Abstract
Influenza A virus interacts with specific types of sialic acid during attachment and entry into susceptible cells. The precise amino acids in the hemagglutinin protein that control sialic acid binding specificity and affinity vary among antigenic subtypes. For H3 subtypes, amino acids 226 and 228 are critical for differentiating between α2,3- and α2,6-linked forms of sialic acid (SA). We demonstrate that position 190 of the HA from A/Udorn/307/72 (H3N2) plays an important role in the recognition of α2,3-SA, as changing the residue from a glutamic acid to an aspartic acid led to alteration of red blood cell hemagglutination and a complete loss of replication in differentiated, murine trachea epithelial cell cultures which express only α2,3-SA. This amino acid change had a minimal effect on virus replication in MDCK cells, suggesting subtle changes in receptor recognition by the H3 hemagglutinin can lead to significant alterations in cell and species tropism.
Keywords: influenza, sialic acid, receptors, hemagglutinin, tropism, respiratory epithelium
Introduction
Influenza A virus is a significant human pathogen that causes annual epidemics in the human population (Thompson et al., 2004; Thompson et al., 2003). While only two antigenic subtypes of influenza A virus circulate in humans (H3N2 and H1N1), a number of different antigenic subtypes exist and circulate widely in avian populations (Dugan et al., 2008). Through reassortment of virus gene segments, influenza A virus strains containing hemagglutinin (H or HA) and neuraminidase (N or NA) antigenic subtypes that are novel to humans can emerge, leading to influenza A virus pandemics that are associated with significantly higher morbidity and mortality (Rajagopal and Treanor, 2007).
In order for an avian influenza A virus strain to replicate and transmit efficiently in humans, a number of barriers must be overcome (Parrish and Kawaoka, 2005). One of the critical barriers appears to be the recognition of appropriate forms of sialic acid (SA), the influenza A virus receptor, by the HA protein. Human influenza A virus strains preferentially recognize sialic acid linked via an α-2,6 glycosidic linkage 2,6 SA) to the penultimate carbohydrate while avian influenza A viruses preferentially recognize α-2,3 SA (2,3 SA) (Rogers et al., 1983) (Nicholls et al., 2008). The recognition of SA by HA is in fact, much more complex than simply differentiating between a 2-3 and 2-6 glycosidic linkage. There are very important interactions of HA with saccharides outside the terminal SA and modifications of the SA and other saccharides can also affect the ability of HA to recognize SA containing carbohydrates (Russell et al., 2006; Stevens et al., 2006b).
While the receptor binding pocket of the influenza A virus HA protein is structurally conserved among a number of HA subtypes the precise amino acids which dictate receptor binding specificity and affinity vary to some degree (Russell et al., 2006; Stevens et al., 2006a). Amino acids 226 and 228 of H3 subtypes of influenza HA have been implicated as critical residues important for differentiating between 2,3 SA and 2,6 SA receptors (Vines et al., 1998). However, a number of other H3 amino acids can modulate receptor recognition or affinity (Martin et al., 1998; Matrosovich et al., 2000; Meisner et al., 2008; Nakajima et al., 2003; Suzuki et al., 2000) (Busch et al., 2008; Lu et al., 2006; Lu et al., 2005; Medeiros et al., 2001; Medeiros et al., 2004; Widjaja et al., 2006). In particular, amino acid 190 has been associated with changes in 2,3 versus 2,6 SA recognition in H3 subtype viruses (Martin et al., 1998; Nobusawa et al., 2000; Yassine et al., 2007). A glutamic acid (E) is present in many avian influenza virus strains while an aspartic acid (D) is often found in human H3 viruses at this position (Matrosovich et al., 2000; Stevens et al., 2006a). The A/Udorn/307/72 influenza virus strain has affinity for both 2,3 SA and 2,6 SA (Matrosovich et al., 2000; Suzuki et al., 2000) and has amino acids associated with 2,6 SA receptor binding at positions 226 and 228 but not at position 190.
Since receptor recognition is believed to be one, if not the, key barrier for cross-species transmission of influenza A viruses, we initiated a study to determine if recognition of 2,3 SA could account for the ability of influenza A/Udorn/307/72 to productively infect murine tracheal epithelial cell (mTEC) cultures. Our results suggest that amino acid changes outside of positions 226 and 228 of H3 HA proteins, in particular position 190, can profoundly impact the ability of influenza A virus to recognize and utilize 2,3 SA as a virus receptor.
Results
The influenza A/Udorn/307/72 (rUdorn) virus HA protein can recognize 2,6 and 2,3 SA and is able to replicate in mTEC cultures (Ibricevic et al., 2006; Newby et al., 2007). The mTEC cultures express only 2,3 SA and minimally tissue culture passaged influenza virus strains that recognize only 2,6 SA are not able to replicate in mTEC cultures (Ibricevic et al., 2006; Newby et al., 2007), suggesting the recognition of 2,3 SA is key for influenza virus replication in these cultures. The rUdorn HA protein contains consensus residues at positions 226 and 228 which confer binding to 2,6 SA, however, it encodes an E at position 190 which has been shown to increase HA affinity for 2,3 SA (Martin et al., 1998; Matrosovich et al., 2000; Nobusawa et al., 2000; Yassine et al., 2007).
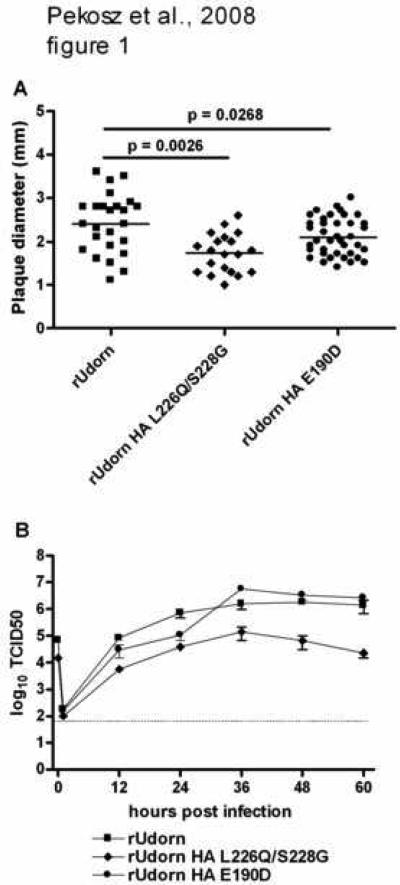
Site directed mutagenesis was used to introduce amino acid changes that would alter the SA recognition of the rUdorn HA to solely 2,3 SA (HA L226Q/S228G). A mutation at position 190 (E190D) was introduced in order to confer greater selectivity for 2,6 SA. Recombinant influenza viruses encoding these mutations were rescued and characterized for their replication in MDCK cells since this cell type is routinely used to propagate a number of different influenza A virus strains. The recombinant virus encoding HA L226Q/S228G had significantly smaller plaques (p=0.0026) than the rUdorn wt virus while the virus encoding HA E190D formed plaques that were not significantly different in size (p=0.0268) from those of rUdorn wt (Fig. 1A). This data indicated the rUdorn HA L226Q/S228G virus was not as efficient at infecting MDCK cells as its parental virus.
Figure 1.
Replication of rUdorn viruses encoding HA proteins with mutations in the receptor binding site. A) The indicated recombinant viruses were analyzed by plaque assay on MDCK cells. The plaque diameter was measured with a micrometer, average plaque diameter (n=25 for rUdorn; n=20 for rUdorn HA L226Q/S228G; n=39 for rUdorn HA E190D) calculated and statistical significance determined by student's t test. B) Virus replication in MDCK cells after infection at an MOI=0.01. Infected cell supernatants were harvested at the indicated times and TCID50 titers determined on MDCK cells. The mean and standard error of the mean are graphed.
The viruses were then characterized for their replication in MDCK cells (Fig. 1B) after a low multiplicity of infection (MOI). The rUdorn HA E190D virus replicated to similar titers as the rUdorn wt virus while the rUdorn HA L226Q/S228G virus replicated to titers that were consistently 10 to 100 fold lower. MDCK cells express higher levels of 2,3 SA when compared to 2,6 SA (Matrosovich et al., 2003; Oh et al., 2008) so the reduced replication of the rUdorn HA L226Q/S228G virus in both plaque assays and after low MOI infection implies that these mutations have a detrimental effect for virus replication in the rUdorn genetic background.
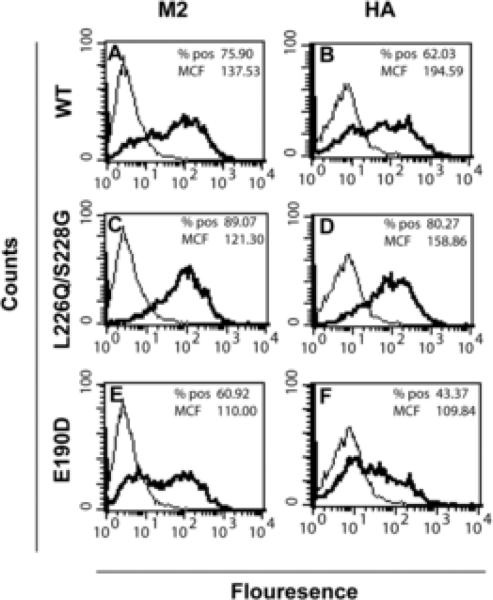
To determine if the rUdorn HA L226Q/S228G virus had altered levels of viral protein expression, MDCK cells were infected at an MOI of 5, harvested 6 hpi and the levels of HA and M2 protein expression on the plasma membrane quantified by flow cytometry (Fig. 2). The numbers of virus infected cells was equivalent for rUdorn wt and rUdorn HA L226Q/S228G viruses when the number of M2 (75.90% versus 89.07%) or HA (62.03% versus 80.27%) positive cells was determined. The amount of cell surface HA and M2 (as judged by the MCF) was also comparable, indicating that the expression level of these viral proteins was not altered by the introduction of the HA mutations at L226Q/S228G. The rUdorn E190D virus had slightly lower numbers of antigen positive cells and protein expression when compared to rUdorn (Fig. 2 E and F), suggesting that slight changes in viral protein expression levels did not alter virus replication (Figure 1).
Figure 2.
Viral protein expression. MDCK cells were infected at an MOI of approximately 5 and harvested for flow cytometry at 6 hpi. The cells were immunostained for the M2 (A, C and E) or HA (B, D, F) protein and analyzed by flow cytometry. The percent positive (% pos) and mean channel fluorescence (MCF) were determined using CellQuest software. The data shown are representative of results from 4 independent experiments. The dark traces represent virus-infected cells while the light traces represent mock-infected cells.
Since the introduced mutations should alter the receptor binding of the recombinant viruses, the hemagglutination activity of the viruses against red blood cells (RBCs) from several species was determined (Table 1). Equivalent infectious units of all viruses were tested for their ability to hemagglutinate human, chicken, horse or swine RBCs. Human, chicken and swine RBCs possess both 2,3 and 2,6 SA while horse RBCs express primarily 2,3 SA (Ito et al., 1997a). The rUdorn and rUdorn HA L226Q/S228G viruses were able to hemagglutinate all the RBCs to similar extents. In contrast, the rUdorn HA E190D virus was not able to hemagglutinate human or horse RBC, but did hemagglutinate chicken and swine RBCs, although the titer was significantly lower than that observed with the other viruses. This data indicate that the introduction of the E190D mutation to the Udorn HA not only altered receptor binding specificity, but also affected the receptor binding affinity for various RBCs. It is interesting to note that position 98 of H3 subtype influenza viruses has recently been shown to alter virus-receptor interactions (Meisner et al., 2008). Mutations at position 98 had little effect on virus replication in MDCK cells but severely attenuated replication in the mouse respiratory tract. This data, together with the data presented here, suggest that alterations in influenza receptor specificity may have more dramatic effects on virus replication in relevant tissue culture systems or animal models than are seen in standard cell lines.
Table 1.
Hemagglutination titers of rUdorn HA mutants
| RBCsb | ||||
|---|---|---|---|---|
| Virusa | Human | Chicken | Horse | Swine |
| rUdorn | 64 | 256 | 256 | 256 |
| rUdorn HA L226Q/S228G | 256 | 512 | 512 | 512 |
| rUdorn HA E190D | <4 | 64 | <4 | 32 |
All viruses were diluted to a concentration of 106 TCID50 before the dilution series was made.
The numbers represent the inverse of the last dilution at which hemagglutination was observed and are representative of 5 independent experiments.
The data from Fig. 1 and Table 1 indicate that the ability to replicate effectively in MDCK cells does not correlate with the ability to hemagglutinate RBCs, as the rUdorn and rUdorn HA L226Q/S228G viruses both hemagglutinated RBCs efficiently, but produced significantly different amounts of infectious virus after infection of MDCK cells. In contrast, the rUdorn HA E190D virus was less efficient at RBC hemagglutination than rUdorn, but replicated to similar titers on MDCK cells.
Altering the receptor binding specificity of rUdorn led to unexpected changes in virus replication on MDCK cells. In order to assess the effects of altering receptor binding specificity on virus replication in other cell types, the recombinant rUdorn viruses were used to infect the human lung epithelial cell line CaLu3 at a low MOI (Fig. 3a). CaLu3 cells express both 2,3 and 2,6 SA and have been used to study the replication of a number of human influenza virus strains including the 1918 and contemporary H5N1 viruses (Tumpey et al., 2005; Zeng et al., 2007). Virus replication in CaLu3 cells was significantly different than replication in MDCK cells for the rUdorn HA E190D virus, as it replicated to lower initial titers than either rUdorn wt or the rUdorn HA L226Q/S228G. In agreement with the data from MDCK cells, the rUdorn HA L226Q/S228G virus replicated to titers 10-100 fold lower than rUdorn wt. The data indicate that a change at position 190 of the H3 HA protein can lead to attenuated virus replication on a human respiratory epithelial cell line.
Figure 3.
Virus infection of respiratory epithelial cells. The indicated viruses were used to infect A) human CaLu3 cells or B) differentiated mTEC cultures at an MOI=0.001. Infected cell supernatants were harvested at the indicated times and TCID50 titers determined on MDCK cells. The mean and standard error of the mean are graphed.
Virus replication in primary, differentiated mTEC cultures was then assessed. These cultures express only 2,3 SA and furthermore, the 2,3 SA is expressed only on the ciliated cells (Ibricevic et al., 2006; Newby et al., 2006). Virus replication mirrored that seen in CaLu3 cells, with the rUdorn HA E190D virus being more attenuated than either rUdorn wt or rUdorn HA L226Q/S228G. In fact, no infectious virus was detected from rUdorn HA E190D infected cells at any time point tested. Again, the rUdorn HA L226Q/S228G virus showed a replication rate that was consistent with its replication in MDCK cells (10-100 fold lower than rUdorn wt). Taken together, the data indicate that altering the receptor binding specificity of the rUdorn HA at position 190, has profound effects on virus replication in murine epithelial cell cultures.
Since the rUdorn HA E190D virus was extremely attenuated after infection of mTEC cultures, the ability of the virus to infect these cells was investigated further. The mTEC cultures were infected with the rUdorn viruses and the cultures were analyzed for viral protein expression at 48 hpi (Fig. 4). Viral antigen positive ciliated cells were detected in mTEC cultures infected with either rUdorn wt or rUdorn HA L226Q/S228G. In two independent experiments, only one cell infected with rUdorn E190D was identified, suggesting that this virus was unable to establish an infection in mTEC cultures. When the number of virus infected cells was quantified (Fig. 4J), the numbers of infected cells was consistent with the extent of virus replication in the cultures (Fig. 3B). Taken together, the data indicate that the amino acid at position 190 of H3 subtype HA proteins plays an important role in virus replication in human and murine epithelial cell cultures.
Figure 4.
Virus infection of mTEC cultures. At 48 hpi, mTEC cultures were fixed and processed for confocal microscopy using antibodies to the ciliated cell protein β-tubulin IV and the HA (subtype H3) protein. Confocal images were acquired using a 63x objective and are magnified by a factor of 2x (A-F) or 1x (G, H, I). The image is reconstituted from a series of z-stacks. J) The virus infected cells in 21-23 randomly chosen fields, in two independent experiments were counted. The individual values and means are shown. Statistical significance determined by student's t test.
Discussion
The interaction of influenza A virus from non-human hosts with cell receptors on human cells is undoubtedly an important factor in determining whether a particular influenza A virus strain is capable of infecting humans (Suzuki et al., 2000). While this paradigm is often portrayed as simply an issue of differential recognition of 2,3 versus 2,6 SA, in reality the interactions of influenza virus with host cell receptors are much more complex (Nicholls et al., 2008; Stevens et al., 2006b).
Sialic acid residues can be linked to a number of different N- and O-linked carbohydrate chains in a cell- or species- specific manner (Varki, 2007; Varki and Varki, 2007). Influenza virus entry into susceptible cells appears to be dependent on SA residues attached to N-linked carbohydrates (Chu and Whittaker, 2004), therefore there may be a distinction between SA residues that allow for binding of influenza and SA residues that can mediate efficient entry of the virus. Other carbohydrate residues besides the terminal SA can contribute significant interactions with HA that can stabilize and facilitate virus binding (Nicholls et al., 2008; Stevens et al., 2006b; Suzuki, 2005).
In addition to the complexity of the carbohydrates and SA present on the cell surface, a number of amino acids that surround the HA receptor binding pocket play important roles in stabilizing virus-cell interactions. Position 190 of H1 HA subtypes (Glaser et al., 2005; Stevens et al., 2006a) and positions 226 and 228 of H3 HA subtypes (Connor et al., 1994; Vines et al., 1998) have critical roles in differentiating 2,3 from 2,6 SA. The receptor binding pocket of HA is a cavity at the tip of the protein which consists of a number of different regions of the protein including but not limited to the ones identified as key for discriminating 2,3 and 2,6 SA (Russell et al., 2006). A number of studies focusing on influenza A virus adaption to replication in tissue culture (Asaoka et al., 2006; Govorkova et al., 1999; Rocha et al., 1993) or embryonated hen eggs (Ito et al., 1997a; Ito et al., 1997b; Lu et al., 2006; Lu et al., 2005; Medeiros et al., 2001; Widjaja et al., 2006) have suggested that amino acid changes at a number of different residues in HA can contribute to replication and/or alteration of receptor recognition. Finally, the glycosylation of HA can have profound effects on receptor recognition (Deom et al., 1986; Ohuchi et al., 1997).
The complexities evident in receptor expression and recognition no doubt play an important but poorly defined role in influenza virus infection in vivo. It is important to note the differences in virus replication observed in the three culture systems analyzed in this study. Recombinant viruses encoding HA proteins with altered receptor binding pockets replicated to different extents, depending on the culture system uses. In MDCK cells, the rUdorn HA E190D virus replicated to levels that were nearly identical to that of rUdorn wt (Fig. 1) but the same virus was severely attenuated in CaLu3 and mTEC cultures (Fig. 3). This data indicates that receptor utilization can have important implications on the cell tropism of influenza virus.
While 2,6 SA is not detectable in mTEC cultures (Ibricevic et al., 2006; Newby et al., 2007), both 2,3 SA and 2,6 SA are present on CaLu3 cells (Zeng et al., 2007), suggesting the change in position 190 of H3 HA proteins may also alter interactions with other carbohydrate residues present on the 2,6 SA containing carbohydrate chains. These kinds of interactions have been discerned in X-ray crystallographic structures (Russell et al., 2006) and inferred from a number of studies involving egg or tissue culture adaption of H3 influenza viruses (Lu et al., 2006; Lu et al., 2005; Martin et al., 1998; Medeiros et al., 2001; Medeiros et al., 2004; Meisner et al., 2008; Nakajima et al., 2003). This data serves to emphasize the role that infections of relevant cell types or culture systems can play in elucidating the biologically important effects of altering receptor binding specificity on influenza virus infection.
A role in receptor recognition for position 190 in H3 HA subtypes has been suggested in a number of studies, as this position comes in contact with the penultimate carbohydrate in several models of HA-receptor interactions (Matrosovich et al., 2000; Russell et al., 2006; Stevens et al., 2006a). A change from E to D at position 190 altered the ability of influenza A/Aichi/51/92 (H3N2) to agglutinate chicken red blood cells and increased recognition of 2,6 SA (Nobusawa et al., 2000), which is consistent with our data (Table 1). A substitution of E190A in the A/Aichi/5/68 (H3N2) HA altered binding to human erythrocytes and sensitivity to inhibition with horse serum, suggesting an altered receptor binding activity (Martin et al., 1998). A D190A mutation in A/turkey/Ohio/313053/04 H3N2 was shown to be critical for transmission of this virus from turkeys to swine, efficient virus replication in ducks and transmission in chickens (Yassine et al., 2007). In contrast, an E190G substitution in the A/Aichi/2/68 did not significantly alter agglutination of chicken RBCs (Nakajima et al., 2003), suggesting that the nature of the amino acid substitution at position 190 is important for receptor recognition. Since our results with A/Udorn/307/72 indicate alterations at position 190 have profound effects on RBC hemagglutination and infection of respiratory epithelial cells, it may be that the importance of position 190 is dependent upon the identity of other amino acids in the particular HA protein under study.
The dramatic reduction in rUdorn HA E190D replication in mTEC cultures illustrates the importance of 2,3 SA recognition in the mouse model of influenza virus infection. Influenza viruses with strictly 2,6 SA recognition did not infect mice or mTEC cultures while viruses with 2,3 SA or mixed SA recognition replicated to varying degrees (Ibricevic et al., 2006). Eliminating 2,3 SA recognition of rUdorn abolished virus replication in mTEC cultures. Since the influenza NA activity has been shown to be important for virus release from infected cells, it is possible that part of the replication defects observed with our recombinant viruses result from a specificity mismatch of HA receptor recognition versus NA receptor destroying activity.
While mice express both 2,3 and 2,6 sialyltransferases, there is contradictory data on the expression pattern of 2,3 versus 2,6 SA in the mouse respiratory tract. Ibricevic, et al, demonstrated 2,3 SA but no detectable 2,6 SA in mouse lung and influenza A viruses that recognized only 2,6 SA were not able to replicate in mice (Ibricevic et al., 2006). Cultures of mTECs from C57Bl6 or Balb/c mice contain only 2,3 SA (Ibricevic et al., 2006; Newby et al., 2007). In contrast, Glaser et al., demonstrated the expression of both forms of SA (Glaser et al., 2007) in the mouse respiratory tract and viruses that recognize 2,6 SA could replicate in C57Bl6 mice, as well as mice that lacked the gene for 2,6 sialyltransferase, suggesting that human influenza viruses can replicate in the absence of 2,6 SA (Glaser et al., 2007). Since some influenza A virus strains can replicate in cell lines devoid of 2,6 SA (Kumari et al., 2007) it may be that there are other, non SA receptors for influenza A virus (Rapoport et al., 2006). Since these studies utilized lectins for SA detection, a resolution of this discrepancy must await the use of other techniques for SA characterization and quantification.
Materials and Methods
Cells
Madin-Darby canine kidney cells (MDCK), human embryonal kidney cells (293T) and human epithelial cells (CaLu3) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. The HB-64 and HB-65 hybridomas (ATCC) were cultured in DMEM containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. For antibody collection, the cells were grown to high density, the media was replaced with RPMI 1640 media containing 10% hybridoma enhancing supplement (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin and the cells were cultured for 5-7 days before cell free supernatants were collected and stored at -20°C.
Murine trachea epithelial cell (mTEC) cultures from Balb/c mice were established on 0.4 µM pore, 0.33 cm2 Transwell-Clear membranes using air-liquid interface (ALI) conditions as previously described (Rowe et al., 2004; You et al., 2002). When mature (after at least 7 days of ALI), the mTEC cultures were composed of 30 to 50% ciliated cells (Newby et al., 2007).
Viruses
The influenza A viruses used in this study were derived from the A/Udorn/307/72 virus (rUdorn) reverse genetics system (Takeda et al., 2002). The rUdorn virus encodes an H3 hemagglutinin (HA) protein which recognizes both α2,3- and α2,6-linked sialic acid. In order generate an HA that preferentially recognizes α2,3-linked sialic acid, PCR mutagenesis was used to introduce amino acid substitutions at positions 226 (L to Q) and 228 (S to G) in plasmid pHH21 Udorn HA (Takeda et al., 2002).To generate an HA that preferentially recognizes α2,6-linked sialic acid, PCR mutagenesis was used to introduce an amino acid at position 190 (E to D). Viruses encoding these HA proteins (rUdorn HA L226Q/S228G and rUdorn HA E190D) were generated entirely from cDNA using the 12 plasmid rescue system (Takeda et al., 2002). The HA sequence of the rescued viruses was confirmed by sequencing the coding region of the HA segment.
Viral stocks were generated by infecting MDCK cells at a multiplicity of infection (MOI) of 0.01 plaque forming units (PFU) per cell and the infected cell supernatant harvested 72 hours post-infection. Infectious virus titers were determined by plaque assay or 50% tissue culture infectious dose (TCID50) as described previously (McCown et al., 2003; McCown and Pekosz, 2006). The diameters of plaques visible in MDCK monolayers stained with Napthol blue black (Sigma) was measured with a micrometer (Scienceware).
Hemagglutination Assay
Chicken, horse, swine and human red blood cells (RBCs, Cocalico Biologicals) were diluted to a 20% v/v solution in Alsever's Solution. In a 96 well plate, containing 100 μl of PBS per well, a 2 fold dilution series of virus was prepared. Equivalent TCID50 units were used. Red blood cells from each species were diluted to 0.5% and 100 μl was added to each well of the dilution series. Hemagglutination (HA) units for each virus was determined by comparing RBCs that settled out of solution to those that remained suspended after an overnight incubation at 4°C.
Infectious virus production
The kinetics of infectious virus production on MDCK and CaLu3 cells were determined by infecting with the indicated virus at an MOI of 0.01 in DMEM containing 4 μg/ml N-acetyl trypsin, penicillin, and streptomycin. The cells were rocked at room temperature for 1 hour, washed with PBS, and incubated in DMEM with 4 μg/ml N-acetyl trypsin, 0.1% bovine serum albumin, penicillin, and streptomycin. At the indicated times post infection, infected cells were collected and stored at −70°C. Infectious virus titers were determined by TCID50.
Cultures of mTECs were infected via the apical chamber with approximately 3,600 PFU of virus diluted in warm DMEM containing penicillin-streptomycin in a total volume of 100 µl. If all cells in the culture were susceptible to influenza virus infection, this would correspond to a MOI of approximately 0.01. The cells were incubated with virus at 37°C for 1 hr, the inoculum was removed, and cells were washed three times with 200 μl of DMEM containing penicillin-streptomycin. After washing, 100 μl of DMEM containing penicillin-streptomycin and 500 μl of TEC MM (Rowe et al., 2004; You et al., 2002) was placed in the apical and basolateral chambers, respectively. Apical supernatants were collected at the indicated times post infection and stored at −70°C. Infectious virus titers were determined by TCID50.
Immunofluorescence confocal microscopy
At 48 hrs post-infection, mTECs were washed three times with phosphate buffered saline (PBS, GIBCO Inc., Carlsbad, CA), and fixed in PBS containing 2% paraformaldehyde for 15 min at room temperature. Cells were washed three times and permeabilized with PBS containing 0.2% Triton-X 100 and 0.1% sodium citrate for ten minutes at room temperature. Cells were washed with PBS and incubated in PBS containing 3% normal goat or normal donkey serum and 0.5% bovine serum albumin (blocking buffer) for 30 min at room temperature. Cells were washed, and incubated with mouse anti-β Tubulin IV (1:100 dilution; BioGenex, San Ramon, CA) and goat anti-A/Aichi/2/68 H3 sera (1:250 immunofluorescence; NIH/NIAD reference reagent V314-591-157). After washing, the cells were incubated with goat anti-mouse (1:500 dilution, Alexa Flour 488, Molecular Probes), and TO-PRO-3 for 45 min. The wash solution for all steps is PBS with 0.2% Tween-20. Transwell-Clear membranes were mounted using 10uL of Molecular Probes ProLong antifade (Molecular Probes), and slides were imaged using a Zeiss LSM 510 Meta confocal microscope. All images were obtained with a 63x oil objective. All images presented are a flattened composite of Z-stack images collected with LSM software. Random fields were chosen and the numbers of antigen positive cells per field were counted.
Flow Cytometry
MDCK cells were infected with virus at an MOI=5, then removed from the tissue culture plate with trypsin at 6 hrs post-infection. The trypsin was inactivated by addition of FBS to the solution at a final concentration of 10%. Cells were washed three times in PBS and immunostained for M2 or HA surface expression using the mouse monoclonal antibody 14C2 (1:500 dilution; anti-influenza A virus M2 protein), and the goat anti-A/Aichi/2/68 H3 sera (1:250 dilution). The secondary antibodies used were goat anti-mouse (1:500 dilution; Alexa Flour 488, Molecular Probes) and donkey anti-goat IgG (1:500 dilution; Alexa Fluor 647; Molecular Probes). Cells were stained live to maximize cell surface staining. All antibody dilutions were made in blocking buffer and washes were made in PBS. The specific staining was quantified using a FACSCalibur dual laser flow cytometer (Becton-Dickinson) and data was collected with Cell Quest software.
Acknowledgements
We thank the members of the Pekosz lab for insightful discussions and comments. We acknowledge and thank Robert A. Lamb and Makoto Takeda for their generous gift of the reverse genetics system for A/Udorn/307/72. All confocal microscopy was performed at the Molecular Microbiology Imaging Center at Washington University in St. Louis. Flow cytometry was performed at the Becton Dickinson Immune Function Laboratory. We acknowledge support from R01 AI053629, the Eliasberg Foundation and the Marjorie Gilbert Foundation.
References
- Asaoka N, Tanaka Y, Sakai T, Fujii Y, Ohuchi R, Ohuchi M. Low growth ability of recent influenza clinical isolates in MDCK cells is due to their low receptor binding affinities. Microbes and Infection. 2006;8(2):511–519. doi: 10.1016/j.micinf.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Busch MG, Bateman AC, Landolt GA, Karasin AI, Brockman-Schneider RA, Gern JE, Suresh M, Olsen CW. Identification of amino acids in the HA of H3 influenza viruses that determine infectivity levels in primary swine respiratory epithelial cells. Virus Research. 2008;133(2):269–279. doi: 10.1016/j.virusres.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Chu VC, Whittaker GR. Influenza virus entry and infection require host cell N-linked glycoprotein. PNAS. 2004;101(52):18153–18158. doi: 10.1073/pnas.0405172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor Specificity in Human, Avian, and Equine H2 and H3 Influenza Virus Isolates. Virology. 1994;205(1):17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- Deom CM, Caton AJ, Schulze IT. Host Cell-Mediated Selection of a Mutant Influenza A Virus That Has Lost a Complex Oligosaccharide from the Tip of the Hemagglutinin. PNAS. 1986;83(11):3771–3775. doi: 10.1073/pnas.83.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. The Evolutionary Genetics and Emergence of Avian Influenza Viruses in Wild Birds. PLoS Pathogens. 2008;4(5):e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser L, Conenello G, Paulson J, Palese P. Effective replication of human influenza viruses in mice lacking a major [alpha]2,6 sialyltransferase. Virus Research. 2007;126(1-2):9–18. doi: 10.1016/j.virusres.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. A Single Amino Acid Substitution in 1918 Influenza Virus Hemagglutinin Changes Receptor Binding Specificity. J. Virol. 2005;79(17):11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorkova EA, Matrosovich MN, Tuzikov AB, Bovin NV, Gerdil C, Fanget B, Webster RG. Selection of receptor-binding variants of human influenza A and B viruses in baby hamster kidney cells. Virology. 1999;262(1):31–8. doi: 10.1006/viro.1999.9892. [DOI] [PubMed] [Google Scholar]
- Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. Influenza Virus Receptor Specificity and Cell Tropism in Mouse and Human Airway Epithelial Cells. J. Virol. 2006;80(15):7469–7480. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y. Receptor Specificity of Influenza A Viruses Correlates with the Agglutination of Erythrocytes from Different Animal Species. Virology. 1997a;227(2):493–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Takada A, Kawamoto A, Otsuki K, Masuda H, Yamada M, Suzuki T, Kida H, Kawaoka Y. Differences in sialic acid-galactose linkages in the chicken egg amnion and allantois influence human influenza virus receptor specificity and variant selection. J. Virol. 1997b;71(4):3357–3362. doi: 10.1128/jvi.71.4.3357-3362.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, Air GM. Receptor binding specificity of recent human H3N2 influenza viruses. Virol J. 2007;4(1):42. doi: 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Zhou H, Chan W, Kemble G, Jin H. Single amino acid substitutions in the hemagglutinin of influenza A/Singapore/21/04 (H3N2) increase virus growth in embryonated chicken eggs. Vaccine. 2006;24(44-46):6691–6693. doi: 10.1016/j.vaccine.2006.05.062. [DOI] [PubMed] [Google Scholar]
- Lu B, Zhou H, Ye D, Kemble G, Jin H. Improvement of Influenza A/Fujian/411/02 (H3N2) Virus Growth in Embryonated Chicken Eggs by Balancing the Hemagglutinin and Neuraminidase Activities, Using Reverse Genetics. J. Virol. 2005;79(11):6763–6771. doi: 10.1128/JVI.79.11.6763-6771.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Wharton SA, Lin YP, Takemoto DK, Skehel JJ, Wiley DC, Steinhauer DA. Studies of the Binding Properties of Influenza Hemagglutinin Receptor-Site Mutants. Virology. 1998;241(1):101–111. doi: 10.1006/viro.1997.8958. [DOI] [PubMed] [Google Scholar]
- Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk H-D. Overexpression of the {alpha}-2,6-Sialyltransferase in MDCK Cells Increases Influenza Virus Sensitivity to Neuraminidase Inhibitors. J. Virol. 2003;77(15):8418–8425. doi: 10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early Alterations of the Receptor-Binding Properties of H1, H2, and H3 Avian Influenza Virus Hemagglutinins after Their Introduction into Mammals. J. Virol. 2000;74(18):8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown M, Diamond MS, Pekosz A. The utility of siRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology. 2003;313(2):514–524. doi: 10.1016/s0042-6822(03)00341-6. [DOI] [PubMed] [Google Scholar]
- McCown MF, Pekosz A. Distinct Domains of the Influenza A Virus M2 Protein Cytoplasmic Tail Mediate Binding to the M1 Protein and Facilitate Infectious Virus Production. J. Virol. 2006;80(16):8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros R, Escriou N, Naffakh N, Manuguerra JC, van der Werf S. Hemagglutinin residues of recent human A(H3N2) influenza viruses that contribute to the inability to agglutinate chicken erythrocytes. Virology. 2001;289(1):74–85. doi: 10.1006/viro.2001.1121. [DOI] [PubMed] [Google Scholar]
- Medeiros R, Naffakh N, Manuguerra J-C, Werf S. v. d. Binding of the hemagglutinin from human or equine influenza H3 viruses to the receptor is altered by substitutions at residue 193. Archives of Virology. 2004;149(8):1663–1671. doi: 10.1007/s00705-003-0287-2. [DOI] [PubMed] [Google Scholar]
- Meisner J, Szretter KJ, Bradley KC, Langley WA, Li Z-N, Lee B-J, Thoennes S, Martin J, Skehel JJ, Russell RJ, Katz JM, Steinhauer DA. Infectivity Studies of Influenza Virus Hemagglutinin Receptor Binding Site Mutants in Mice. J. Virol. 2008;82(10):5079–5083. doi: 10.1128/JVI.01958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Nobusawa E, Tonegawa K, Nakajima S. Restriction of Amino Acid Change in Influenza A Virus H3HA: Comparison of Amino Acid Changes Observed in Nature and In Vitro. J. Virol. 2003;77(18):10088–10098. doi: 10.1128/JVI.77.18.10088-10098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby CM, Rowe RK, Pekosz A. Influenza A virus infection of primary differentiated airway epithelial cell cultures derived from Syrian golden hamsters. Virology. 2006;354(1):80–90. doi: 10.1016/j.virol.2006.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby CM, Sabin L, Pekosz A. The RNA Binding Domain of Influenza A Virus NS1 Protein Affects Secretion of Tumor Necrosis Factor Alpha, Interleukin-6, and Interferon in Primary Murine Tracheal Epithelial Cells. J. Virol. 2007;81(17):9469–9480. doi: 10.1128/JVI.00989-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls JM, Chan RWY, Russell RJ, Air GM, Peiris JSM. Evolving complexities of influenza virus and its receptors. Trends in Microbiology. 2008;16(4):149–157. doi: 10.1016/j.tim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Nobusawa E, Ishihara H, Morishita T, Sato K, Nakajima K. Change in Receptor-Binding Specificity of Recent Human Influenza A Viruses (H3N2): A Single Amino Acid Change in Hemagglutinin Altered Its Recognition of Sialyloligosaccharides. Virology. 2000;278(2):587–596. doi: 10.1006/viro.2000.0679. [DOI] [PubMed] [Google Scholar]
- Oh DY, Barr IG, Mosse JA, Laurie KL. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J Clin Microbiol. 2008;46(7):2189–94. doi: 10.1128/JCM.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi M, Ohuchi R, Feldmann A, Klenk HD. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J. Virol. 1997;71(11):8377–8384. doi: 10.1128/jvi.71.11.8377-8384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish CR, Kawaoka Y. The Origins of New Pandemic Viruses: The Acquisition of New Host Ranges by Canine Parvovirus and Influenza A Viruses. Annual Review of Microbiology. 2005;59(1):553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- Rajagopal S, Treanor J. Pandemic (avian) influenza. Semin Respir Crit Care Med. 2007;28(2):159–70. doi: 10.1055/s-2007-976488. [DOI] [PubMed] [Google Scholar]
- Rapoport E, Mochalova L, Gabius HJ, Romanova J, Bovin N. Search for additional influenza virus to cell interactions. Glycoconjugate Journal. 2006;23(1-2):115–125. doi: 10.1007/s10719-006-5444-x. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Xu X, Hall HE, Allen JR, Regnery HL, Cox NJ. Comparison of 10 influenza A (H1N1 and H3N2) haemagglutinin sequences obtained directly from clinical specimens to those of MDCK cell- and egg-grown viruses. The Journal Of General Virology. 1993;74(Pt 11):2513–2518. doi: 10.1099/0022-1317-74-11-2513. [DOI] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- Rowe RK, Brody SL, Pekosz A. Differentiated Cultures of Primary Hamster Trachealairway Epithelial Cells. In Vitro Cell Dev Biol Anim. 2004;40(10):303–311. doi: 10.1290/0408056.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RJ, Stevens DJ, Haire LF, Gamblin SJ, Skehel JJ. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconjugate Journal. 2006;V23(1):85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. Glycan Microarray Analysis of the Hemagglutinins from Modern and Pandemic Influenza Viruses Reveals Different Receptor Specificities. Journal of Molecular Biology. 2006a;355(5):1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Paulson JC, Wilson IA. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Micro. 2006b;4(11):857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. Sialobiology of influenza: molecular mechanism of host range variation of influenza viruses. Biol Pharm Bull. 2005;28(3):399–408. doi: 10.1248/bpb.28.399. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ito T, Suzuki T, Holland RE, Jr., Chambers TM, Kiso M, Ishida H, Kawaoka Y. Sialic Acid Species as a Determinant of the Host Range of Influenza A Viruses. J. Virol. 2000;74(24):11825–11831. doi: 10.1128/jvi.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J Virol. 2002;76(3):1391–9. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-Associated Hospitalizations in the United States. JAMA. 2004;292(11):1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. Jama. 2003;289(2):179–86. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A. Characterization of the Reconstructed 1918 Spanish Influenza Pandemic Virus. Science. 2005;310(5745):77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446(7139):1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest. 2007;87(9):851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vines A, Wells K, Matrosovich M, Castrucci MR, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998;72(9):7626–31. doi: 10.1128/jvi.72.9.7626-7631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja L, Ilyushina N, Webster RG, Webby RJ. Molecular changes associated with adaptation of human influenza A virus in embryonated chicken eggs. Virology. 2006;350(1):137–145. doi: 10.1016/j.virol.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Yassine H, Al-Natour M, Lee C-W, Saif Y. Interspecies and intraspecies transmission of triple reassortant H3N2 influenza A viruses. Virology Journal. 2007;4(1):129. doi: 10.1186/1743-422X-4-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol. 2002;283(6):L1315–1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
- Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, Tumpey TM, Katz JM. Highly Pathogenic Avian Influenza H5N1 Viruses Elicit an Attenuated Type I Interferon Response in Polarized Human Bronchial Epithelial Cells. J. Virol. 2007;81(22):12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]