miRNAs are thought to repress target mRNA expression by recruiting the CCR4–NOT deadenylase complex that accelerates poly(A) tail shortening and subsequent decapping. This article shows that the decapping activator DDX6 binds directly to a subunit of the NOT complex, CNOT1. This NOT, recruited by miRISC, in turn recruits DDX6.
Keywords: microRNA, deadenylation, CCR4–NOT, DDX6, decapping, mRNA decay
Abstract
MicroRNAs (miRNAs) play critical roles in a variety of biological processes through widespread effects on protein synthesis. Upon association with the miRNA-induced silencing complex (miRISC), miRNAs repress target mRNA translation and accelerate mRNA decay. Degradation of the mRNA is initiated by shortening of the poly(A) tail by the CCR4–NOT deadenylase complex followed by the removal of the 5′ cap structure and exonucleolytic decay of the mRNA. Here, we report a direct interaction between the large scaffolding subunit of CCR4–NOT, CNOT1, with the translational repressor and decapping activator protein, DDX6. DDX6 binds to a conserved CNOT1 subdomain in a manner resembling the interaction of the translation initiation factor eIF4A with eIF4G. Importantly, mutations that disrupt the DDX6–CNOT1 interaction impair miRISC-mediated gene silencing in human cells. Thus, CNOT1 facilitates recruitment of DDX6 to miRNA-targeted mRNAs, placing DDX6 as a downstream effector in the miRNA silencing pathway.
INTRODUCTION
miRNAs have emerged as key mediators of post-transcriptional control, and regulate a significant proportion of the mammalian proteome (Selbach et al. 2008; Sun and Lai 2013). After being processed from a primary transcript precursor, one strand of the mature miRNA duplex is loaded onto an Argonaute (Ago) protein and imperfectly base-pairs with sequences primarily found in the 3′ untranslated regions (3′ UTRs) of target mRNAs (Fabian and Sonenberg 2012). Direct recruitment of the GW182 protein by Ago forms the minimal miRISC, which facilitates translational inhibition and destabilization of miRNA targets (Fabian and Sonenberg 2012). Gene silencing is established through a sequential mechanism, whereby translation is inhibited prior to mRNA decay (Mathonnet et al. 2007; Fabian et al. 2009; Bazzini et al. 2012; Bethune et al. 2012; Djuranovic et al. 2012). Most data are consistent with inhibition of translation initiation by miRNAs (Humphreys et al. 2005; Pillai et al. 2005; Mathonnet et al. 2007; Iwasaki et al. 2009; Zdanowicz et al. 2009; Walters et al. 2010; Fukaya and Tomari 2012; Moretti et al. 2012; Meijer et al. 2013; Ricci et al. 2013). Translation initiation is a complex process whereby the 40S ribosomal subunit is recruited to the mRNA and positioned at the initiation codon by several eukaryotic initiation factors (Sonenberg and Hinnebusch 2009). The eukaryotic translation initiation factor 4F (eIF4F) complex recognizes the 5′ cap structure (m7GpppN, where N is any nucleotide) present on all nuclear-transcribed eukaryotic mRNAs. eIF4F is composed of the cap-binding subunit eIF4E, the DEAD-box RNA helicase eIF4A, and the scaffolding factor eIF4G (Sonenberg and Hinnebusch 2009). eIF4F associates with the 40S ribosomal subunit via eIF3, and eIF4A unwinds RNA secondary structures in the mRNA 5′ UTR to facilitate ribosomal scanning for the start codon. While the presence of a 5′ cap structure on the mRNA, the association of eIF4F with the 5′ cap, and the eIF4A helicase have been demonstrated to be important for miRNA-mediated gene silencing, the exact mechanism is not known (Humphreys et al. 2005; Pillai et al. 2005; Mathonnet et al. 2007; Zdanowicz et al. 2009; Walters et al. 2010; Fukaya and Tomari 2012; Meijer et al. 2013).
After translation arrest, the miRISC promotes mRNA deadenylation (i.e., removal of the poly(A) tail), decapping and 5′-3′ degradation. miRNA-mediated deadenylation is initiated by the CCR4–NOT complex, a multisubunit deadenylase machinery that is recruited by the miRISC (Fabian and Sonenberg 2012). CCR4–NOT associates with the miRISC through a direct interaction between GW182 and the large, scaffolding subunit of CCR4–NOT, CNOT1 (Braun et al. 2011; Chekulaeva et al. 2011; Fabian et al. 2011). Deadenylation is followed by removal of the 5′ cap by the decapping enzyme, DCP2, and its associated stimulatory factors, including DCP1, EDC3, EDC4, DDX6, and Pat1b (Eulalio et al. 2007; Chen et al. 2009). Finally, decapped miRNA targets are degraded by the 5′–3′ exonuclease, Xrn1 (Rehwinkel et al. 2005; Chen et al. 2009). Coupling between deadenylation and decapping in eukaryotes occurs through an ill-understood molecular mechanism, although DDX6 and Pat1b are thought to functionally link both processes (Coller et al. 2001; Haas et al. 2010; Ozgur et al. 2010). The contribution of DDX6 and Pat1b to the transition from mRNA deadenylation to decapping, as well as the protein–protein contacts that link the deadenylation and decapping complexes, remain unclear.
DDX6 belongs to the DEAD-box family of proteins, and plays an evolutionarily conserved role in translational repression and the activation of mRNA decapping (Coller et al. 2001; Nakamura et al. 2001; Fischer and Weis 2002; Minshall and Standart 2004; Coller and Parker 2005; Fenger-Gron et al. 2005; Carroll et al. 2011; Ernoult-Lange et al. 2012). Mechanistic effects of the yeast DDX6 ortholog, Dhh1p, on gene expression have been well studied, with translation being repressed at the initiation phase in a nutrient-responsive manner (Coller and Parker 2005). Dhh1p also associates with ribosomes, and inhibits mRNA translation concomitant with the elongation step when tethered to the mRNA (Sweet et al. 2012). With respect to mRNA decapping, DDX6 is important for assembly of the decapping complex (Nissan et al. 2010), and may stimulate DCP2 activity (Fischer and Weis 2002). Both facets of DDX6-dependent inhibition of protein synthesis (i.e., translation inhibition and mRNA decapping) are thought to converge in the miRNA-silencing pathway, where efficient miRISC-dependent repression requires DDX6 (Chu and Rana 2006; Beilharz et al. 2009; Su et al. 2011). DDX6 associates with Ago and GW182 proteins in human cells (Chu and Rana 2006; Ozgur and Stoecklin 2013). These interactions were suggested to occur through binding to Pat1b (Ozgur and Stoecklin 2013). Thus, while DDX6 is required for miRNA silencing, the precise manner by which DDX6 is recruited to miRNA targets remains poorly understood.
Studies in Saccharomyces cerevisiae reported genetic and physical associations between the DDX6 ortholog, Dhh1p, and the CCR4–NOT complex, via an N-terminal fragment of the yeast CNOT1 ortholog, Not1p (Hata et al. 1998; Maillet and Collart 2002). Based on these observations, as well as the documented association of CNOT1 with the miRISC (Braun et al. 2011; Chekulaeva et al. 2011; Fabian et al. 2011), we wished to investigate whether human DDX6 and CNOT1 interact directly. Here, we establish a direct interaction between human DDX6 and CNOT1. Notably, our results indicate that the DDX6–CNOT1 complex adopts a conformation that is structurally similar to the eIF4A–eIF4G co-crystal structure (Schutz et al. 2008). Mutations at the DDX6–CNOT1 interface that disrupt binding also result in impaired miRISC-mediated gene silencing. Taken together, we describe a physical basis for the functional coupling between deadenylation and decapping, and provide insight into the manner by which DDX6 contributes to miRNA-directed control of gene expression.
RESULTS
DDX6 binds directly to CNOT1
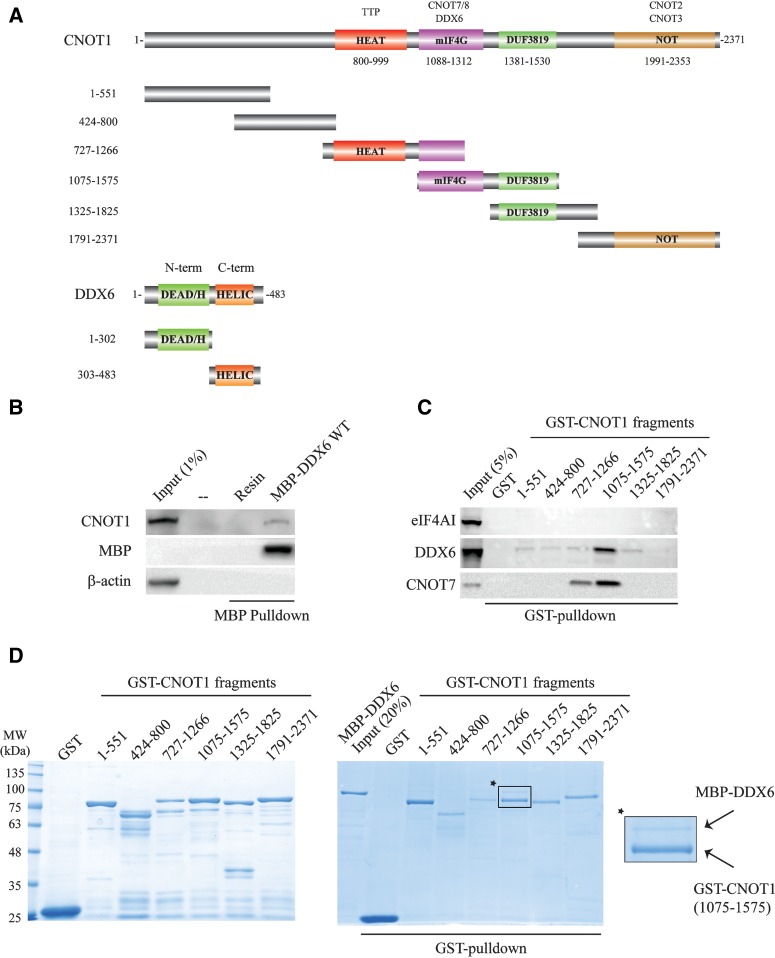
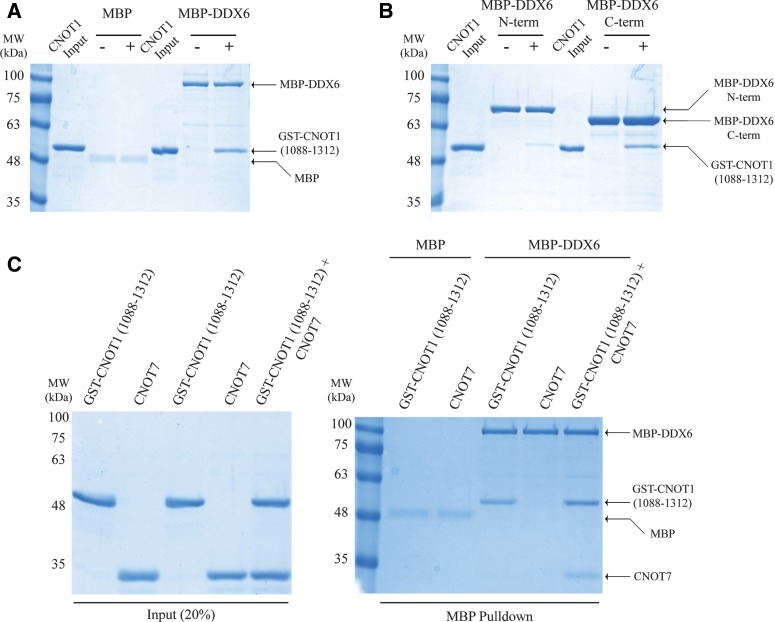
To determine whether human DDX6 and CNOT1 interact, we incubated recombinant full-length maltose-binding protein (MBP)-tagged human DDX6 (Fig. 1A) in an RNase A-treated HEK293T cell lysate and performed MBP-pulldown experiments (Fig. 1B). MBP-DDX6 weakly coprecipitated human CNOT1 (<1% input), but not β-actin, indicating that human DDX6 and CNOT1 are capable of associating with one another (Fig. 1B). To delineate the regions of CNOT1 that mediate the binding to DDX6, we incubated recombinant glutathione-S-transferase (GST)-tagged CNOT1 protein fragments (Fig. 1A) in an RNase A-treated HEK293T cell lysate and performed GST-pulldown experiments (Fig. 1C). Human CNOT7 interacted with a CNOT1 fragment encompassing residues 1075–1575 as expected (Basquin et al. 2012; Petit et al. 2012), whereas eIF4AI did not associate with any GST–CNOT1 fragment (Fig. 1C). In addition to CNOT7, CNOT11075–1575 efficiently coprecipitated DDX6 (Fig. 1C). To determine whether the interaction between DDX6 and CNOT1 was direct, we carried out in vitro GST-pulldown assays using the GST–CNOT1 fragments and recombinant MBP-tagged human DDX6. We detected a specific interaction between CNOT11075–1575 and MBP–DDX6 (Fig. 1D), and identified the CNOT1 mIF4G (middle domain of eIF4G) domain (residues 1088–1312) as sufficient for binding to DDX6 (Fig. 2A). We next sought to delineate the region(s) in DDX6 that interact with CNOT1. We generated two MBP-fused DDX6 protein fragments that divided DDX6 into its N-terminal (residues 1–302) and C-terminal (residues 303–483) lobes (Fig. 1A), and performed MBP-pulldown experiments with the GST–CNOT11088–1312 protein fragment. While both the N- and C-terminal lobes of DDX6 contributed to the interaction with CNOT11088–1312, the C-terminal fragment coprecipitated CNOT11088–1312 more efficiently (Fig. 2B). The CNOT1 mIF4G domain directly binds the catalytic subunit of the CCR4–NOT complex, CNOT7 (Basquin et al. 2012; Petit et al. 2012). Significantly, MBP-pulldown experiments showed that recombinant MBP–DDX6, GST–CNOT11088–1312, and CNOT7 are capable of forming a ternary complex (Fig. 2C), demonstrating that CNOT1 interactions with DDX6 and CNOT7 are not mutually exclusive. Taken together, these data establish human DDX6 and CNOT1 as direct binding partners, and define the CNOT1 mIF4G domain as the interaction site for the DDX6 C-terminal and, to a lesser extent, N-terminal lobes.
FIGURE 1.
Human DDX6 and CNOT1 interact directly. (A) Schematic representations of the CNOT1 and DDX6 protein fragments used in pulldown and coprecipitation experiments. Conserved elements are indicated within diagrams, while known interacting partners are labeled above the respective domains with which they associate. (HEAT) Huntington, elongation factor 3, protein phosphatase 2A, TOR domain; (mIF4G) middle domain of eIF4G; (DUF3819) domain of unknown function 3819; (NOT) NOT superfamily homology domain; (DEAD/H) DEAD/DEAH-box helicase domain; N-term domain; (HELIC) helicase C-terminal domain; C-term domain. (B) MBP-tagged DDX6 was immobilized on amylose resin and incubated in RNase A-treated HEK293T cell lysate. Precipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting using the specified antibodies. Dashed line above indicates empty lane. (C) Indicated GST-fused CNOT1 fragments, or GST alone as a negative control, were immobilized on Glutathione–Sepharose resin and incubated in RNase A-treated HEK293T cell lysate. Precipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting using the specified antibodies. (D) Coomassie-stained gels of GST–CNOT1 protein fragments (left) and GST-pulldown reactions (right). GST-fused CNOT1 fragments incubated with MBP-tagged full-length human DDX6 in the presence of Glutathione–Sepharose beads. GST alone was used as a negative control. Inset corresponds to enlarged image to the right, depicting the association between MBP–DDX6 and GST–CNOT11075–1575.
FIGURE 2.
The two lobes of DDX6 interact with the CNOT1 mIF4G domain. (A) Coomassie-stained gels of in vitro MBP-pulldown reactions. MBP–DDX6 was immobilized on an amylose resin and incubated with the GST-tagged CNOT1 mIF4G domain (residues 1088–1312). MBP was used as a negative control. (B) Coomassie-stained gels of in vitro MBP-pulldown reactions. MBP–DDX6 N-term (residues 1–302) or MBP–DDX6 C-term (residues 303–483) were immobilized on an amylose resin and incubated with GST–CNOT11088–1312. For A and B, inputs were 20%, (−) and (+) indicate pulldown reactions in the absence or presence of GST-CNOT11088–1312, respectively, and arrows indicate positions of respective proteins. (C) MBP–DDX6, GST-CNOT11088–1312, and CNOT7 form a stable ternary complex. Coomassie-stained gels of MBP-pulldown reactions. Wild-type MBP–DDX6 was immobilized on an amylose resin and incubated in the presence of GST-CNOT11088–1312 and/or untagged CNOT7. MBP served as a negative control.
Sequence analysis and in silico modeling predicts mutations that impair DDX6 binding to CNOT1
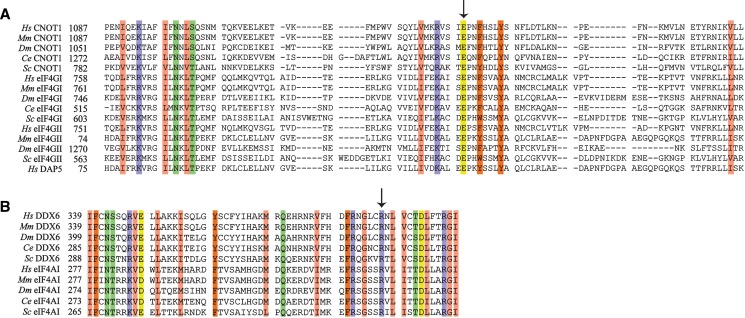
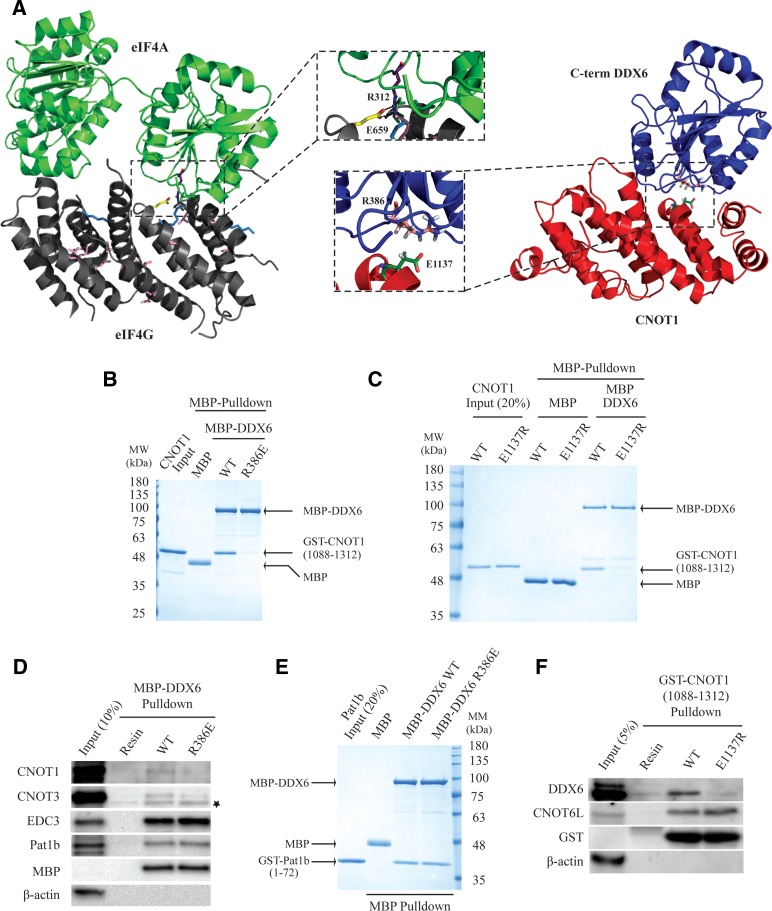
Multiple examples of DEAD-box proteins binding to different mIF4G domains have been described (Schutz et al. 2008; Montpetit et al. 2011; Buchwald et al. 2013), such as the prototypical complex of eIF4A and eIF4G. eIF4A binding to eIF4G is primarily mediated by an interaction with the mIF4G domain in eIF4G (Imataka and Sonenberg 1997). The co-crystal structure of yeast eIF4A and eIF4GmIF4G depicts a large interface between the C-terminal lobe of eIF4A and the N terminus of the eIF4G mIF4G domain, while contact between the N-terminal lobe of eIF4A with eIF4G is less extensive (Oberer et al. 2005; Schutz et al. 2008). Disruption of the eIF4A–eIF4G complex in vitro can be accomplished by mutating several conserved residues in the eIF4A C-terminal lobe (Dominguez et al. 2001; Oberer et al. 2005; Schutz et al. 2008). Based on the observation that the DDX6 C-terminal lobe bound to the CNOT1 mIF4G domain more efficiently than the DDX6 N-terminal lobe (Fig. 2B), as well as structural homology with eIF4A and eIF4G, we hypothesized that a DDX6–CNOT1 complex may adopt a quaternary structure that resembles the eIF4A–eIF4G complex. A sequence alignment of mIF4G domains from the eIF4G paralogs eIF4GI and eIF4GII with CNOT1 proteins identified only two conserved residues in eIF4G proteins that are important for binding to eIF4A (Glu659 and Thr618 in yeast eIF4GI, corresponding to Glu1137 and Ser1102 in human CNOT1) (Fig. 3A). Glu659 in yeast eIF4GI forms a salt bridge with Arg312 in eIF4A (Fig. 4A; left; Schutz et al. 2008), and charge reversal of Glu659 abolishes the interaction between the proteins in vitro (Dominguez et al. 2001; Schutz et al. 2008). To gain insight into how DDX6 and CNOT11088–1312 physically associate, crystal structures of the human DDX6 C-terminal domain (Tritschler et al. 2009) (PDB ID: 2WAX) and human CNOT1 mIF4G domain (Petit et al. 2012) (PDB ID: 4GML) were positioned in silico using the eIF4A–eIF4GI complex as a template, and docking simulations were performed using the RosettaDock online server (Lyskov and Gray 2008). Analysis of the lowest energy DDX6–CNOT1 model suggested that Glu1137 in CNOT1 is capable of forming a salt bridge with Arg386 in DDX6 (Fig. 4A; right). Notably, Arg386 is conserved both in sequence and position among DDX6 and eIF4A proteins throughout evolution (Figs. 3B, 4A). Thus, sequence alignment and in silico modeling indicate that a salt bridge between Arg386 in DDX6 and Glu1137 in CNOT1 could be important for their association.
FIGURE 3.
Sequence alignment of CNOT1 with eIF4G and DDX6 with eIF4A. (A) Sequence alignment of mIF4G domains from CNOT1, eIF4GI, and eIF4GII proteins. Alignments were generated using DIALIGN (Subramanian et al. 2008) and the BioEdit software. Proteins from the following species were used: Hs (Homo sapiens), Mm (Mus musculus), Dm (Drosophila melanogaster), Ce (Caenorhabditis elegans), Sc (Saccharomyces cerevisiae). Numberings indicate corresponding amino acid residues. Coloring highlights residues that are conserved among all proteins, and use the following scheme: salmon (hydrophobic, nonaromatic), purple (basic), green (polar), yellow (acidic), dark orange (aromatic). Position of conserved glutamic acid residue mutated in this study is indicated by an arrow. (B) Sequence alignment of DDX6 and eIF4AI C-terminal domains. Alignments were generated and labeled as in A. Position of the conserved arginine residue that was mutated in this study is shown by the arrow.
FIGURE 4.
Point mutations at the predicted DDX6-CNOT1 interface abolish their interaction. (A, left) Co-crystal structure of yeast eIF4A (light green) bound to eIF4G (gray) (PDB ID: 2VSO) (Schutz et al. 2008). Residues conserved among CNOT1 and eIF4G mIF4G domains are depicted, using the color scheme from Figure 3. Inset shows enlargement of the ionic salt bridge formed between eIF4A-R312 and eIF4G-E659. (Right) Lowest energy in silico model generated by the RosettaDock online server (Lyskov and Gray 2008). The human DDX6 C-terminal domain (C-term) (blue, PDB ID: 2WAX) (Tritschler et al. 2009) with FDF-peptide omitted and the human CNOT1 mIF4G domain (red, PDB ID: 4GML) (Petit et al. 2012) were superpositioned onto the eIF4A–eIF4G co-crystal structure using PyMOL, separated by ∼15 Å to reduce bias in the initial docking steps, and submitted to the RosettaDock server. Protein models were docked by performing an initial rigid body search, followed by optimization of side-chain contacts. Inset depicts position of conserved amino acid residues DDX6–R386 (pink) and CNOT1–E1137 (green). Putative salt bridge formed between DDX6–R386 and CNOT1–E1137 based on in silico modeling is not indicated. (B) Coomassie-stained gel of in vitro MBP-pulldown reactions. Wild-type (WT) and mutant (R386E) MBP-DDX6 proteins were immobilized on amylose resin and incubated with GST–CNOT11088–1312. MBP alone was used as a negative control. (C) Coomassie-stained gels of in vitro MBP-pulldown reactions using wild-type (WT) and mutant (E1137R) GST–CNOT11088–1312. MBP-tagged wild-type DDX6 or MBP alone (negative control) were immobilized on amylose resin and incubated with the indicated GST–CNOT11088–1312 proteins. (D) Indicated MBP–DDX6 proteins were incubated in RNase A-treated HeLa cell lysates, and precipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting using the indicated antibodies. Asterisk indicates nonspecific band. (E) Wild-type (WT) MBP–DDX6 and mutant (R386E) MBP–DDX6 were immobilized on amylose beads and incubated with purified GST-fused Pat1b peptide (residues 1–72). Coprecipitating proteins were separated by SDS-PAGE and visualized by Coomassie staining. MBP acted as a negative control. (F) Wild-type and E1137R GST–CNOT11088–1312 proteins were immobilized on Glutathione–Sepharose and incubated in RNase A-treated HeLa cell lysate. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and probed using the indicated antibodies. For D and F, amylose or Glutathione–Sepharose resins were used as negative controls, respectively.
To investigate whether these amino acids contribute to the DDX6–CNOT1 interaction, we mutated Arg386 in DDX6 and Glu1137 in CNOT1 in order to reverse their charges (i.e., Arg386Glu and Glu1137Arg), and performed in vitro pulldown experiments. Indeed, the interaction between MBP–DDX6 and GST–CNOT11088–1312 was disrupted by substituting Arg386 in DDX6 with glutamic acid (Fig. 4B), while mutating Glu1137 to arginine in GST–CNOT11088–1312 impaired binding to wild-type MBP-DDX6 (Fig. 4C). Moreover, while wild-type recombinant MBP–DDX6 pulled down CCR4–NOT components when incubated in an RNase A-treated HeLa cell lysate, a recombinant MBP–DDX6 protein harboring an Arg386Glu mutation could not (Fig. 4D). In stark contrast to the defect in CNOT subunit association with MBP–DDX6–Arg386Glu, the association of the mutant protein with Pat1b and EDC3, factors that directly interact with the DDX6 C-terminal lobe (Tritschler et al. 2009; Sharif et al. 2013), were not impaired (Fig. 4D,E). Taken together, these data suggest that the Arg386Glu mutation did not compromise general DDX6 folding, but rather specifically impaired the DDX6–CNOT1 interaction. GST-pulldown experiments from HeLa cell lysates using recombinant GST–CNOT11088–1312 as bait demonstrated that mutating Glu1137Arg in CNOT1 impaired its interaction with DDX6 without affecting its interaction with a CCR4–NOT component, CNOT6L (Fig. 4F). Collectively, these results demonstrate that DDX6 and CNOT1 directly associate via a conserved binding mode with eIF4A and eIF4G, and identify point mutations that disrupt the interaction between DDX6 and CNOT1.
The DDX6–CNOT1 interaction is required for optimal miRNA-mediated gene silencing in human cells
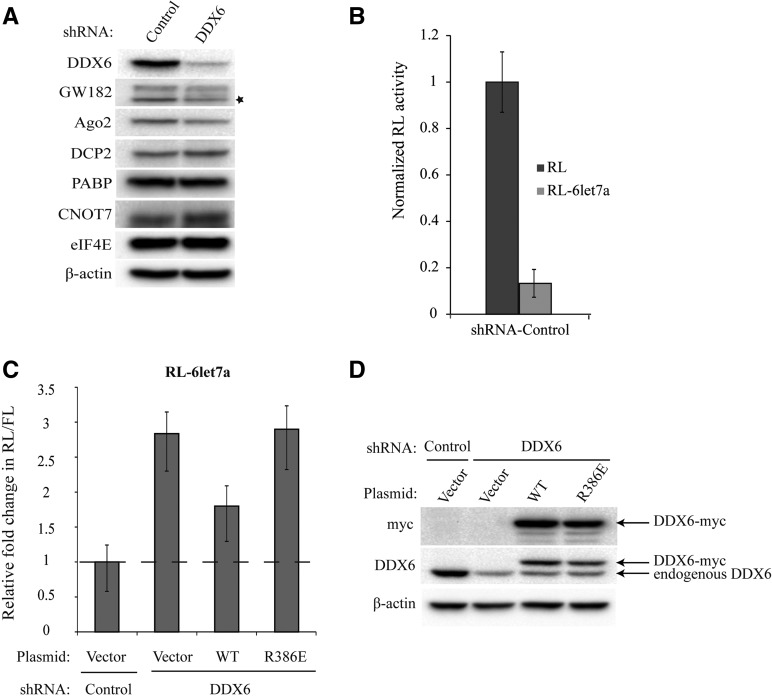
To investigate the importance of the DDX6–CNOT1 interaction in miRNA-mediated gene silencing, we performed complementation assays using HeLa cells from which DDX6 was stably depleted via shRNA. Knocking down DDX6 levels did not appreciably codeplete other miRISC factors (i.e., Ago and GW182), or proteins involved in translational regulation or mRNA decay (Fig. 5A). DDX6-depleted cells were transiently cotransfected with a Renilla luciferase construct either lacking (RL), or containing six bulged let7a miRNA binding sites in its 3′ UTR (RL-6let7a), together with a firefly luciferase construct (FL) as a transfection control. Normalized RL activity was markedly reduced by the presence of let7a-binding sites, with ∼90% repression in HeLa cells expressing a control shRNA against GFP (Fig. 5B). In agreement with previous observations (Chu and Rana 2006; Eulalio et al. 2007; Beilharz et al. 2009; Su et al. 2011), depletion of DDX6 resulted in a 2.8-fold derepression of let7a-mediated silencing (Fig. 5C). Restoring DDX6 levels in DDX6-depleted cells, via transient transfection of a plasmid coding for myc-tagged DDX6 that is refractory to shRNA, partially rescued the defect in miRNA-mediated gene silencing observed in DDX6 knock-down cells (1.8-fold vs. 2.8-fold derepression). However, expression of a myc-tagged DDX6 mutant (Arg386Glu) that cannot bind CNOT1 failed to rescue miRNA silencing (Fig. 5C,D).
FIGURE 5.
Impairment of the DDX6–CNOT1 interaction diminishes gene silencing by the let7a miRNA in HeLa cells stably depleted of DDX6. (A) Stable depletion of DDX6 from HeLa cells is specific. Lysates from HeLa cells expressing shRNAs against GFP (Control) or DDX6 were separated by SDS-PAGE and analyzed by immunoblotting using antibodies against the indicated proteins. The asterisk indicates nonspecific band. (B) HeLa cells expressing a control shRNA were cotransfected with RL or RL-6let7a plasmids, along with an FL construct to account for variations in transfection efficiency. Values represent relative normalized Renilla luciferase activity (Renilla/Firefly), with normalized RL readings set equal to 1. Error bars indicate ±SEM (n = 3). (C) HeLa cells expressing control or DDX6 shRNAs were transfected with empty vector (Vector), WT DDX6-myc, or R386E DDX6-myc. Two days later, cells were cotransfected with RL-6let7a or RL, along with FL. Values represent fold changes in normalized (Renilla/Firefly) RLUs. Silencing of the RL-6let7a reporter in control cells (∼90% repression relative to normalized RL) was set as a fold change of 1. Error bars indicate ±SEM (n = 3). (D) WT and R386E DDX6-myc protein expression levels were similar. Lysates from C were analyzed by immunoblotting with the indicated antibodies.
The interaction of DDX6 with CNOT1 contributes to gene silencing engendered by the GW182 silencing domain
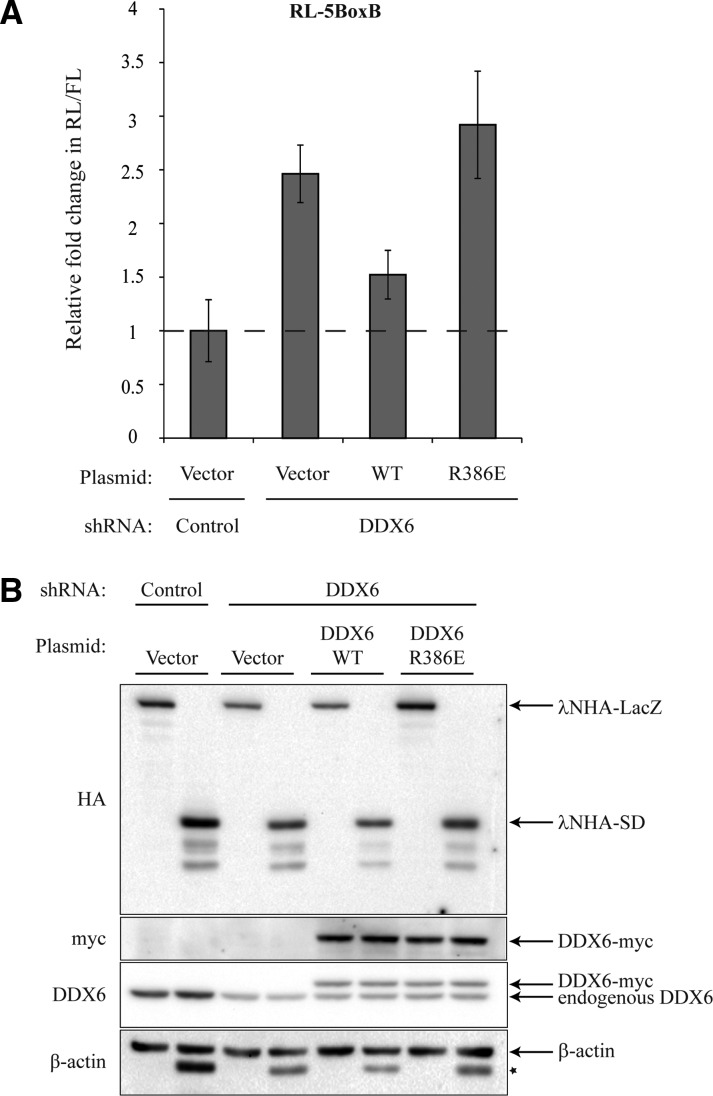
We next sought to dissect the role of the DDX6–CNOT1 interaction in miRNA silencing by using a tethered function approach. GW182 is recruited to miRNA targets via Ago proteins, and the GW182 C-terminal “silencing” domain (SD) is necessary for miRNA-mediated repression, and sufficient to inhibit gene expression when tethered to reporter mRNAs in human and Drosophila S2 cells (Chekulaeva et al. 2009; Eulalio et al. 2009; Zekri et al. 2009; Zipprich et al. 2009; Huntzinger et al. 2010; Jinek et al. 2010; Fabian et al. 2011). Direct tethering of the GW182 SD using the λN-BoxB system accelerates target mRNA deadenylation and decay, and induces translational repression (Fabian et al. 2011; Fukaya and Tomari 2012; Moretti et al. 2012). Expression of human λNHA-SD (residues 1382–1690 of the human GW182 paralog, TNRC6C) in HeLa cells expressing a control shRNA results in ∼90% repression of Renilla luciferase activity from a construct containing five BoxB elements, relative to tethering of β-galactosidase (λNHA-LacZ) (Chekulaeva et al. 2011). Depletion of DDX6 impaired λNHA-SD-mediated silencing by ∼2.5-fold, while expression of wild-type DDX6-myc partially restored reporter activity (Fig. 6A). Consistent with the effects on the RL-6let7a reporter, DDX6 knock-down cells expressing DDX6–Arg386Glu-myc remained incapable of maximal GW182-mediated repression (Fig. 6A). λNHA-LacZ and λNHA-SD, as well as DDX6-myc proteins, were expressed to comparable levels (Fig. 6B). Because GW182 recruits the CCR4–NOT complex via the CNOT1 subunit, our results demonstrate that DDX6 function, at least in part, lies downstream from GW182 in the miRNA silencing pathway.
FIGURE 6.
DDX6 acts downstream from GW182 in the miRNA silencing pathway. (A) HeLa cells expressing control or DDX6 shRNAs were transfected with empty vector (Vector), WT DDX6-myc, or R386E DDX6-myc. Two days later, cells were cotransfected with RL-5BoxB, FL, and either λNHA-LacZ or λNHA-Silencing Domain (SD). Values represent fold changes in normalized (Renilla/Firefly) RLUs. Silencing of the RL-5BoxB reporter in control cells expressing λNHA-SD (∼90% repression relative to cells expressing λNHA-LacZ) was set as a fold change of 1. Error bars indicate ±SEM (n = 3). (B) Lysates from A were analyzed by immunoblotting with the indicated antibodies. Asterisk indicates residual signal from HA blot.
DISCUSSION
While this manuscript was under review, two reports describing co-crystal structures of human DDX6 bound to the human CNOT1 mIF4G domain were published (Chen et al. 2014; Mathys et al. 2014), which demonstrate that the DDX6–CNOT1 complex is important for effecting silencing by miRNAs. Although we suggested a putative salt bridge between Arg386 in DDX6 and Glu1137 in CNOT1 based on structural homology with the eIF4A–eIF4G co-crystal structure, the latter publications have found that, while these residues are important for DDX6 binding to CNOT1, they do not form a salt bridge. Instead, Arg386 from DDX6 forms contacts with a conserved patch of residues in CNOT1 including Phe1098, Leu1101, and Phe1140, while Glu1137 in CNOT1 hydrogen bonds with Gln333 from DDX6. Our findings are consistent with those of Chen et al. (2014) and Mathys et al. (2014), in that mutation of Arg386 impairs gene silencing by miRNAs.
We provide evidence for a direct interaction between the DEAD-box protein, DDX6, and the scaffolding subunit of the CCR4–NOT deadenylase complex, CNOT1. Moreover, we demonstrate that DDX6 interacts with the mIF4G domain present within CNOT1, in a manner that remarkably resembles the binding of the translation factor eIF4A with eIF4G. Our data also indicate that the DDX6–CNOT1 interaction is the only means by which CCR4–NOT can recruit DDX6 to miRNA-targeted mRNAs, as a DDX6 mutant that cannot bind to CNOT1 is unable to associate with CNOT subunits in cell lysates (Fig. 4B,D). This DDX6 mutant can still interact with other decapping factors, such as Pat1b and EDC3, indicating that this mutation does not result in global misfolding of DDX6 (Fig. 4D,E). The association of recombinant DDX6 with endogenous CNOT1 is weak (<1% input) (Figs. 1B, 4D), potentially due to interference of complex formation by the MBP tag fused to DDX6. It is also possible that the decapping machinery associates with the CCR4–NOT deadenylase complex in a transient manner, and may only occur in the presence of a deadenylated mRNA that is destined for decapping. Thus, DDX6 and CNOT1 may not form a stable complex at all times in vivo. In addition, DDX6 protein copy numbers in mammalian cells are ∼10-fold greater than CNOT1 (Schwanhausser et al. 2011), which could also explain the small fraction of endogenous CNOT1 that was coprecipitated with MBP–DDX6.
DDX6 is an important effector of miRNA silencing in human cells. In accordance with published observations (Chu and Rana 2006; Eulalio et al. 2007; Beilharz et al. 2009; Su et al. 2011), depleting DDX6 from HeLa cells using an shRNA strategy impaired mRNA silencing conferred by either the let-7a miRNA or by the GW182 silencing domain (Figs. 5, 6). Importantly, DDX6 binding to CNOT1 is critical for maintaining optimal miRISC silencing in human cells. Rescuing DDX6 protein levels in DDX6-depleted HeLa cells via expression of a DDX6 point mutant (Arg386Glu) that cannot bind CNOT1 failed to rescue miRISC-dependent mRNA silencing (Figs. 5, 6). At steady-state, an increase in mRNA levels of the Renilla luciferase reporters when DDX6 was knocked-down accounted for the derepression of miRISC-dependent silencing that we observed (Figs. 5, 6; data not shown). This is in agreement with the function of DDX6 in decapping activation and mRNA decay (Coller et al. 2001; Fischer and Weis 2002; Fenger-Gron et al. 2005; Eulalio et al. 2007). However, due to the established ability of DDX6 to act as a translational repressor (Coller and Parker 2005; Carroll et al. 2011), as well as the observation that miRNA targets are subject to translational inhibition prior to mRNA destabilization (Mathonnet et al. 2007; Fabian et al. 2009; Bazzini et al. 2012; Bethune et al. 2012; Djuranovic et al. 2012), it is likely that DDX6 contributes to initial effects on mRNA translation by miRNAs. Recent work using DDX6 knockdown cells expressing miRNA-targeted luciferase mRNAs in the presence of catalytically inactive deadenylase subunits supports this notion (Mathys et al. 2014).
We present biochemical evidence to support a direct interaction between DDX6 and CNOT1 that is similar to that of the eIF4A–eIF4G co-crystal structure (Schutz et al. 2008). Notably, several reports have described DEAD-box proteins that directly bind to mIF4G domain-containing factors, all through a shared architecture (Schutz et al. 2008; Montpetit et al. 2011; Buchwald et al. 2013). Biophysical studies of these complexes have ascribed modulatory roles for mIF4G domains in regulating RNA-binding and release, ATPase activity, and helicase function of their respective DEAD-box protein partners (Weirich et al. 2006; Schutz et al. 2008; Montpetit et al. 2011; Buchwald et al. 2013). Thus far, DDX6 has been characterized as a weak ATPase with a high affinity for RNA (Dutta et al. 2011; Ernoult-Lange et al. 2012), and yeast Dhh1p does not detectably unwind RNA secondary structures in vitro (Dutta et al. 2011). It is possible that the CNOT1 mIF4G domain is capable of stimulating a latent helicase activity within DDX6. This may be brought about by structural rearrangements within DDX6 that improve ATPase rates and RNA release, as the mIF4G-containing proteins eIF4G and Gle1 do for their respective helicase binding partners, eIF4A and Dbp5 (Oberer et al. 2005; Weirich et al. 2006; Schutz et al. 2008; Montpetit et al. 2011). Indeed, the CNOT1 interaction has recently been shown to enhance the ATPase activity of DDX6, further supporting the notion that CNOT1 may affect the ability of DDX6 to bind and/or unwind RNA (Mathys et al. 2014).
DDX6 associates with multiple decapping factors in eukaryotes, and stimulates mRNA decapping (Coller et al. 2001; Fischer and Weis 2002; Coller and Parker 2005; Fenger-Gron et al. 2005; Carroll et al. 2011). In this report, we establish CNOT1, and by extension the CCR4–NOT deadenylase complex, as a direct binding partner of DDX6. In vivo work in yeast and mammalian cells has reported functional coupling between deadenylation and decapping, with the former typically preceding cap hydrolysis and 5′–3′ decay of the mRNA (Muhlrad et al. 1994; Yamashita et al. 2005). However, evidence defining the molecular factors that physically link these two processes has been lacking. Dhh1p and metazoan Pat1 proteins have been likely candidates due to their abilities to enhance decapping and associate with CCR4–NOT (Coller et al. 2001; Fischer and Weis 2002; Haas et al. 2010; Nissan et al. 2010; Ozgur et al. 2010). Pat1 directly binds to DDX6 (Sharif et al. 2013), suggesting that it may associate indirectly with CCR4–NOT via DDX6. Interestingly, Pat1 and the decapping enhancer EDC3 both compete for the same binding site on DDX6 (Sharif et al. 2013). Based on these observations we posit that, upon completion of miRNA-mediated deadenylation by the CCR4–NOT complex, direct recruitment of DDX6, and perhaps Pat1b, assists in dissociation of the cap-binding complex. Subsequent rearrangements of the repressive mRNP would then allow for recruitment of decapping factors associated with DDX6 and Pat1b to commit the mRNA for degradation.
We and others have identified several CNOT1-interacting proteins (e.g., Tristetraprolin, GW182, and Roquin) that bring about silencing of distinct classes of mRNAs, including those containing AU-rich elements (ARE) and miRNA-binding sites (Braun et al. 2011; Chekulaeva et al. 2011; Fabian et al. 2011, 2013; Leppek et al. 2013). Collectively, these data establish CNOT1 as a versatile molecular scaffold that coordinates gene silencing by interacting with various ribonucleoprotein platforms. Recruitment of DDX6 by CCR4–NOT provides another mechanism by which CCR4–NOT post-transcriptionally regulates gene expression. Indeed, efficient repression of ARE-containing mRNAs, mRNAs containing Roquin-binding sites, and miRNA-targeted mRNAs requires DDX6 (Fenger-Gron et al. 2005; Chu and Rana 2006; Glasmacher et al. 2010). Conservation of the CNOT1 mIF4G domain in Saccharomyces cerevisiae, which is present within the N-terminal fragment of Not1p that was previously shown to immunoprecipitate Dhh1p (Maillet and Collart 2002), suggests that at least certain post-transcriptional control pathways in yeast likely use a DDX6–CNOT1 complex. Notably, tethering of a deadenylase-dead mutant of CCR4–NOT to reporter mRNAs in Xenopus laevis oocytes and human cells brings about silencing in a deadenylation-independent manner (Cooke et al. 2010; Chekulaeva et al. 2011). Moreover, expression of a CNOT1 fragment containing the mIF4G domain that directly binds DDX6 derepressed let-7-mediated inhibition of a reporter mRNA in HeLa cells (Huntzinger et al. 2013). Thus, our results are consistent with the notion that DDX6 acts as an effector protein that contributes to the inhibitory potential of the CCR4–NOT complex on eukaryotic gene expression. Whether DDX6 brings about silencing directly, or whether its role is to recruit other effector molecules to CCR4–NOT remains to be established. Future work dissecting the involvement of DDX6 in CCR4–NOT-mediated gene silencing should provide important mechanistic insights into several paradigms of post-transcriptional control.
MATERIALS AND METHODS
DNA constructs
Full-length (residues 1–483), N-terminal (residues 1–302), and C-terminal (residues 303–483) human DDX6 (AAH65007) were PCR amplified and subcloned into the NcoI and NotI sites of pMAL-c5x with six C-terminal histidines. For mammalian expression, full-length DDX6 was subcloned into the pcDNA4.1 mycHis-B vector using the BamHI and XhoI sites. GST–CNOT1 fragments were described previously (Fabian et al. 2013). Numbering of CNOT1 residues correspond to the sequence of isoform C (NP_001252541). CNOT1 (residues 1088–1312) was subcloned into pGEX6p1 using the SalI and NotI restriction sites and contained an additional six C-terminal histidine residues. RL-5BoxB was described (Pillai et al. 2004). λNHA-LacZ and λNHA-GW182 (TNRC6C, residues 1382–1690) were described (Chekulaeva et al. 2011).
Protein expression and purification
Proteins were expressed using Rosetta-2(DE3) Escherichia coli cells (EMD Biosciences) and purified using Ni-NTA resin (Qiagen). Bacterial cultures were induced using 0.5 mM IPTG for 4–5 h at 30°C, and cell pellets were resuspended in Buffer A (50 mM HEPES-KOH at pH 7.8, 5% glycerol, 500 mM NaCl, 2 mM β-mercaptoethanol). Cells were lysed by sonication, cleared by centrifugation at 4°C (35 min at 35,000g) and incubated with Ni-NTA resin for 30 min. The resin was washed three times with Buffer A containing 30 mM imidazole, and proteins were eluted in Buffer A supplemented with 500 mM imidazole.
GST and MBP pulldown experiments
A total of 42 pmol of MBP, MBP-tagged DDX6 proteins, or GST-tagged CNOT1 fragments were incubated in Binding Buffer (25 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.1% NP40, 2 mM MgCl2) for 2 h at 4°C with 20 μL of washed Amylose resin (New England Biolabs) or Glutathione Sepharose-4B (GE Life Sciences). Beads were then washed with 1 mL of Binding Buffer five times, resuspended in 35 μL Laemmli sample buffer, and boiled for 10 min. Proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. For coprecipitation experiments from cell lysates, HEK293T or HeLa cell pellets were resuspended in Lysis Buffer (25 mM HEPES-KOH at pH 7.3, 200 mM KCl, 2 mM MgCl2, 75 mM KOAc, 0.3% NP40) supplemented with 1X EDTA-free protease inhibitor cocktail (Roche), and cleared by centrifugation at 20,000g for 20 min at 4°C. 2 mg of cell lysate were pre-cleared with 20 μL of washed resin for 1 h at 4°C, and incubated overnight with 100 pmol of recombinant MBP- or GST-tagged proteins, 10 μg of RNase A, and 20 μL of washed resin. Proteins were separated by SDS-PAGE and visualized by Western blotting.
Cell culture and lentiviral transduction
Cells were incubated at 37°C at 5% CO2, in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 100 units/mL penicillin/streptomycin (Invitrogen). Lentiviral vectors were obtained from Sigma. The human DDX6 shRNA vector accession number is TRCN0000074694 (Sigma), and the SHC002 shRNA (Sigma) targeting Turbo GFP was used as a control. Cotransfection of shRNA vectors with lentiviral packaging plasmids (PLP1, PLP2, PLP-VSVG, from Invitrogen) was carried out using HEK293T cells. Supernatants were collected 72 h post-transfection, passed through a 0.45-μM nitrocellulose filter, supplemented with 5 μg/mL polybrene, and applied to HeLa cells at ∼20% confluency. Cells were reinfected the following day, and selected with puromycin (1 μg/mL, Sigma) for 5 d.
Transfections and luciferase assays
A total of 200–400 ng of empty vector (pcDNA4.1 mycHis-B) or pcDNA4.1 mycHis-B-DDX6 (WT or Arg386Glu) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Two days after transfection, cells were cotransfected with 20 ng of RL plasmid DNA (RL, RL-6let7a, or RL-5BoxB) and 20 ng FL plasmid. For tethering assays, 250 ng of λNHA-LacZ or λNHA-SD were included in the cotransfection reactions. Medium was changed the following day, and cells were processed for luciferase assays using the Dual Luciferase Reporter Assay System (Promega) following the manufacturer's recommendations.
Antibodies
Antibodies used in this study were as follows: rabbit polyclonal anti-DDX6 (Bethyl Laboratories), rabbit polyclonal anti-Pat1b (Bethyl Laboratories), rabbit polyclonal anti-DCP2 (Bethyl Laboratories), mouse monoclonal anti-EDC3 (Abcam), rabbit polyclonal anti-GW182 (Bethyl Laboratories), mouse monoclonal anti-eIF4E (BD Transduction Laboratories), rabbit polyclonal anti-PABP (Abcam), rabbit monoclonal anti-Ago2 (Cell Signaling), mouse monoclonal anti-myc (BioShop Canada Inc.), mouse monoclonal anti-β-actin (Sigma). Rabbit polyclonal antibodies against CNOT1 and CNOT7 were kindly provided by T. Yamamoto.
ACKNOWLEDGMENTS
We thank S. Perreault and M. Vipparti for assistance, and Zoya Padamsi for technical help. Research was funded by Canadian Institutes of Health Research grants to N.S. (MOP-93607) and M.R.F. (MOP-130425). M.M. is supported by a Canadian Diabetes Association Postdoctoral fellowship. N. Siddiqui was supported by a fellowship from the Cole Foundation. C.R. is a recipient of an internal fellowship from the McGill Department of Biochemistry and a PhD award from the Groupe de Recherche Axe sur la Structure des Proteines (GRASP).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045302.114.
REFERENCES
- Basquin J, Roudko VV, Rode M, Basquin C, Seraphin B, Conti E 2012. Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol Cell 48: 207–218 [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Humphreys DT, Clancy JL, Thermann R, Martin DI, Hentze MW, Preiss T 2009. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS One 4: e6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune J, Artus-Revel CG, Filipowicz W 2012. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep 13: 716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Buchwald G, Schussler S, Basquin C, Le Hir H, Conti E 2013. Crystal structure of the human eIF4AIII-CWC22 complex shows how a DEAD-box protein is inhibited by a MIF4G domain. Proc Natl Acad Sci 110: E4611–E4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Munchel SE, Weis K 2011. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol 194: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W, Parker R 2009. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA 15: 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W 2011. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Zheng D, Xia Z, Shyu AB 2009. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol 16: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E 2014. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 54: 737–750 [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TM 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A, Prigge A, Wickens M 2010. Translational repression by deadenylases. J Biol Chem 285: 28506–28513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Kislig E, Altmann M, Trachsel H 2001. Structural and functional similarities between the central eukaryotic initiation factor (eIF)4A-binding domain of mammalian eIF4G and the eIF4A-binding domain of yeast eIF4G. Biochem J 355: 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Zheng S, Jain D, Cameron CE, Reese JC 2011. Intermolecular interactions within the abundant DEAD-box protein Dhh1 regulate its activity in vivo. J Biol Chem 286: 27454–27470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernoult-Lange M, Baconnais S, Harper M, Minshall N, Souquere S, Boudier T, Benard M, Andrey P, Pierron G, Kress M, et al. 2012. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA 18: 1702–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E 2007. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 21: 2558–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E 2009. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA 15: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N 2012. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19: 586–593 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. 2011. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 18: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N 2013. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol 20: 735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Fischer N, Weis K 2002. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J 21: 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Tomari Y 2012. MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol Cell 48: 825–836 [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Hoefig KP, Vogel KU, Rath N, Du L, Wolf C, Kremmer E, Wang X, Heissmeyer V 2010. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol 11: 725–733 [DOI] [PubMed] [Google Scholar]
- Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E 2010. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol 189: 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A 1998. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T 2005. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci 102: 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J 29: 4146–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kuzuoglu-Ozturk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E 2013. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res 41: 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Sonenberg N 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol 17: 6940–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y 2009. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell 34: 58–67 [DOI] [PubMed] [Google Scholar]
- Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA 2010. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol 17: 238–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G 2013. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 153: 869–881 [DOI] [PubMed] [Google Scholar]
- Lyskov S, Gray JJ 2008. The RosettaDock server for local protein–protein docking. Nucleic Acids Res 36: W233–W238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Collart MA 2002. Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem 277: 2835–2842 [DOI] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. 2007. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317: 1764–1767 [DOI] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W 2014. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in MicroRNA repression. Mol Cell 54: 751–765 [DOI] [PubMed] [Google Scholar]
- Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M 2013. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340: 82–85 [DOI] [PubMed] [Google Scholar]
- Minshall N, Standart N 2004. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res 32: 1325–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K 2011. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472: 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW 2012. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol 19: 603–608 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes Dev 8: 855–866 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S 2001. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128: 3233–3242 [DOI] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, Parker R 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell 39: 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberer M, Marintchev A, Wagner G 2005. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev 19: 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Stoecklin G 2013. Role of Rck-Pat1b binding in assembly of processing-bodies. RNA Biol 10: 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Chekulaeva M, Stoecklin G 2010. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol 30: 4308–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit AP, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O 2012. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res 40: 11058–11072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W 2004. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309: 1573–1576 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11: 1640–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci EP, Limousin T, Soto-Rifo R, Rubilar PS, Decimo D, Ohlmann T 2013. miRNA repression of translation in vitro takes place during 43S ribosomal scanning. Nucleic Acids Res 41: 586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U 2008. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein–protein interactions. Proc Natl Acad Sci 105: 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sharif H, Ozgur S, Sharma K, Basquin C, Urlaub H, Conti E 2013. Structural analysis of the yeast Dhh1-Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic Acids Res 41: 8377–8390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Meng S, Lu Y, Trombly MI, Chen J, Lin C, Turk A, Wang X 2011. Mammalian hyperplastic discs homolog EDD regulates miRNA-mediated gene silencing. Mol Cell 43: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian AR, Kaufmann M, Morgenstern B 2008. DIALIGN-TX: greedy and progressive approaches for segment-based multiple sequence alignment. Algorithms Mol Biol 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Lai EC 2013. Adult-specific functions of animal microRNAs. Nat Rev Genet 14: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet T, Kovalak C, Coller J 2012. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol 10: e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O 2009. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell 33: 661–668 [DOI] [PubMed] [Google Scholar]
- Walters RW, Bradrick SS, Gromeier M 2010. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA 16: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol 8: 668–676 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darzynkiewicz E, Hentze MW 2009. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell 35: 881–888 [DOI] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstadt S, Izaurralde E 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol 29: 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W 2009. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA 15: 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]