required for regulating rod photoreceptor size. BMC Cell Biol. 2010;11:60. doi: 10.1186/1471-2121-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–54. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- Koontz LM, Liu-Chittenden Y, Yin F, Zheng Y, Yu J, Huang B, Chen Q, Wu S, Pan D. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupperman E, An S, Osborne N, Waldron S, Stainier DY. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 2000;406:192–5. doi: 10.1038/35018092. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–99. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–7. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney WM, Jr, Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388:217–25. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Raya A, Callol-Massot C, Kawakami Y, Oishi I, Rodriguez-Esteban C, Izpisua Belmonte JC. miles-apart-Mediated regulation of cell-fibronectin interaction and myocardial migration in zebrafish. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl 1):S77–82. doi: 10.1038/ncpcardio0764. [DOI] [PubMed] [Google Scholar]
- Miller E, Yang J, DeRan M, Wu C, Su AI, Bonamy GM, Liu J, Peters EC, Wu X. Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem Biol. 2012;19:955–62. doi: 10.1016/j.chembiol.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Omori Y, Malicki J. oko meduzy and Related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr Biol. 2006;16:945–57. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- Osborne N, Brand-Arzamendi K, Ober EA, Jin SW, Verkade H, Holtzman NG, Yelon D, Stainier DY. The spinster homolog, two of hearts, is required for sphingosine 1-phosphate signaling in zebrafish. Curr Biol. 2008;18:1882–8. doi: 10.1016/j.cub.2008.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–69. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Paquet D, Bhat R, Sydow A, Mandelkow EM, Berg S, Hellberg S, Falting J, Distel M, Koster RW, Schmid B, Haass C. A zebrafish model of tauopathy allows in vivo imaging of neuronal cell death and drug evaluation. J Clin Invest. 2009;119:1382–95. doi: 10.1172/JCI37537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellissier LP, Alves CH, Quinn PM, Vos RM, Tanimoto N, Lundvig DM, Dudok JJ, Hooibrink B, Richard F, Beck SC, Huber G, Sothilingam V, Garcia Garrido M, Le Bivic A, Seeliger MW, Wijnholds J. Targeted ablation of CRB1 and CRB2 in retinal progenitor cells mimics Leber congenital amaurosis. PLoS Genet. 2013;9:e1003976. doi: 10.1371/journal.pgen.1003976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati AV, Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14:390–8. doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. A POU factor binding site upstream of the Chx10 homeobox gene is required for Chx10 expression in subsets of retinal progenitor cells and bipolar cells. Dev Biol. 2005;281:240–55. doi: 10.1016/j.ydbio.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Rowan S, Chen CM, Young TL, Fisher DE, Cepko CL. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–52. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Kikuchi Y, Kuroiwa A, Takeda H, Stainier DY. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development. 2006;133:4063–72. doi: 10.1242/dev.02581. [DOI] [PubMed] [Google Scholar]
- Sawada A, Nishizaki Y, Sato H, Yada Y, Nakayama R, Yamamoto S, Nishioka N, Kondoh H, Sasaki H. Tead proteins activate the Foxa2 enhancer in the node in cooperation with a second factor. Development. 2005;132:4719–29. doi: 10.1242/dev.02059. [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–8. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Skouloudaki K, Puetz M, Simons M, Courbard JR, Boehlke C, Hartleben B, Engel C, Moeller MJ, Englert C, Bollig F, Schafer T, Ramachandran H, Mlodzik M, Huber TB, Kuehn EW, Kim E, Kramer-Zucker A, Walz G. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci U S A. 2009;106:8579–84. doi: 10.1073/pnas.0811691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6:371–82. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell. 2010;19:831–44. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–71. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, Mills GB, Guan KL. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–91. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–87. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The arrow shown in the movie signifies an area where a mesenchymal cell will start migrating away from the lateral mesoderm and subsequently up-regulate 4xGTIIC:d2GFP expression. Acquisition interval equals 20 minutes/frame.
The arrows shown in the movie indicate the coalescing of 4xGTIIC:d2GFP positive cardiac progenitor cells to form the tubular heart. Acquisition interval equals 20 minutes/frame.
4xGTIIC:d2GFP and cmlc2:eGFP heart expression in wild-type and nls-yapDN injected embryos. Acquisition interval equals 0.7 second/frame.
(A–C) The expression pattern of each transgene is similar at 2 dpf. Each transgene image was captured using the same optical settings for comparison of the relative levels of expression between each transgenic line. The d2GFP transgenic line expresses in the same tissues as both the eGFP and mCherry embryos but the levels of expression are lower due to the more rapid turnover.
(A′A‴) crb2a +/? embryos maintain polarity at 48 hpf. Arrows represent intact junctions in the neural retina marked by phalloidin staining. (B–B‴) crb2a −/− embryos lose polarity at 48 hpf. Arrows represent ectopic junctions marked by phalloidin staining. (C–D) crb2a −/− embryos have RPE cells that appear abnormal in shape and pigment distribution compared to crb2a +/? Asterisk marks an area in the RPE layer that lacks pigmentation. (E–F) En face phalloidin staining at the level of the RPE. Note the disorganization in crb2a−/− mutants. Asterisks mark examples of apical neuroepithelia endfeet visible underlying some RPE cells (G) Quantification of RPE cell size in eyes of crb2a +/? (n=38 cells, <10 embryos) and crb2a −/− (n=42 cells, <10 embryos). Error bars represent S.E.M. p=unpaired t-test with equal S.D.
Supplemental Table 1. List of transgenic lines/plasmids generated and used for this study.