Abstract
Deregulation of cell polarity proteins has been linked to the processes of invasion and metastasis. TRIM62, is a regulator of cell polarity and a tumor suppressor in breast cancer. Here, we demonstrate that human non-small cell lung cancer lesions show a step-wise loss of TRIM62 levels during disease progression, which was associated with poor clinical outcomes. To directly examine the role of Trim62 in development of lung cancer, we deleted Trim62 in a mutant K-Ras mouse model of lung cancer. In this context, haploinsufficiency of TRIM62 synergized with a K-RasG12D mutation to promote invasiveness and disrupt three-dimensional morphogenesis, both of which are associated with epithelial-mesenchymal transitions. Re-expression of TRIM62 reverted these phenotypes in tumor cell lines. Thus, TRIM62 loss cooperates with K-Ras mutation in tumorigenesis and metastasis in vivo indicating that decreased levels of TRIM62 may play an important role in the evolution of lung cancer.
Keywords: EMT, metastasis, anoikis, mouse models
INTRODUCTION
Carcinomas are epithelial cancers that constitute 90% of human malignancies and are fatal due to their metastases.1 Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related deaths in Western countries.2 No curative therapies are available for patients with metastatic NSCLC, whose median overall survival is approximately 12 months.3 Metastasis is a complex process whereby neoplastic epithelial cells lose their ability to adhere to each other and to the basal membrane, become motile, penetrate the vascular endothelium, survive passage through the circulatory system, extravasate, colonize sites distant, and generate secondary macroscopic outgrowths.1, 4 Genes that disrupt cell polarity and epithelial architecture contribute to invasive and metastatic phenotypes.5
The epithelial-mesenchymal transition (EMT) is a process whereby epithelial cells lose polarity and cell-cell contacts.5 Central to this process is down regulation of E-cadherin.1, 4 In fact, several EMT inducers are transcriptional repressors of E-cadherin expression, including Snail, Slug, Twist, and Zeb2,5, 6 which are highly expressed by metastatic cells.1 By contrast, the mechanisms whereby cells are locked in an epithelia state are largely unknown.
The tripartite motif-containing (TRIM) proteins harbor RING, B-box, and coiled-coil domains, and are highly conserved throughout the metazoan kingdom.7, 8 The RING domain of several TRIM proteins exhibits ubiquitin E3 ligase-mediated protein degradation.9–13 The physiological role of most TRIM family proteins remains unknown. Thus far, only TRIM19 (PML), involved in the fusion protein PML-RARα that causes acute promyelocytic leukemia,14–16 and TRIM24, implicated in liver and breast cancer,17, 18 have been linked to human cancer.
TRIM62, also known as Dear1, displays loss of heterozygosity at high frequency in human carcinomas.19–23 TRIM62 regulates apical-basal epithelial polarity in vitro23 and undergoes copy number losses in breast, lung, pancreas, and colon tumors.24 TRIM62 is highly expressed in normal epithelia and mutations were identified in breast carcinomas and other epithelial cancers.23, 24
In this study, we found that TRIM62 loss is an early and frequent event in NSCLC that is coupled with induction of EMT. To directly investigate the role of TRIM62 in lung cancer, we crossed mice lacking TRIM62 with a mutant K-Ras driven lung cancer mouse model that recapitulates cardinal features of human lung cancer.25 TRIM62-deficient K-RasLA1 mice developed lung adenocarcinomas with invasive and metastatic phenotypes.
MATERIALS AND METHODS
Mice
TRIM62+/− (C57BL/6J and Balb/c background) mice were crossed with K-RasLA1 (129/Sv) or p53+/− (C57BL/6J) mice.24, 25 All mice used were littermates and were cared for according to IACUC guidelines.
Immunohistochemistry
Immunohistochemistry was performed using TRIM62 23 (Novus Biol, Inc. Littleton.CO), Vimentin (Thermo Scientific, Fremont, CA), E-cadherin (Santa Cruz Biotechnology; Santa Cruz, CA), CD44 (BD Pharmingen, San Jose, CA), and Twist (Santa Cruz Biotechnology; Santa Cruz, CA) antibodies and visualized with Vectastain ABC and DAB kits from (Vector, Burlingame, CA). Tissue microarrays were previously described.26 NSCLC specimens were examined using E-cadherin (1:100, Santa Cruz Biotechnology; G-10: SC-8426, Santa Cruz, CA) and TRIM62 antibodies23 (1:100, Bethyl Laboratories, Montgomery, Texas).
Western blots
TRIM62 (Bethyl Laboratories, Montgomery, Texas), β-actin (Sigma), or tubulin (Sigma, St Louis, MO) antibodies were used.
Real-time RT-PCR
cDNA was synthesized from RNA (2 µg) with First-Strand cDNA Synthesis Kit (GE Bioscience, Piscataway, NJ). Resulting cDNA was subjected to real-time PCR with the SYBR green fast kit (Applied Biosystems). Relative gene expression was compared to RPLP0 as control. Three experiments for each condition were averaged.
Immunofluorescence
Immunofluorescence was performed using SPC (sc-7705, Santa Cruz Biotechnology; WRAB-SPC, Seven Hills Bioreagents) and CCSP (07-623, Upstate Biochemicals) antibodies.
Anchorage-independent cell viability assays
Cell lines were plated in triplicate in ultra-low attachment 60 mm flat-bottom plates (Corning, NY, USA) in DMEM (Sigma, St. Louis, MO) supplemented with 10% (vol/vol) FBS (Gibco, Lifetechnologies), penicillin (200 µg/mL, Mediatech, Manassas, VA), and streptomycin (200 µg/mL, Mediatech, Manassas, VA), at 5% CO2 and 37°C for 7 days. Culture media were renewed every two days. Suspended cells were collected daily and the percentage of dead cells was determined by Trypan blue exclusion assays (Gibco, Lifetechnologies).
Culture assays
Migration assays were performed as published.27 For invasion assays, 5x105 cells were plated in the top chamber of Matrigel-coated membranes (8-µm pore size, BD Biosciences, San Jose, CA). Five high-power fields/filter and triplicate filters/sample were counted. Assays were repeated at least twice. Three-dimensional cultures were performed according to Debnath et al.28 MISSION shRNA lentiviral vectors expressing GFP or TRIM62 shRNAs and packaging vectors were from Sigma-Aldrich (NM_018207). Stable clones were selected by adding puromycin (2 µg/ml) to medium. Mouse TRIM62 cDNA (Origene, Rockville, MD) was cloned into pBabe-puro and transfected into Phoenix cells with lipofectAMINE (Life Technologies, Inc., Rockville, MD).
Statistical analysis
Data are presented as mean±s.e.m. Student’s t test (two-tailed) was used to compare two groups unless otherwise indicated (χ2 test). Survival curves were estimated by Kaplan–Meier and compared by means of the log-rank (Mantel-Cox) test. Correlations between TRIM62 and E-cadherin expression were made using the Pearson correlation test. All p values were two-sided and statistical significance was set at <0.05.
RESULTS
Loss of TRIM62 expression in NSCLC
NSCLC is characterized by aberrant drivers of proliferation such as mutant oncogenes (e.g. K-Ras), and by mutations in tumor suppressors (e.g. p53). Since TRIM62 regulates epithelial morphogenesis and is an independent predictor of survival in breast cancer,23 we examined TRIM62 levels in NSCLC cells and lung tumors. TRIM62 was present in the immortalized normal bronchial epithelial cell line BEAS-2B (Figure 1a). However, TRIM62 was absent or markedly decreased in 66% (6/9) of NSCLC cell lines assessed, including several derived from highly invasive and/or metastatic NSCLC (i.e. Calu-3, NCI-H661, NCI-H1819, HCC-4006). We next examined tissue microarrays for TRIM62 levels. The microarray included 214 cases of NSCLC, of which 129 were stage I, 40 stage II, 39 stage III, and 6 stage IV (Table S1). Staining revealed that 86% of samples exhibited complete loss of TRIM62 (Figure 1b). Given that TRIM62 expression is frequently focal,23 it is possible that tissue microarray analysis, where only a small tumor area is examined, may overestimate the number of samples with loss of TRIM62. To examine this possibility, we analyzed TRIM62 protein levels in full tumor sections obtained from a separate cohort of 72 patients with NSCLC (stage I, n=62; stage II, n=10) (Table S2). TRIM62 was completely gone in 46 (64%) cases, suggesting that TRIM62 loss is an early event in the pathogenesis of NSCLC. Of those 72 samples, we stained 22 paired consecutive full tumor sections for both TRIM62 and E-cadherin. For each, three areas of normal bronchial epithelium, atypical adenomatous hyperplasia, and non-small cell lung cancer were scored for expression of TRIM62 and E-cadherin (Figure S1). TRIM62 levels decreased stepwise during progression, with atypical adenomatous hyperplasia exhibiting TRIM62 levels intermediate between normal bronchial epithelium and NSCLC (Figure 1c-d). We detected a clear correlation between decreased TRIM62 levels and markedly decreased or absent E-cadherin expression in areas of invasive NSCLC, suggesting an association between TRIM62 loss and advanced histological stages of human NSCLC (Figure 1e). A similar correlation between loss of TRIM62 and E-cadherin was observed when adenocarcinomas were compared to squamous cell carcinomas and when well-differentiated NSCLC were compared to moderately- or poorly-differentiated NSCLC (Figure S1). To determine the impact of TRIM62 loss on long-term outcomes of patients with early stage NSCLC, we examined the cohort of 72 patients. Patients with early stage NSCLC and loss of TRIM62 had a significantly shorter time to relapse compared to those with TRIM62 expression (5.1 vs 2.87 years, HR 0.60; 95%CI 0.25-1.41, p=0.049)(Figure 1f). A trend towards worse 5-year relapse-free survival (Figure 1g) and overall survival (Figure 1h) was also observed for patients lacking TRIM62 levels as compared to patients expressing TRIM62. Thus, TRIM62 loss is an early event in the pathogenesis of invasive NSCLC in humans and a clinical predictor of relapse.
Figure 1. Loss of TRIM62 is a frequent event in human non-small cell lung cancer.
a, Immunoblot analysis of TRIM62 levels in immortalized bronchial epithelial cells (BEAS-2B) and in non-small cell lung cancer (NSCLC) cell lines; α-Tubulin levels were used as loading control. b, Immunostaining of 214 primary NSCLC sections with an antibody against TRIM62. Examples of present (top panel) and absent (bottom panel) immunohistochemical cytoplasmic expression of TRIM62 in cells from lung adenocarcinomas are shown. c, Immunostaining of lesions present in full lung sections of 22 human NSCLC specimens with antibodies against TRIM62 and E-cadherin. Staining was scored in normal bronchial epithelium (NBE), atypical adenomatous hyperplasia (AAH), and adenocarcinoma lesions. d, Full sections of NSCLC specimens were stained with anti- TRIM62 (left panel) or anti-E-cadherin antibodies (right panel). Black arrows denote preserved staining in the normal lung epithelium (internal control). Red arrows denote loss of TRIM62 or E-cadherin immunostaining in areas involved by adenocarcinoma. e, The weighted histoscore was used to calculate the Pearson correlation between levels of TRIM62 and E-cadherin staining. f-h, Kaplan-Meier plots of time to relapse (f), five-year relapse-free survival (g), and overall survival (h) of 72 patients with non-small cell lung cancer (NSCLC) stratified by TRIM62 protein expression. NSCLC tissues were scored as TRIM62-positive (red curves) if they retained TRIM62 expression or TRIM62-negative (blue curves) if the expression of TRIM62 was absent. The expression of TRIM62 was assessed by immunohistochemistry using an anti- TRIM62 antibody. Differences in survival were calculated according to the log-rank test.
Trim62 deficiency promotes epithelial malignancies
Trim62-deficient (lacking one or both Trim62 alleles) mice are susceptible to tumor development at a low frequency and at long latency in a C57BL6/129Sv background that is generally resistant to epithelial cancers such as breast cancer.24 We therefore intercrossed these mice with Balb/c mice to generate mice in a mixed genetic background more likely to be susceptible to epithelial cancers. We confirmed absence of Trim62 protein in several tissues by immunohistochemistry (Figure 2a-b). In this mixed background, Trim62+/− and Trim62−/− littermates were viable, born at the expected Mendelian ratio, fertile, and did not exhibit any significant morphologic, behavioral, or hematological phenotypes. No significant differences were observed in survival between Trim62+/− and Trim62−/− mice (740 vs 676 days, p=0.29; the hazard ratio [HR] for death in the Trim62−/− group was 0.75, 95% confidence interval [CI] 0.45-1.26) (Figure 2c). However, survival of both cohorts was significantly shorter than wild-type littermates (814 days, p=0.01; HR=1.97, 95% CI 1.13-3.44) (Figure 2c). Examination of 43 moribund Trim62+/− or Trim62−/− mice (20-24 months old) revealed tumors in 91% of animals (Table 1), compared to none in a cohort of 11 age-matched controls (p=0.0001, Fisher’s exact test) and compared to 16% in Trim62-deficient mice in a C57BL/6J/129Sv background.24 Epithelial tumors predominated in Trim62 deficient mice (Table 1). Invasive adenocarcinomas of the lung, liver, and breast were present in 23%, 21%, and 14% of mice, respectively. Furthermore, 7% of mice had metastasis, and 44% harbored more than one tumor. Therefore, Trim62 insufficiency in this genetic background was most frequently associated with adenocarcinomas, which suggests an important role for Trim62 in epithelial homeostasis.
Figure 2. Trim62 loss in mice on a mixed genetic background impacts survival.
a-b, Lack of expression of the Trim62 protein assessed by immunohistochemistry in pulmonary (a) and cardiac (b) tissues obtained from Trim62−/− mice. c, Kaplan-Meier plots depicting the survival of Trim62 mutant mice.
Table 1.
Tumor spectrum of Trim62 mutant mice
| Whole cohort (n=43) |
Trim62+/− (n=29) |
Trim62−/− (n=14) |
|
|---|---|---|---|
| No. mice with tumors (%) | 39 (91) | 27 (93) | 12 (86) |
| Organ/Tumor type | |||
| Liver | |||
| Hepatocellular carcinoma (%) | 10 (23) | 8 (28) | 2 (14) |
| Hepatic adenoma (%) | 10 (23) | 8 (28) | 2 (14) |
| Lung | |||
| Bronchioalveolar carcinoma (%) | 9 (21) | 5 (17) | 4 (29) |
| Bronchioalveolar adenoma (%) | 12 (28) | 5 (17) | 7 (50) |
| Breast | |||
| Breast adenocarcinoma (%) | 6 (14) | 5 (17) | 1 (7) |
| Hematopoietic | |||
| Lymphoma (%) | 6 (14) | 4 (14) | 2 (14) |
| Leukemia (%) | 1 (2) | 0 | 1 (7) |
| Soft tissue | |||
| Spindle cell sarcoma (%) | 2 (5) | 2 (7) | 0 |
| Miscellanea | |||
| Pituitary gland carcinoma (%) | 1 (2) | 1 (3) | 0 |
| Harderian gland adenoma (%) | 1 (2) | 1 (3) | 0 |
| Squamous cell papilloma (%) | 2 (5) | 2 (7) | 0 |
| No. mice with no tumors (%) | 4 (9) | 2 (7) | 2 (14) |
| No. mice with metastasis (%) | 3 (7) | 3 (10) | 0 |
| No. mice with ≥2 tumors (%) | 19 (44) | 13 (45) | 6 (43) |
Trim62 deficiency cooperates with K-RasG12D
K-Ras activating mutations are found in 25-50% of human lung adenocarcinomas29 and >90% of spontaneous and chemically induced lung tumors in mice.30 K-RasLA1 mice inherit a latent K-Ras allele with an activating glycine to aspartic acid mutation at codon 12 (G12D), which undergoes spontaneous recombination in vivo.25 K-RasLA1 mice die at a median of 300 days due to excessive lung tumor burden consisting of non-invasive hyperplastic adenomas arising from alveolar type II cells.25 K-RasLA1 lung tumors are early-stage lesions devoid of invasive features that rarely, if ever, metastasize.
Given the proclivity of Trim62-deficient mice to develop adenocarcinomas, we intercrossed Trim62−/− mice with K-RasLA1 mice. Notably, Trim62-deficient K-RasLA1 mice had reduced life spans compared with those of K-RasLA1 single mutant littermates (median survival 186 days vs 291 days, p<0.0001; HR=0.28, 95% CI 0.17-0.46) (Figure 3a). The survival of Trim62+/−:K-RasLA1 mice was not significantly different compared to Trim62−/−:K-RasLA1 mice (median survival 192 days vs 181 days, p=0.58). We also crossed K-RasLA1 mice to mice with deletion of one p53 allele, a tumor suppressor frequently altered in human lung cancer. The survival of p53+/−:K-RasLA1 mice was similar to that previously reported25, 31 but notably, significantly longer than that of Trim62-deficient K-RasLA1 mice (240 days vs 186 days, p=0.01; HR=2.0, CI 1.1-3.7).
Figure 3. Cooperation of Trim62 deficiency and mutant K-Ras in tumorigenesis.
a, Kaplan-Meier plots depicting the survival of K-RasLA1, p53+/−:K-RasLA1, Trim62+/−:K-RasLA1, and Trim62−/−:K-RasLA1 mice. The survival of Trim62+/−:K-RasLA1 and Trim62−/−:K-RasLA1 mice was significantly reduced compared with that of K-RasLA1 and p53+/−:K-RasLA1 mice. b, Type and number of lung lesions found on necropsy of K-RasLA1, Trim62+/−:K-RasLA1, and Trim62−/−:K-RasLA1 mice. Adenocarcinomas and metastasis were only present in K-RasLA1 mice lacking one or both Trim62 alleles. AAH denotes adenomatous atypical hyperplasia. c, Disruption of the lung architecture by multiple bilateral advanced tumors in a 119-day-old Trim62-deficient:K-RasLA1 mouse. d, Hematoxilin and eosin staining of a coronal section of the lungs of a 147-day-old mouse. e, Parietal pleural metastasis. f, Renal metastasis. g-j, Clara cell-specific marker (CCA) and prosurfactant apoprotein-C (SP-C) immunofluorescent staining of p53 null mouse embryonic fibroblasts, cells not of lung origin used as a negative control staining (g), and Trim62+/−:K-RasLA1 cells derived from a renal metastasis (h, CCA; i, Pro-SP-C; j, CCA and Pro-SP-C overlay). k-p, Microphotographs showing the spectrum of lesions found in Trim62+/−:K-RasLA1 and Trim62−/−:K-RasLA1 mouse lungs: k, bronchial structures showing atypical papillary bronchial hyperplasia; l, solid adenoma; m, solid adenoma with atypical changes; n, adenocarcinoma with bronchioloalveolar features; o, invasive adenocarcinoma with solid and papillary features; p, metastatic adenocarcinoma in the kidney. (H&E, magnification 100x). q-r, Microphotographs showing loss of expression (head arrows) of Trim62 in tumor cells in (q) adenoma and (r) adenocarcinoma. Alveolar hyperplasia shows strong Trim62 immunostaining (black arrows) (magnification 200x).
Trim62-deficient K-RasLA1 mice featured a higher number of lung lesions compared to K-RasLA1 mice (Figure 3b-d). More importantly, lung tumors in Trim62:K-RasLA1 mice displayed a full malignant phenotype compared to K-RasLA1 tumors, including dysplastic and anaplastic changes, vascular invasion, inflammatory infiltrates, and marked cellular pleomorphism. In addition, 3 of 21 (15%) animals had multifocal lung adenocarcinoma renal metastases (Figure 3e-j). In contrast, lung lesions in K-RasLA1 (n=24) mice consisted exclusively of adenomas without metastases while compound mutant mice exhibited a wide spectrum of histological lung lesions including adenocarcinomas (Figure 3k-p). Additionally, tumors arising in Trim62+/−:K-RasLA1 mice exhibited minimal to no expression of TRIM62 protein (Figure 3q-r). These results establish Trim62 as a haploinsufficient tumor suppressor and indicate a cooperative effect between loss of Trim62 expression and mutant K-Ras in the development of invasive lung cancer.
Trim62 deficiency enhances invasion of K-RasLA1 cells
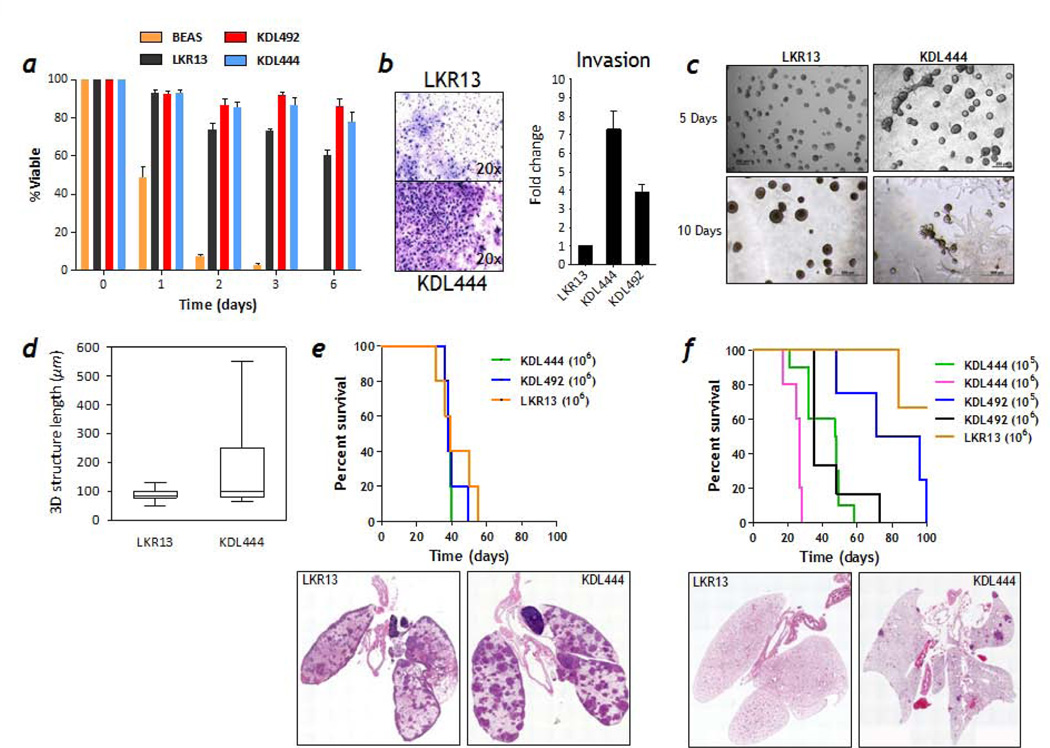
Trim62+/−:K-RasLA1 and Trim62−/−:K-RasLA1 mice had a significantly shorter life span than K-RasLA1 mice as a consequence of rapidly growing intrathoracic tumor burden resulting in respiratory failure. We hypothesized that the relatively low incidence of metastases (15%) observed in Trim62-deficient K-RasLA1 mice was due to the dramatic reduction in life span that hampered the ability of lung cancer cells to fully realize their metastatic potential. To test this hypothesis, we established 3 cell lines from different Trim62+/−:K-RasLA1 primary lung cancers: K-Ras-Trim62-Lung 444 (KDL444) was derived from a mouse that died with bilateral kidney metastases, and cell lines KDL492 and KDL436 were derived from primary lung tumors in mice that died with no macroscopic evidence of metastases. We next examined potential changes in cell behavior following Trim62 loss. Anoikis is a form of programmed cell death that befalls epithelial cells upon losing attachment to the extracellular matrix.32 We tested the anoikis sensitivity of Trim62+/−:K-RasLA1 lung cancer cell lines and compared to LKR13, a published cell line derived from a K-RasLA1 mouse lung tumor that lacks metastasis (Figure 4a).33, 34 Another control used was BEAS-2B cells, an immortalized human bronchial epithelial cell line, whose viability in suspension culture decreased dramatically over 48 hours, reflecting anchorage-independent cell death characteristic of non-transformed epithelial cells. The viability of LKR13 cells was 60% while the viability of KDL444 and KDL492 cells was >80% after 6 days in culture. BEAS-2B cells were not viable at 6 days. In Boyden chamber assays, KDL444, KDL492, and LKR13 cells were more motile than BEAS-2B cells (data not shown), but KDL444 and KDL492 cells exhibited a 7-fold and 4-fold increase in invasive properties through fibronectin-supplemented reconstituted basement membrane (Matrigel)-coated membranes compared to LKR13, respectively (Figure 4b).
Figure 4. Trim62 deficiency endows mutant K-Ras cells with invasive and metastatic potential.
a, K-RasLA1 (LKR13) and Trim62+/−:K-RasLA1 (KDL444 and KDL492) mutant cells, but not normal human bronchial epithelial cells transformed by SV40 T-antigen cells (BEAS-2B), exhibit resistance to anoikis. b, Trim62+/−:K-RasLA1 (KDL444) cells exhibited a higher invasion potential than K-RasLA1 (LKR13) in invasion assays done using transwell/Boyden chambers. c, Phase contrast micrographs demonstrate that KDL444 cells, but not LKR13 cells, exhibit an invasive phenotype in three-dimensional Matrigel/collagen-based cultures (magnification 200x, top panels; 500x, bottom panels). d, LKR13 and KDL444 cells were seeded in collagen 1-supplemented Matrigel and the size of at least 25 acini was quantified. e, KDL444, KDL492, and LKR13 cells produce similar number of metastatic foci in the lungs of syngeneic littermates when injected intravenously. Survival of mice injected with each cell type is shown. f, Trim62+/−:K-RasLA1 (KDL444 and KDL492) cells, but not K-RasLA1 (LKR13) cells, produce metastatic foci in the lungs of syngeneic wild-type mice when injected subcutaneously. Survival of mice injected with KDL444, KDL492, and LKR13 cells is shown.
Next, we examined the effects of Trim62 loss in 3D culture (Figure 4c-d). KDL444 and LKR13 cells were grown in collagen 1-supplemented Matrigel. Despite similar proliferation rates (data not shown), both cell lines exhibited marked differences in 3D culture. LKR13 cells formed spherical structures characterized by excess proliferation and absent lumens with a median size of 80 µm in diameter after 10 days in culture. However, KDL444 cells formed spherical structures that, after only 5 days in 3D culture, displayed invasive features and organized as large invasive structures (median size, 105 µm in diameter after 10 days in culture). In aggregate, these observations suggest that Trim62 loss synergizes with mutant K-Ras to promote resistance to anoikis and invasiveness.
Trim62-deficient K-RasLA1 cells are metastasis competent in vivo
We next tested whether the enhanced invasive phenotype observed in Trim62 deficient cells in vitro promotes a metastatic phenotype in vivo. We injected Trim62+/−:K-RasLA1 KDL444 and KDL492 cells, and K-RasLA1 LKR13 cells (1x106) into immunocompetent syngeneic wild-type littermates via tail vein (Figure 4e). The overall survival of the four groups was similar (~38 days, p=0.55). Analysis of mice sacrificed four weeks post-injection, revealed the presence of colonies in lungs of 11/11 mice injected with KDL444, 5/6 mice injected with KDL492, and 5/5 mice injected with LKR13 cells. Thus, Trim62 knockdown had little effect on the number and size of lung colonies after transplantation through the tail vein. These experiments however, obviate the critical early metastatic steps of tissue invasion and intravasation. We therefore injected 1x105 or 1x106 KDL444 or KDL492 cells subcutaneously in the flanks of wild-type, syngeneic, immunocompetent littermate mice (Figure 4f). Control mice injected with 1x106 LKR13 cells developed subcutaneous tumors with similar latencies and growth rates, and never became sick albeit some were sacrificed due to excessive tumor burden. On pathological examination, none of 6 LKR13-injected mice had metastases. This was confirmed in 24 other LKR13-injected animals used as controls in other experiments, confirming the inability of LKR13 cells to metastasize from subcutaneous tissue. On the other hand, all mice injected subcutaneously with either 1x105 or 1x106 KDL444 or KDL492 cells became moribund, albeit at different rates depending on the number of cancer cells injected (sooner in animals injected with 106 cells) and the cell type (sooner in animals injected with KDL444). These mice developed highly vascularized flank tumors and 8/10 KDL444-injected mice and 6/10 KDL492-injected mice developed lung metastases. These data suggest that Trim62 deficiency facilitates evasion from innate immune surveillance, entry and survival into the systemic circulation, and colonization at distant sites.
Trim62-deficiency imparts metastatic potential to KRASLA1 cells
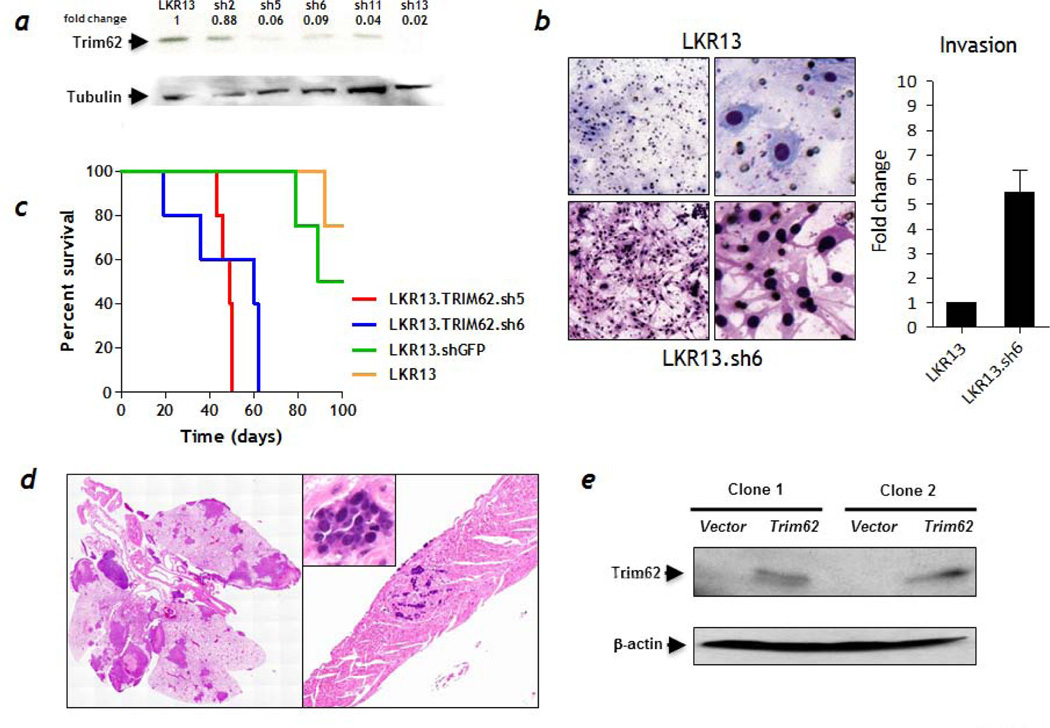
The experiments described above could not rule out the possibility that KDL444 and KDL492 cells might have acquired additional genetic abnormalities that contributed to metastasis. To examine this possibility, we generated several LKR13 cell lines stably expressing short hairpin RNAs (shRNA) from five different Trim62 sequences. The efficacy of Trim62 knockdown was confirmed by immunoblotting and RT-PCR (Figure 5a and S2). Trim62 knockdown did not affect proliferation rates of infected LKR13 cells compared with parental LKR13 cells (data not shown). In a transwell Matrigel-based invasion assay, LKR13 cells with knockdown of Trim62 exhibited 5-fold higher invasive capacity compared to LKR13 cells (Figure 5b) and these cells (LKR13sh5 and LKR13sh6) were also more resistant to anoikis than LKR13 cells (Figure S3). Subcutaneous injections in syngeneic immunocompetent mice showed the median survival of LKR13sh5- and LKR13sh6-injected mice (49 and 60 days) was markedly shortened compared to the survival of LKR13shGFP- or LKR13-injected mice (median survival not reached; p=0.001) (Figure 5c). While all animals developed flank tumors, 2/7 LKR13sh5 and 3/8 LKR13sh6 mice exhibited metastasis to different organs such as the heart, lungs, and lymph nodes (Figure 5d). By contrast, although all mice injected with LKR13shGFP (n=10) or with LKR13 (n=10) cells developed flank tumors, none harbored metastasis. To further examine the direct impact of Trim62 loss on metastasis, we stably transfected KDL444 cells with a Trim62 expression vector. Trim62 was detected by Western blotting in two independent clones (Figure 5e) which were injected subcutaneously (1x106 cells) into wild-type mice. While 9/10 animals developed subcutaneously tumors, none developed metastatic disease, suggesting that ectopic Trim62 expression suppressed the metastatic potential of KDL444 cells.
Figure 5. Stable depletion of Trim62 in mutant K-Ras cells induces a metastatic phenotype.
a, Western blot analysis of Trim62 using an specific antibody in Trim62 shRNA LKR13 cells compared with control cells (LKR13 parental cell line). α-Tubulin levels were used as loading control. b, Trim62 shRNA LKR13 cells (bottom panels) exhibited a mesenchymal phenotype and higher invasiveness compared with parental LKR13 cells (top panels) in invasion assays done using transwell/Boyden chambers. c, Kaplan-Meier survival plots of mice injected subcutaneously with Trim62 shRNA5, shRNA6, and GFP shRNA (control) LKR13 cells and parental LKR13 cells (control) in syngeneic wild-type mice. d, A fraction of mice injected with Trim62-depleted LKR13 cells developed metastasis involving, among other organs, the lungs and the heart. e, Trim62 protein expression in the KDL444 lung cancer cell line (western blot): Lanes 1 & 3, control (KDL444 infected with vector only); Lanes 2 & 4, (KDL444 infected with pBabe-puro/ Trim62 cDNA expression vector). β-actin levels were used as loading control.
Trim62 is a negative regulator of EMT in K-RASG12D driven lung cancers
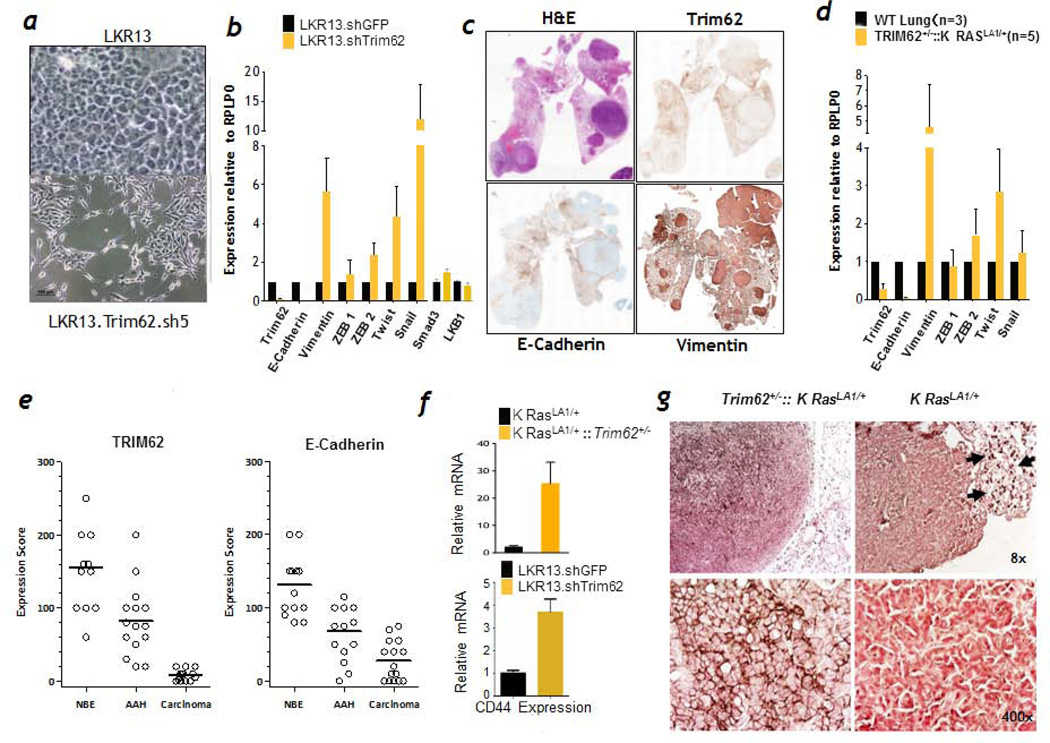
As detailed above, Trim62-deficient K-RasLA1 mice were prone to invasive and metastatic lung cancers. Trim62 also negatively regulates TGF-β mediated epithelial-mesenchymal transition (EMT) in breast cancer cell lines.24 We therefore tested whether Trim62 loss is associated with an EMT signature in K-RasLA1 mice. E-cadherin loss is a hallmark of EMT in invasive human carcinomas which correlates with clinical stage, aggressiveness, and prognosis.35, 36 After Trim62 knockdown, the morphology of LKR13sh5 and LKR13sh6 cells switched from a cobblestone pattern that characterizes parental LKR13 cells as well as LKR13 cells transfected with an empty vector, to a fibroblastic, spindle-like morphology, which characterizes cells undergoing EMT (Figure 6a and data not shown). LKR13sh5 and LKR13sh6 cells also exhibited loss of contact inhibition on plastic (data not shown). To confirm these findings, we measured expression of EMT markers (Figure 6b). Expression of E-cadherin was significantly reduced in Trim62-knockdown LKR13 cells compared with LKR13 cells whereas expression of the mesenchymal marker vimentin, Twist and Snail (master regulators of EMT37), and Smad3 which mediates TGFβ signaling were elevated upon Trim62 knockdown. Expression of LKB1 which encodes a kinase that regulates cell polarity was not altered. In order to confirm our in vitro findings, we analyzed 15 primary lung tumors from Trim62+/−:K-RasLA1 mice. These tumors consistently showed down regulation of Trim62 and E-cadherin and up regulation of vimentin by immunohistochemistry and RT-PCR analyses (Figure 6c and S4). RT-PCR analysis demonstrated a drastic reduction in Trim62 mRNA levels, mirrored by a similar decrease in E-cadherin mRNA (Figure 6d and S5). Conversely, expression of vimentin increased significantly in most lung lesions, confirming the presence of an EMT signature. Twist mRNA levels were elevated in numerous mice, although expression of Twist protein varied significantly in different lung lesions within the same mouse (Figure 6d and S6). Snail expression was even more variable, being markedly high in two tumors, slightly elevated in three, and unchanged compared with normal lung parenchyma in two tumors. In all cases, Trim62 staining was preserved in the surrounding non-tumoral lung parenchyma (Figure 6c, upper right panel), suggesting the presence of selective pressure to lose Trim62 expression in Trim62+/−:K-RasLA1 cells. We next assessed Trim62 levels across the histological spectrum of lung lesions developed by 24 Trim62+/−:K-RasLA1 mice (Figure 6e). High levels of Trim62 staining were observed in normal bronchial epithelium of Trim62+/−:K-RasLA1 mice, whereas down-regulation or, more frequently, loss of Trim62 was observed in the transition from normal epithelium to adenomatous atypical hyperplasia to invasive adenocarcinoma, suggesting a direct role of suppression of Trim62 during progression to malignant transformation.
Figure 6. Trim62 insufficiency induces epithelial mesenchymal transition and CD44 overexpression in K-RasLA1 cells.
a, Phase contrast micrographs of LKR13 (upper panel) and Trim62 shRNA5 LKR13 cells (bottom panel) grown under plastic culture conditions. b, Median mRNA transcript levels of Trim62 and EMT markers in cells stably expressing shRNA5, shRNA6, and shRNA13. Smad3 and LKB1 markers were examined in LKR13.sh5 cells. c, Full lung section stained with hematoxilin & eosin (top left), Trim62 (top right), E-cadherin (bottom left), and vimentin (bottom right). d, Expression levels of genes that regulate epithelial mesenchymal markers in Trim62+/−: K-RasLA1 mouse lung tumors. e, Immunostaining of lesions present in full lung sections of 24 Trim62+/−:K-RasLA1 mice with antibodies against Trim62 and E-cadherin. Staining was scored in normal bronchial epithelium (NBE), atypical adenomatous hyperplasia (AAH), and adenocarcinoma lesions. f, CD44 mRNA levels in tumors obtained from K-RasLA1 and Trim62+/−:K-RasLA1 mice (three tumors of each genotype were examined) and from LKR13.sh5 cells. g, Immunostaining of lung tumors arising in K-RasLA1 and Trim62+/−:K-RasLA1 mice using an anti-CD44 antibody. Arrows (right upper panel) denote CD44-positive mononuclear cells, which serve as positive internal controls.
Trim62 loss is associated with overexpression of CD44
We next determined the involvement of CD44, a type I transmembrane glycoprotein that serves as a hyaluronan receptor that mediates adhesion to the extracellular matrix.38–40 CD44 also fosters anchorage-independent growth in vitro as well as tumor growth and metastasis in murine tumor models.41–43 CD44 mRNA levels were remarkably higher in Trim62+/−:K-RasLA1 tumors (n=3) compared to K-RasLA1 tumors (n=3) by RT-PCR and in LKR13sh5 cells (Figure 6f). The levels of CD44 protein in tumors from K-RasLA1 (four lung adenocarcinomas), Trim62+/−:K-RasLA1 (four lung adenocarcinomas), and Trim62+/− (one lung and two breast adenocarcinomas) mice showed high levels of CD44 in Trim62-deficient mice, regardless of K-Ras mutational status and tumor type (Figure 6g and S7). These results implicate Trim62 in the regulation of CD44 expression and suggest that up regulation of CD44 may act in concert with EMT to promote a metastatic phenotype in tumors lacking Trim62.
DISCUSSION
Deregulation of polarity promotes dysplastic and neoplastic changes in epithelia by disrupting morphogenesis, inhibiting apoptosis, and synergizing with oncogenes.44 To test whether Trim62 loss would render a context of epithelial vulnerability for carcinoma development upon oncogenic stimulus, we crossed Trim62 deficient mice with K-RasLA1 mice.25 Survival of Trim62+/− or Trim62−/− mice with K-RasG12D expression was drastically reduced compared to that of K-RasLA1 indicating that Trim62 is a potent tumor suppressor. Unlike K-RasLA1 mice, Trim62 deficient:K-RasLA1 mice harbored carcinomas of the lung with invasive features and metastatic potential. Lung tumor cells from Trim62 deficient:K-RasLA1 mice that died without evidence of metastasis produced metastatic lesions when implanted subcutaneously into syngeneic littermates, indicating that Trim62 deficient:K-RasLA1 cells are innately metastasis competent. These results are highly significant as most metastasis models involve the introduction of tumor cells directly into the systemic circulation of immunocompromised mice, thus obviating the critical metastatic steps of tissue invasion, intravasation, and evasion of immune surveillance. The low frequency of metastasis in Trim62 deficient:K-RasLA1 mice likely relates the presence of excessive lung tumor burden and local invasion that kills the host before tumor cells can metastasize. Indeed, Trim62 knockdown in metastasis-incompetent K-RasLA1–positive LKR13 cells resulted in a metastatic phenotype following subcutaneous injection into syngeneic immunocompetent mice. Thus, Trim62 loss endows K-RasLA1 cells with the ability to overcome the initial steps of metastasis, such as migration and invasion in vivo as well as with the ability to evade immune surveillance. Together, Trim62 loss compromises cell polarity and K-Ras mutation provides abnormal growth signals, a lethal combination in lung tumorigenesis.
Metastasis of Trim62-deficient tumors is directly linked to induction of EMT, a process also associated with resistance to chemotherapy and the acquisition of stem cell properties by cancer cells.6 Transcription factors such as Twist, Snail, and Zeb-1/2 promote EMT.5, 6 Transcriptional activators of EMT appear to operate in a hierarchical manner, with Snail being activated at the onset of EMT while Slug, Twist, and Zeb proteins are induced subsequently to maintain a mesenchymal motile phenotype.6 Since Twist expression was elevated in our Trim62 deficient:K-RasLA1 lung tumors, Twist activation might represent a late event in tumors already undergoing EMT primarily elicited by Trim62 loss. Notably, this process appears to be abetted by overexpression of CD44, a glycoprotein reported to promote metastasis through different mechanisms,38–40 including binding to the TGFβ receptor, a canonical inducer of EMT.5 In fact, a recent report has shown that high levels of CD44 are associated with an EMT signature and stem-cell like traits in NSCLC cell lines.45 Thus, Trim62 loss cooperates with mutant K-Ras in the pathogenesis of lung cancer by promoting invasion and metastasis by deregulating the expression of several molecules that control EMT. Further insights into the regulation of proteins that maintain the apical–basal axis of polarity in epithelia and their interplay with oncogenes are warranted to improve the currently dismal outcomes of patients with carcinomas.
Supplementary Material
ACKNOWLEDGEMENTS
Animal studies were supported by the Cancer Center Support Grant CA016672 and a grant CA82577 to GL both from the National Institutes of Health.
Footnotes
The authors have no conflicts of interest.
AUTHOR CONTRIBUTIONS
All animal and tissue culture experiments were performed by AQC, SMP, SX and PY. LMS and IIW performed the analyses of human samples. NC and AMK provided reagents. GL contributed to discussions and revised manuscript.
REFERENCES
- 1.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Cataldo VD, Gibbons DL, Perez-Soler R, Quintas-Cardama A. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N Engl J Med. 2011;364:947–955. doi: 10.1056/NEJMct0807960. [DOI] [PubMed] [Google Scholar]
- 4.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 7.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 8.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Short KM, Hopwood B, Yi Z, Cox TC. MID1 and MID2 homo- and heterodimerise to tether the rapamycin-sensitive PP2A regulatory subunit, alpha 4, to microtubules: implications for the clinical variability of X-linked Opitz GBBB syndrome and other developmental disorders. BMC Cell Biol. 2002;3:1. doi: 10.1186/1471-2121-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niikura T, Hashimoto Y, Tajima H, et al. A tripartite motif protein TRIM11 binds and destabilizes Humanin, a neuroprotective peptide against Alzheimer's disease-relevant insults. Eur J Neurosci. 2003;17:1150–1158. doi: 10.1046/j.1460-9568.2003.02553.x. [DOI] [PubMed] [Google Scholar]
- 11.Takahata M, Bohgaki M, Tsukiyama T, Kondo T, Asaka M, Hatakeyama S. Ro52 functionally interacts with IgG1 and regulates its quality control via the ERAD system. Mol Immunol. 2008;45:2045–2054. doi: 10.1016/j.molimm.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci U S A. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 14.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 15.Kakizuka A, Miller WH, Jr, Umesono K, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 16.Goddard AD, Borrow J, Freemont PS, Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- 17.Khetchoumian K, Teletin M, Tisserand J, et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39:1500–1506. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- 18.Tsai WW, Wang Z, Yiu TT, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borg A, Zhang QX, Olsson H, Wenngren E. Chromosome 1 alterations in breast cancer: allelic loss on 1p and 1q is related to lymphogenic metastases and poor prognosis. Genes Chromosomes Cancer. 1992;5:311–320. doi: 10.1002/gcc.2870050406. [DOI] [PubMed] [Google Scholar]
- 20.Millikan RC, Ingles SA, Diep AT, et al. Linkage analysis and loss of heterozygosity for chromosome arm 1p in familial breast cancer. Genes Chromosomes Cancer. 1999;25:354–361. [PubMed] [Google Scholar]
- 21.Ragnarsson G, Eiriksdottir G, Johannsdottir JT, Jonasson JG, Egilsson V, Ingvarsson S. Loss of heterozygosity at chromosome 1p in different solid human tumours: association with survival. Br J Cancer. 1999;79:1468–1474. doi: 10.1038/sj.bjc.6690234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy BA, Etkin LD, Freemont PS. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17:344–345. doi: 10.1016/0968-0004(92)90308-v. [DOI] [PubMed] [Google Scholar]
- 23.Lott ST, Chen N, Chandler DS, et al. DEAR1 is a dominant regulator of acinar morphogenesis and an independent predictor of local recurrence-free survival in early-onset breast cancer. PLoS Med. 2009;6:e1000068. doi: 10.1371/journal.pmed.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen N, Balasenthil S, Reuther J, et al. DEAR1 Is a Chromosome 1p35 Tumor Suppressor and Master Regulator of TGF-beta-Driven Epithelial-Mesenchymal Transition. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-12-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 26.Solis LM, Behrens C, Dong W, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong S, Grijalva R, Zhang L, et al. Up-regulation of vascular endothelial growth factor in breast cancer cells by the heregulin-beta1-activated p38 signaling pathway enhances endothelial cell migration. Cancer Res. 2001;61:1727–1732. [PubMed] [Google Scholar]
- 28.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 29.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 30.Malkinson AM. Molecular comparison of human and mouse pulmonary adenocarcinomas. Exp Lung Res. 1998;24:541–555. doi: 10.3109/01902149809087385. [DOI] [PubMed] [Google Scholar]
- 31.Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26:6896–6904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- 32.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Wislez M, Spencer ML, Izzo JG, et al. Inhibition of mammalian target of rapamycin reverses alveolar epithelial neoplasia induced by oncogenic K-ras. Cancer Res. 2005;65:3226–3235. doi: 10.1158/0008-5472.CAN-04-4420. [DOI] [PubMed] [Google Scholar]
- 34.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimazui T, Schalken JA, Giroldi LA, et al. Prognostic value of cadherin-associated molecules (alpha-, beta-, and gamma-catenins and p120cas) in bladder tumors. Cancer Res. 1996;56:4154–4158. [PubMed] [Google Scholar]
- 36.Kase S, Sugio K, Yamazaki K, Okamoto T, Yano T, Sugimachi K. Expression of E-cadherin and beta-catenin in human non-small cell lung cancer and the clinical significance. Clin Cancer Res. 2000;6:4789–4796. [PubMed] [Google Scholar]
- 37.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 38.Gunthert U, Hofmann M, Rudy W, et al. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65:13–24. doi: 10.1016/0092-8674(91)90403-l. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann M, Rudy W, Zoller M, et al. CD44 splice variants confer metastatic behavior in rats: homologous sequences are expressed in human tumor cell lines. Cancer Res. 1991;51:5292–5297. [PubMed] [Google Scholar]
- 40.Yu Q, Stamenkovic I. Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes Dev. 1999;13:35–48. doi: 10.1101/gad.13.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbour AP, Reeder JA, Walsh MD, Fawcett J, Antalis TM, Gotley DC. Expression of the CD44v2-10 isoform confers a metastatic phenotype: importance of the heparan sulfate attachment site CD44v3. Cancer Res. 2003;63:887–892. [PubMed] [Google Scholar]
- 42.Weber GF, Bronson RT, Ilagan J, Cantor H, Schmits R, Mak TW. Absence of the CD44 gene prevents sarcoma metastasis. Cancer Res. 2002;62:2281–2286. [PubMed] [Google Scholar]
- 43.Yu Q, Toole BP, Stamenkovic I. Induction of apoptosis of metastatic mammary carcinoma cells in vivo by disruption of tumor cell surface CD44 function. J Exp Med. 1997;186:1985–1996. doi: 10.1084/jem.186.12.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan L, Rosenberg A, Bergami KC, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung EL, Fiscus RR, Tung JW, et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One. 5:e14062. doi: 10.1371/journal.pone.0014062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.