There has been incremental evidence of miRNA binding sites not only being restricted to the 3′ UTR of mRNAs, but being consistently present in both coding regions and fewer examples in 5′ UTR regions. In this manuscript, the authors use a predictive algorithm to identify two putative miR-103a-3p in the 5′ UTR of the GPRC5A mRNA. The target mRNA is of interest in cancer progression, acting as either a tumor suppressor or an oncogene in different cancers. They clearly demonstrate that ectopic expression of this miRNA leads to a decrease in the levels of GPRC5A mRNA and protein in pancreatic cells. Interestingly, the authors show that miR-103a-3p targeting of the 5′ UTR but not the CDS of GPRC5A results in decreased levels of the corresponding protein.
Keywords: microRNAs, miRNAs, 5′ UTR targeting, GPRC5A, miR-103a
Abstract
MicroRNAs (miRNAs) are short noncoding RNAs that regulate the expression of their targets in a sequence-dependent manner. For protein-coding transcripts, miRNAs regulate expression levels through binding sites in either the 3′ untranslated region (3′ UTR) or the amino acid coding sequence (CDS) of the targeted messenger RNA (mRNA). Currently, for the 5′ untranslated region (5′ UTR) of mRNAs, very few naturally occurring examples exist whereby the targeting miRNA down-regulates the expression of the corresponding mRNA in a seed-dependent manner. Here we describe and characterize two miR-103a-3p target sites in the 5′ UTR of GPRC5A, a gene that acts as a tumor suppressor in some cancer contexts and as an ongocene in other cancer contexts. In particular, we show that the interaction of miR-103a-3p with each of these two 5′ UTR targets reduces the expression levels of both GPRC5A mRNA and GPRC5A protein in one normal epithelial and two pancreatic cancer cell lines. By ectopically expressing “sponges” that contain instances of the wild-type 5′ UTR targets we also show that we can reduce miR-103a-3p levels and increase GPRC5A mRNA and protein levels. These findings provide some first knowledge on the post-transcriptional regulation of this tumor suppressor/oncogene and present additional evidence for the participation of 5′ UTRs in miRNA driven post-transcriptional regulatory control.
INTRODUCTION
MiRNAs comprise a group of short noncoding RNAs that post-transcriptionally regulate gene expression in multicellular organisms in a sequence-dependent manner (Bartel 2004). The “seed” region of a miRNA, defined as the sequence spanning bases 2 through 7 inclusive from the 5′ end of the miRNA, determines a miRNA's spectrum of targets (Miranda et al. 2006; Bartel 2009; Rigoutsos and Tsirigos 2010; Xia et al. 2012). So far, more than 17,000 mature miRNA sequences from 140 different species have been identified (Kozomara and Griffiths-Jones 2011). With regard to target cardinality, a single miRNA can simultaneously target multiple mRNAs, thusly decreasing, to varying degrees, the abundance of the corresponding protein (Miranda et al. 2006; Baek et al. 2008; Selbach et al. 2008).
MiRNA research began more than 20 years ago (Lee et al. 1993; Wightman et al. 1993; Hamilton and Baulcombe 1999; Reinhart et al. 2000) and efforts since then have revealed that the identification of miRNA targets is an inherently difficult problem (Rigoutsos and Tsirigos 2010). Nonetheless, the field has made great advances during this time and numerous miRNA targets have been described in the literature to date with the majority of these targets being located in the 3′ UTR of the targeted mRNAs (Bartel 2009). In recent years, others and we have shown that miRNAs can also target mRNAs within their CDS and decrease the corresponding protein's abundance (Duursma et al. 2008; Forman et al. 2008; Lal et al. 2008; Shen et al. 2008; Tay et al. 2008; Rigoutsos 2009; Brest et al. 2011; Hao et al. 2011; Nelson et al. 2011; Sauna and Kimchi-Sarfaty 2011; Gartner et al. 2013; Hausser et al. 2013; Radhakrishnan et al. 2013; Shabalina et al. 2013).
In contrast, identifying targets in the 5′ UTR of mRNAs has proven more difficult. In early work, use of artificial constructs containing multiple copies of known miRNA targets showed that from a mechanistic standpoint miRNAs can repress mRNAs through 5′ UTR binding just as efficiently as through 3′ UTR binding (Lytle et al. 2007; Devlin et al. 2010; Moretti et al. 2010). For naturally occurring targets, two subsequent studies reported examples whereby 5′ UTR targeting by the miRNA did not down-regulate the mRNA but instead enhanced protein translation and increased protein levels (Henke et al. 2008; Orom et al. 2008; Tsai et al. 2009; Da Sacco and Masotti 2012). Four subsequent reports described a few examples of 5′ UTR binding sites that led to the down-regulation of the targeted transcript (Jopling et al. 2005; Lee et al. 2009; Grey et al. 2010; Dewing et al. 2012). More recently, a C. elegans study discussed the possibility of a miRNA target in the 5′ UTR of CBP-1's mRNA (Vora et al. 2013).
Below, we report on our validation of two human miR-103a-3p targets in the 5′ UTR of the human GPRC5A gene (ENSG00000013588/ENST00000014914). GPRC5A encodes an orphan G-protein-coupled receptor that was originally reported to be overexpressed in normal lung tissue and underexpressed in lung cancer; since then, GPRC5A's dysregulation has been associated with multiple cancer types: In some cancers, GPRC5A can act as a tumor suppressor whereas in others it can act as an oncogene (Tao et al. 2007; Acquafreda et al. 2009; Cheng et al. 2012). MiR-103a-3p is a notable miRNA in that it is evolutionarily conserved and involved in regulating multiple cellular processes such as cell division, cellular metabolism and stress, angiogenesis, etc. (Finnerty et al. 2010). MiR-103a-3p's dysregulation has been associated with many human diseases including several cancers, Alzheimer's disease, and diabetes (Martello et al. 2010; Yao et al. 2010; Trajkovski et al. 2011).
RESULTS
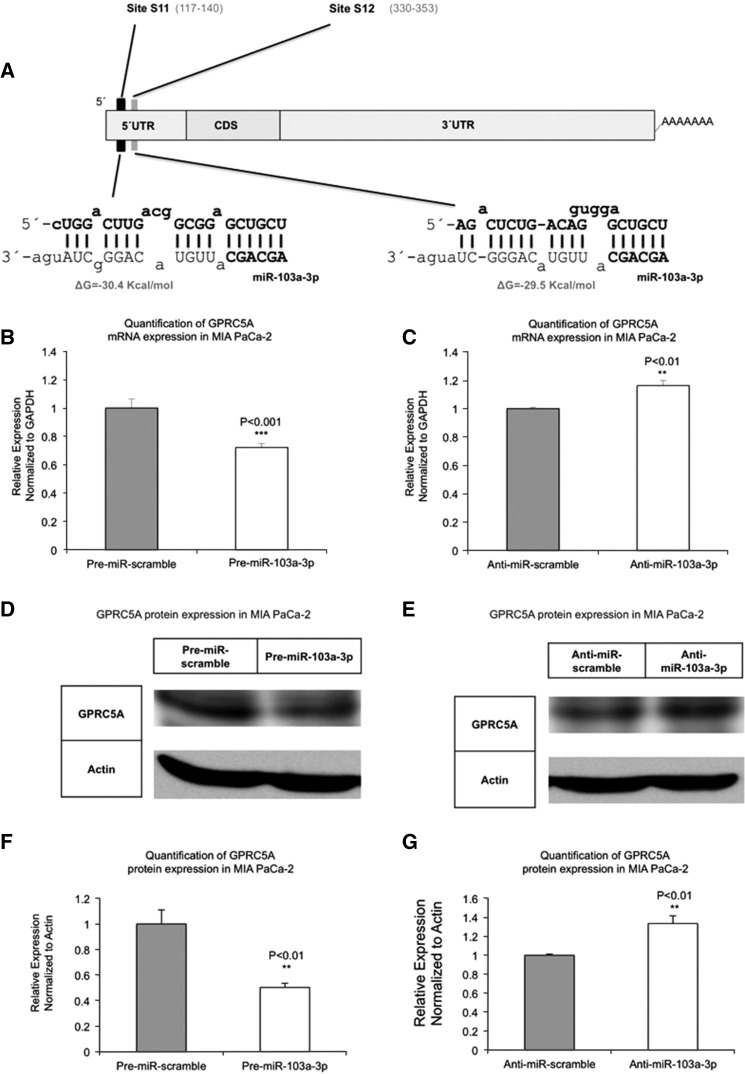
We studied the interactions of miR-103a-3p and GPRC5A, both of which are endogenous to pancreatic cell lines and tissue (both normal and cancer). We focused on two candidate miR-103a-3p targets in the 5′ UTR of GPRC5A. The first putative miR-103a-3p MRE (site S11) is located between nucleotides 117 and 140 inclusive, whereas the second putative MRE (site S12) is located between nucleotides 330 and 353 inclusive (Fig. 1A).
FIGURE 1.
MiR-103a-3p abundance affects both GPRC5A mRNA and protein levels. (A) Analysis using the rna22 algorithm suggests two putative miR-103a-3p binding sites, S11 and S12, respectively, in the 5′ UTR. (B) Ectopic overexpression of miR-103a-3p in MIA PaCa-2 cells reduces GPRC5A mRNA levels. (C) Ectopic overexpression of anti-miR-103a-3p in MIA PaCa-2 cells increases GPRC5A mRNA levels. (D) Ectopic expression of miR-103a-3p reduces GPRC5A protein expression. (E) Overexpression of miR-103a-3p inhibitors increases GPRC5A protein expression. (F) Quantification result of D. (G) Quantification result of E. All numerical data are mean ± SD. (**) P < 0.01; (***) P < 0.001, n = 3. GAPDH, glyceraldehyde-3-phosphate dehydrogenase, GAPDH and Actin are internal controls.
Increase in miR-103a-3p abundance reduces both GPRC5A mRNA and protein levels
We transiently transfected MIA PaCa-2 cells with Pre-miR-103a-3p or Anti-miR-103a-3p at a concentration of 50 nM for 48 h. MIA PaCa-2 Cells transfected with only a scrambled sequence, either Pre-miR-scramble or Anti-miR-scramble, were examined in parallel as controls. Transfection with Pre-miR-103a-3p enhanced the expression of mature miR-103a-3p 900 ± 132-fold (P < 0.001)—see Supplemental Figure 1A—whereas transfection with Anti-miR-103a-3p reduced the expression of mature miR-103a-3p 11.9 ± 2.6-fold (P < 0.001)—see Supplemental Figure 1B. In comparison to Pre-miR-scramble, transfection of MIA PaCa-2 cells with Pre-miR-103a-3p resulted in a 30% (P < 0.001) decrease of GPRC5A mRNA (Fig. 1B). Notably, the decrease in protein levels (50%, P < 0.01) was much higher than the decrease of mRNA levels (Fig. 1D). In addition, transfection of MIA PaCa-2 cells with Anti-miR-103a-3p resulted in up-regulation of GPRC5A mRNA and an increase in GPRC5A protein levels, compared with Anti-miR-scramble treatment group (Fig. 1C,E).
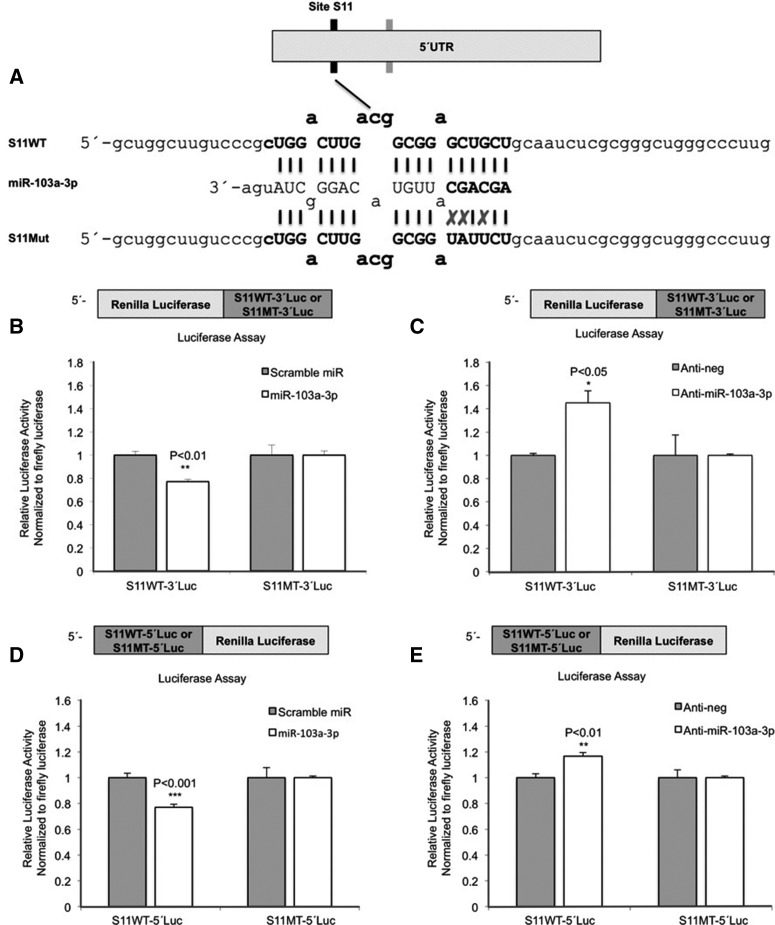
MiR-103a-3p directly interacts with both S11 and S12 in the 5′ UTR of GPRC5A
We constructed separate reporter expression vectors containing the wild-type (WT) or mutant (MT) binding sites, in turn placing them downstream (S11WT-3′Luc, S12WT-3′Luc, S11MT-3′Luc, and S12MT-3′Luc) from and upstream (S11WT-5′Luc, S12WT-5′Luc, S11MT-5′Luc, and S12MT-5′Luc) of the luciferase gene (Fig. 2A; Supplemental Fig. 2A). MIA PaCa-2 cells were cotransfected with Pre-miR-scramble or Pre-miR-103a-3p and a reporter expression vector containing wild-type or mutant binding site, respectively. The S11 site was more responsive to miR-103a-3p treatment than the S12 site: Indeed, pre-miR-103a-3p reduced luciferase activity by 27% ± 6% (P < 0.01) in cells transfected with S11WT-3′Luc and 17% ± 4% (P < 0.05) in cells transfected with S12WT-3′Luc (Fig. 2B; Supplemental Fig. 2B). Repeating the experiments with the 5′ luciferase constructs gave similar results: In cells transfected with S11WT-5′Luc pre-miR-103a-3p reduced luciferase activity by 24% ± 6% (P < 0.01), whereas the reduction was 17% ± 4% (P < 0.05) in cells transfected with S12WT-5′Luc (Fig. 2D; Supplemental Fig. 2D). Introduction of disruptive mutations in each of the two miR-103a-3p sites rescued the inhibitory effect of Pre-miR-103a-3p on luciferase activity and for both the 5′ and the 3′ luciferase constructs (Fig. 2B,D; Supplemental Fig. 2B,D). MIA PaCa-2 cells were also cotransfected with Anti-miR-scramble or Anti-miR-103a-3p and a reporter expression vector containing the WT or MT binding site (separately for the 5′ and 3′ luciferase constructs). Transfection with Anti-miR-103a-3p increased luciferase activity by 45% ± 18% (P < 0.05) and 18% ± 12% (P = 0.06) in cells transfected with S11WT-3′Luc and S12WT-3′Luc, respectively (Fig. 2C; Supplemental Fig. 2C) and by 17% ± 3% (P < 0.01) and 30% ± 6% (P < 0.001) in cells transfected with S11WT-5′Luc and S12WT-5′Luc, respectively (Fig. 2E; Supplemental Fig. 2E). Notably, for each of the two sites S11 and S12, the observed increase in luciferase activity in the presence of anti-miR-103a-3p was concordant with the decrease of luciferase activity in the presence of miR-103a-3p; i.e., the S11 site was more responsive to miR-103a-3p/anti-miR-103a-3p than the S12 site. Lastly, mutations in the two miR-103a-3p sites impaired the induction effect of Anti-miR-103a-3p on luciferase activity (Fig. 2C; Supplemental Fig. 2C).
FIGURE 2.
MiR-103a-3p directly targets two sites in the 5′ UTR of GPRC5A. (A) The scheme indicates the sequences of the predicted miR-103a-3p binding sites (S11) within the 5′ UTR of GPRC5A and the sequences of S11 wild-type (WT, top) and mutant (MT, bottom) used in this study. (B) Luciferase activity in MIA PaCa-2 cells upon transfection of indicated reporter constructs and pre-miR-103a-3p were compared with cells transfected with indicated reporter constructs and pre-miR-scramble. (C) Luciferase activity in MIA PaCa-2 cells upon transfection of indicated reporter constructs and miR-103a-3p inhibitors was compared with cells transfected with indicated reporter constructs and Anti-miR-scramble. (D) Luciferase activity in MIA PaCa-2 cells upon transfection of indicated reporter constructs and pre-miR-103a-3p were compared with cells transfected with the indicated reporter constructs and pre-miR-scramble. (E) Luciferase activity in MIA PaCa-2 cells upon transfection of the indicated reporter constructs and miR-103a-3p inhibitors was compared with cells transfected with the indicated reporter constructs and Anti-miR-scramble. All numerical data are mean ± SD. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; n = 3. S11WT, psiCHECK-2 vector containing miR-103a-3p binding site 1; S11MT, psiCHECK-2 vector containing mutant miR-103a-3p binding site 1.
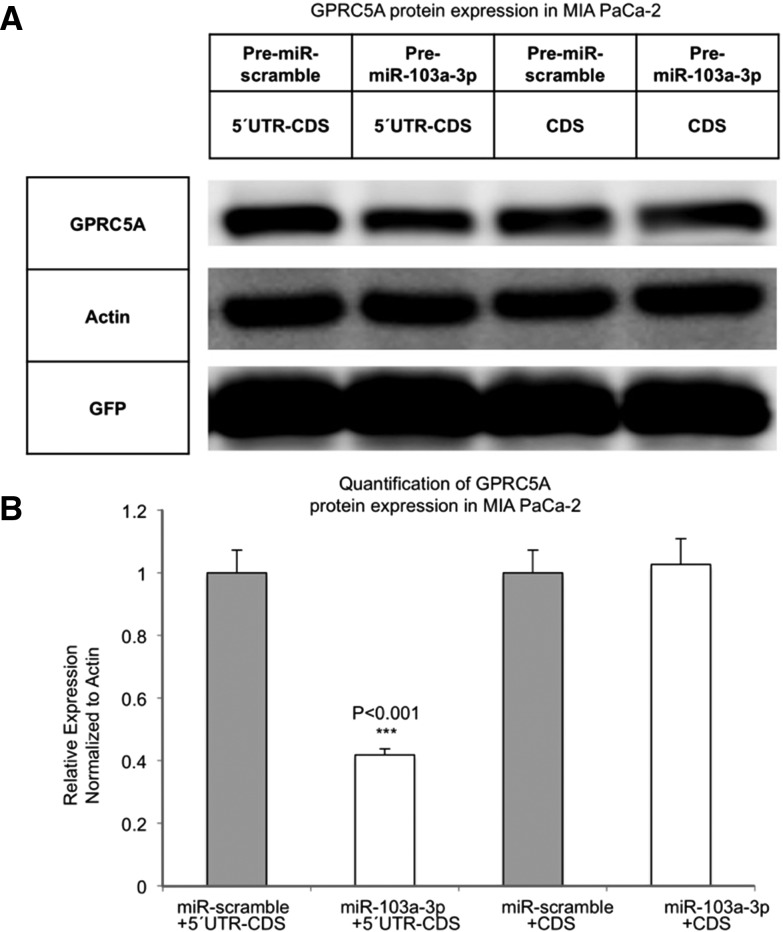
MiR-103a-3p targeting of the 5′ UTR of GPRC5A mRNA decreases GPRC5A protein levels
We next sought to determine whether the interaction of miR-103a-3p with the 5′ UTR of GPRC5A affected GPRC5A protein levels. To this end, we constructed two expression vectors. The first vector, labeled GPRC5A-5′UTR-CDS, contained GPRC5A's wild-type 5′ UTR and CDS regions only (Supplemental Fig. 3A,B); i.e., the vector lacked GPRC5A's 3′ UTR. The second vector, labeled GPRC5A-CDS, lacked both untranslated regions and comprised only GPRC5A's wild-type CDS region (Supplemental Fig. 3C). We cotransfected MIA PaCa-2 cells with a control vector containing GFP open frame region (ORF), Pre-miR-scramble or Pre-miR-103a-3p, and either the GPRC5A-5′UTR-CDS or the GPRC5A-CDS expression vector. Pre-miR-103a-3p reduced GPRC5A expression in cells transfected with the GPRC5A-5′UTR-CDS vector but did not have an inhibitory effect on the GPRC5A-CDS vector (Fig. 3A). This demonstrated that miR-103a-3p down-regulates GPRC5A protein by binding to the 5′ UTR of GPRC5A's mRNA.
FIGURE 3.
MiR-103a-3p targeting of the 5′ UTR of GPRC5A mRNA decreases GPRC5A protein level. (A) GPRC5A protein expression level was determined by Western blots in MIA PaCa-2 cells that were cotransfected with a control vector containing GFP open frame region (OFR), Pre-miR-scramble or Pre-miR-103a-3p, and either the GPRC5A-5′UTR-CDS or the GPRC5A-CDS expression vector. (B) Quantification result of A. (GFP) Green fluorescent protein is transfection control. Actin is internal control. 5′UTR-CDS, pcDNA vector containing GPRC5A 5′ UTR and CDS region; CDS, pcDNA vector containing GPRC5A CDS region. All numerical data are mean ± SD. (***) P < 0.001, n = 3.
Overexpression of the 5′ UTR MRE can increase GPRC5A mRNA and protein levels
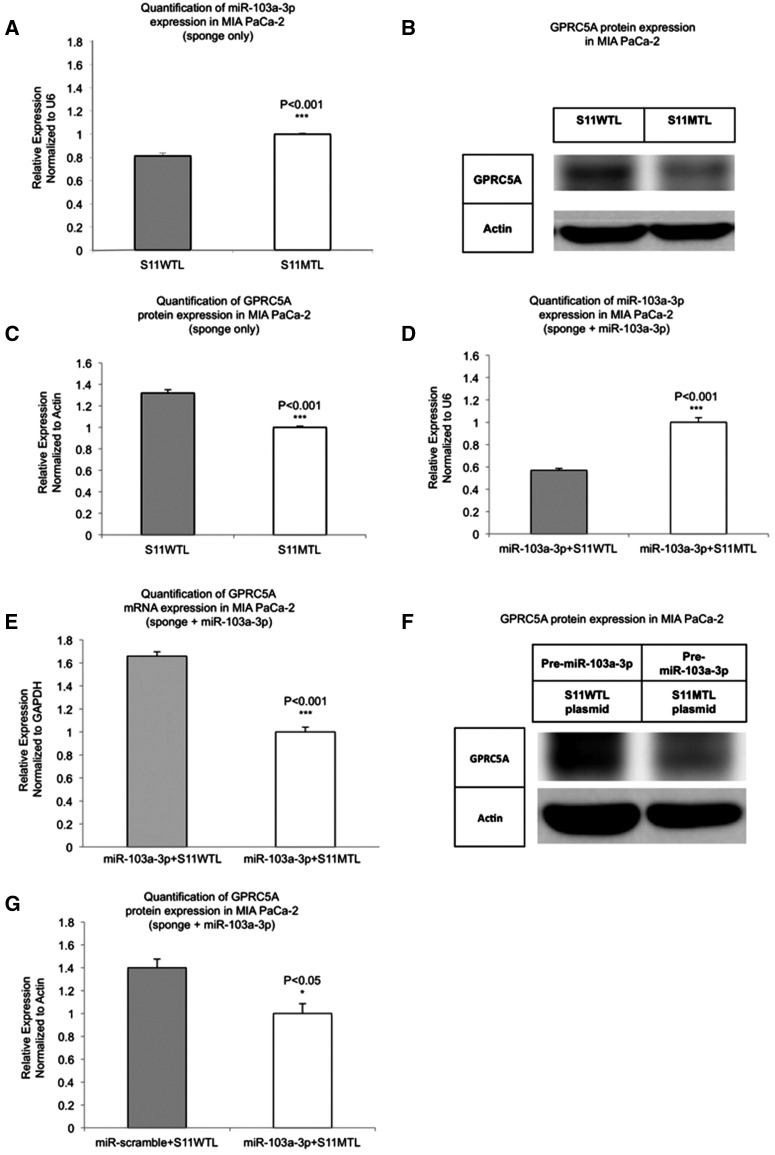
To further corroborate the targeting of GPRC5A's 5′ UTR by miR-103a-3p we made use of the concept of “sponging” or “decoying” (Ebert and Sharp 2010a,b; Poliseno et al. 2010; Tay et al. 2011), which was recently demonstrated to be able to induce observable functional effects (Karreth et al. 2011; Tay et al. 2011; Ala et al. 2013). We focused on the first of the two 5′ UTR MREs (i.e., site S11), which was more responsive to miR-103a-3p/anti-miR-103a-3p treatment than the site S12, and assessed the ability of a sponge comprising 10 tandem copies of the S11 MRE to act as a decoy for GPRC5A. The sponge vector was labeled GPRC5A-S11WTL (Supplemental Fig. 4A,B), whereas the control sponge vector, which contained 10 tandem copies of the mutant miR-103a-3p MRE, was labeled GPRC5A-S11MTL (Supplemental Fig. 4C).
First, we verified that transfection of MIA PaCa-2 cells with the true sponge GPRC5A-S11WTL reduced the expression level of the endogenous mature miR-103a-3p (19% ± 2.6%–P < 0.001) compared with transfection with the control sponge GPRC5A-S11MTL (Fig. 4A). More importantly, transfection with the true sponge GPRC5A-S11WTL up-regulated GPRC5A mRNA (data not shown) and to a larger extent GPRC5A protein (Fig. 4B,C) compared with transfection with the control GPRC5A-S11MTL. We were able to recapitulate the same observations in HPNE, a second pancreas cell line: In HPNE cells, transfection with the GPRC5A-S11WTL sponge up-regulated GPRC5A mRNA (61.6%–P < 0.001) and GPRC5A protein (56%–P < 0.01) compared with transfection with the control sponge GPRC5A-S11MTL (Supplemental Fig. 5A–C). We also tested with a luciferase reporter construct containing the second (S12WT) 5′ UTR binding site of miR-103a-3p and found that a single copy of it suffices to up-regulate GPRC5A mRNA (23%–P < 0.001)—see Supplemental Figure 5D.
FIGURE 4.
Overexpression of the 5′ UTR MRE can increase GPRC5A mRNA and protein levels. (A) MiR-103a-3p expression is inhibited by overexpression of the wild-type sponge (S11WTL) compared with the control sponge (S11MTL) in MIA PaCa-2 cells. (B) GPRC5A protein expression is promoted by overexpression of wild-type miR-103a-3p binding site compared with the mutant site in MIA PaCa-2 cells. (C) Quantification result of B. (D) Taqman miRNA assay was performed to test mature miR-103a-3p expression in MIA PaCa-2 cells cotransfected with pre-miR-103a-3p and S11WTL. Cells cotransfected with pre-miR-103a-3p and S11MTL were used as controls. (E) GPRC5A mRNA expression was tested by RT-PCR in MIA PaCa-2 cells treated with Pre-miR-103a-3p in addition to cotransfecting with GPRC5A-S11WTL or GPRC5A-S11MTL. (F) GPRC5A protein expression was tested by Western blots in MIA PaCa-2 cells treated with Pre-miR-103a-3p in addition to cotransfecting with GPRC5A-S11WTL or GPRC5A-S11MTL. (G) Quantification result of F. All numerical data are mean ± SD. (*) P < 0.05; (***) P < 0.001, n = 3. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GAPDH and Actin are internal controls. S11WTL, pcDNA vector containing 10 tandem copies of miR-103a-3p binding site 1; S11MTL, pcDNA vector containing 10 tandem copies of mutant miR-103a-3p binding site 1.
We repeated the above experiments, this time cotransfecting MIA PaCa-2 and HPNE cells with Pre-miR-103a-3p in addition to cotransfecting with GPRC5A-S11WTL or GPRC5A-S11MTL. The level of miR-103a-3p decreased in MIA PaCa-2 cells that were cotransfected with GPRC5A-S11WTL by 43% ± 2.1% compared with cells cotransfected with control GPRC5A-S11MTL (P < 0.001) (Fig. 4D). This translated to an increase of both GPRC5A mRNA (P < 0.001) and protein levels in MIA PaCa-2 and HPNE cells that were cotransfected with Pre-miR-103a-3p and GPRC5A-S11WTL compared with MIA PaCa-2 and HPNE cells cotransfected with Pre-miR-103a-3p and control GPRC5A-S11MTL (Fig. 4E–G; Supplemental Fig. 6A–C).
Lastly, we tested a third pancreas cell line (Panc-1) as well as a nonpancreas one (HEK-293T) and were able to recapitulate the above findings in both (Supplemental Fig. 7A–C). In particular, we transfected HEK-293T cell with the GPRC5A-S11WTL and the control sponge GPRC5A-S11MTL: We found that, just like with the HPNE and MIA PaCa-2 cell lines, GPRC5A-S11WTL up-regulated GPRC5A protein compared with control, albeit somewhat modestly.
These experiments provide additional evidence that miR-103a-3p regulates GPRC5A by directly interacting with the latter's 5′ UTR. Moreover, they demonstrate that GPRC5A's 5′ UTR can potentially function as a decoy for other miR-103a-3p targets.
DISCUSSION
The potential of miRNA regulation of mRNAs through binding sites that occur in 5′ UTRs was demonstrated early on (Lytle et al. 2007; Moretti et al. 2010). However, only a few validated examples of naturally occurring 5′ UTR miRNA targets exist in the literature to date (Jopling et al. 2005; Lytle et al. 2007; Orom et al. 2008; Lee et al. 2009; Grey et al. 2010; Vora et al. 2013). For two of these few examples, the seed-driven constitutive miRNA interaction with the 5′ UTR of the targeted mRNA promoted protein translation and thus led to an increase (instead of a decrease) of protein levels (Orom et al. 2008; Tsai et al. 2009).
The described work and findings represent one more data point in support of 5′ UTR targeting by endogenous miRNAs whereby the targeting reduced the abundance of both the mRNA and corresponding protein. In particular, using luciferase assays, we provided initial evidence that the putative MREs in the 5′ UTR of GPRC5A were in fact targeted by miR-103a-3p. Additionally, we designed two constructs, GPRC5A-5′UTR-CDS and GPRC5A-CDS, and demonstrated that GPRC5A-5′UTR-CDS, but not GPRC5A-CDS, responded to overexpression of miR-103a-3p, thereby further supporting the finding that the miR-103a-3p MREs were located in GPRC5A's 5′ UTR. By overexpressing a sponge that we constructed to contain 10 tandem copies of the most responsive (site S11) of the two 5′ UTR MREs we were able to reduce the endogenous levels of miR-103a-3p and to up-regulate both GPRC5A mRNA and protein levels.
We also demonstrated that the S11 5′ UTR MRE could function as a decoy of miR-103a-3p in vitro and was able to reduce miR-103a-3p levels and increase GPRC5A mRNA and protein levels. We established these findings in three pancreatic cell lines: the normal epithelial HPNE cell line and the MIA PaCa-2 and the Panc-1 cancer cell lines. These findings have the following important ramification. MiR-103a-3p has been shown to play important roles in cellular processes such as DNA repair, metabolism, cell cycle progression, and cell differentiation (Liu et al. 2009; Yang et al. 2009; Finnerty et al. 2010; Liao and Lonnerdal 2010; Polster et al. 2010) and to be dysregulated in multiple diseases (e.g., cancers) and conditions (e.g., diabetes, Alzheimer's disease, etc.) (Roldo et al. 2006; Xie et al. 2009; Yao et al. 2010). To date only a few targets are known for miR-103a-3p. In light of our decoying finding and given miR-103a-3p's involvement in so many settings it follows that GPRC5A, through its 5′ UTR MRE for miR-103a-3p, could potentially regulate indirectly processes such as DNA repair, metabolism, the cell cycle, etc., by modulating the expression of other mRNAs, in complete analogy to what was recently shown for PTEN (Tay et al. 2011). When one considers that in some cancers GPRC5A has been shown to act as an oncogene, whereas in others as a tumor suppressor, the potential of GPRC5A to act through miR-103a-3p as a competing endogenous RNA or ceRNA (Karreth et al. 2011; Tay et al. 2011; Ala et al. 2013) for other protein-coding transcripts suggests that GPRC5A may be involved in previously unsuspected, currently uncharacterized, and presumably complex gene networks. Studying these possible roles of GPRC5A is currently the topic of ongoing research activity in our laboratory.
MATERIALS AND METHODS
Cell culture
The HEK-293T, MIA PaCa-2, HPNE, and Panc-1 cell lines were obtained from the American Type Culture Collection. All of these cells were grown in DMEM medium (Fisher Scientific) supplemented with 10% fetal bovine serum (Life Technologies), 1% Penicillin and Strep (Fisher Scientific), and 1% glutamine (Fisher Scientific), at 37°C in a humidified atmosphere containing 5% CO2.
Cell transfection
The cells were transfected with 50 nM Pre-miR-103a-3p or 50 nM Anti-miR-103a-3p (Ambion) by the reverse transfection method using the X-tremeGENE siRNA transfection reagent (Roche). Cells transfected with only a scrambled sequence, either Pre-miR-scramble or Anti-miR-scramble (Ambion), were examined in parallel as controls. Cells were then subjected to further assays or to RNA/protein extraction after 2 d. Lipofectamine 2000 (Life Technologies) was used for transfection of the psiCHECK-2 reporter vector (Promega) and pcDNA-3.1 overexpression vector (Life Technologies) and for cotransfection of vectors and Pre-miRs.
RNA isolation and real-time quantitative polymerase chain reaction analysis
Total RNA was extracted using TRIzol reagent (Life Technologies). For the detection of GPRC5A mRNA, first-strand complementary DNA was synthesized from 1000 ng of total RNA in the presence of oligo-dT (12–18) primer (Promega) and MMLV reverse transcriptase according to the manufacturer's instructions (Promega). Human glyceraldehyde 3-phosphate dehydrogenase RNA was amplified in parallel as an internal control. Real-time quantitative polymerase chain reaction (qPCR) was performed with SYBR Green PCR Master Mix (Life Technologies) and 20 ng of templates using a StepOnePlus Real-Time PCR System (Life Technologies). For miR-103a-3p detection, TaqMan MicroRNA Assay is performed with the miR-103a-3p probe (Life Technologies) following the manufacturer's instructions. Human U6 is used as internal control. Eight nanograms of total RNA is used in the RT reaction with 5X RT primers. All primer sequences used for GPRC5A mRNA and miR-103a-3p detection are listed in Supplemental Table 1 (available online). PCRs were performed at 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. ΔCt was calculated by subtracting the Ct of U6 or glyceraldehyde 3-phosphate dehydrogenase mRNA from the Ct of the mRNA of interest. ΔΔCt was then calculated by subtracting the ΔCt of the negative control from the ΔCt of the sample. The fold change in mRNA or miRNA was calculated according to the equation 2ΔΔCt.
Computational prediction of putative targets
Using the rna22 algorithm that we published previously (Miranda et al. 2006) and that has been used by us and others to identify many miRNA targets beyond the 3′ UTR of genes (Duursma et al. 2008; Lal et al. 2008, 2009; Tay et al. 2008; Rigoutsos 2009; Marin-Muller et al. 2013), we identified two candidate targets for miR-103a-3p in GPRC5A's mRNA. In what follows, we will be using the terms “miRNA binding site” and “miRNA response element” (MRE) interchangeably.
DNA vectors
The coding region of the GPRC5A mRNA with and without the 5′ UTR was amplified by PCR from MIA PaCa-2 cDNA. The DNA sequence with 10 tandem repeats of the predicted miR-103a-3p binding sites and the control DNA sequence with 10 tandem repeats of seed-region mutant miR-103a-3p-binding sites were synthesized as fragments (Life Technologies). The fragments were inserted into the pcDNA-3.1 vector between the NheI and NotI sites. The vectors were labeled GPRC5A-5′UTR-CDS, GPRC5A-CDS, GPRC5A-S11WTL, and GPRC5A-S11MTL, respectively.
Reporter vectors
The predicted microRNA binding sites or MREs for microRNA-response element were synthesized as sense and antisense oligomers, annealed, and cloned into a psiCHECK-2 vector. We created two instances: one where the predicted miRNA binding sites were cloned into the psiCHECK-2 vector, directly 3′-downstream from, and a second where they were cloned directly 5′-upstream of Renilla Luciferase. These reporters were labeled S11WT-3′Luc, S11MT-3′Luc, S11WT-5′Luc, and S11MT-5′Luc, S12WT-3′Luc, S12MT-3′Luc, S12WT-5′Luc, and S12MT-5′Luc, respectively (WT: wild type; MT: mutant). All primers used for these constructs are listed in Supplemental Table 1.
Luciferase assay
Each psiCHECK-2 vector containing a reporter construct was cotransfected into HPNE and MIA PaCa-2 cells with Pre-miR-103a-3p or anti-miR-103a-3p by using Lipofectamine 2000 according to the manufacturer's protocol for cotransfection of DNA and pre-miRs. In parallel, each psiCHECK-2 vector containing a reporter construct was also cotransfected into HPNE and MIA PaCa-2 cells with pre-miR-scramble or Anti-miR-scramble as control. Cells were harvested at 48 h after transfection, and the Renilla and Firefly luciferase activities in the cellular lysate were assayed by using the Dual-Glo Luciferase Assay (Promega) according to the manufacturer's protocol. Light intensity for each sample was measured by using Synergy 2 Multi-Mode Microplate Reader (BioTek), and each value from Renilla luciferase was normalized by Firefly luciferase.
Western blots
Transfected cells were lysed on ice in Pierce IP lysis buffer (Thermo Scientific) containing 1X complete protease inhibitor (Roche). Debris was pelletized by centrifugation at 13,200 rpm for 15 min, and protein concentrations were determined using Pierce BCA assay (Thermo Scientific). Lysates were heat-denatured at 100°C for 10 min before separation in 10% sodium dodecyl sulfate–polyacrylamide gels and transferred to nitrocellulose membrane (GE Healthcare). Membranes were blocked with 5% bovine serum albumin (Sigma-Aldrich) in Tris-buffered saline Tween-20 buffer (10 mM Tris at pH 7.6, 150 mM NaCl, and 0.1% Tween-20) and probed with primary antibody in Tris-buffered saline Tween-20 with 5% bovine serum albumin at the recommended dilutions at 4°C. Primary antibodies included GPRC5A antibody (Sigma-Aldrich), β-actin antibody (Cell Signaling Technology), and GFP antibody (Santa Cruz Biotechnology Inc.). Membranes were incubated with secondary antibody (Cell Signaling Technology) diluted in Tris-buffered saline Tween-20 with 5% bovine serum albumin for 1 h at room temperature. The signal was detected with Pierce ECL Western Blotting Substrate (Thermo Scientific) and GE ImageQuant LAS 4000 (GE Healthcare).
Statistical analysis
Statistical analysis was performed using Excel (Microsoft) and SPSS (IBM). Unless otherwise indicated, the level of significance for the difference between data sets was assessed using one-way analysis of variance. Data are expressed as the means ± SD. P-values ≤0.05 were considered statistically significant.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by institutional funds, a William M. Keck Foundation award (I.R.), and a Hirshberg Foundation for Pancreatic Cancer Research award (I.R.). We thank Yi Jing, Kevin Quann, Elena Mogilyansky, Eric Londin, Phillipe Loher, and Jonathan Brody for their assistance with various aspects of this study.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045757.114.
Freely available online through the RNA Open Access option.
REFERENCES
- Acquafreda T, Soprano KJ, Soprano DR 2009. GPRC5A: a potential tumor suppressor and oncogene. Cancer Biol Ther 8: 963–965 [DOI] [PubMed] [Google Scholar]
- Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Leopold V, Tay Y, Provero P, Zecchina R, Pandolfi PP 2013. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci 110: 7154–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier J-F, Hébuterne X, et al. 2011. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet 43: 242–245 [DOI] [PubMed] [Google Scholar]
- Cheng L, Yang S, Yang Y, Zhang W, Xiao H, Gao H, Deng X, Zhang Q 2012. Global gene expression and functional network analysis of gastric cancer identify extended pathway maps and GPRC5A as a potential biomarker. Cancer Lett 326: 105–113 [DOI] [PubMed] [Google Scholar]
- Da Sacco L, Masotti A 2012. Recent insights and novel bioinformatics tools to understand the role of microRNAs binding to 5′ untranslated region. Int J Mol Sci 14: 480–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AH, Thompson P, Robson T, McKeown SR 2010. Cytochrome P450 1B1 mRNA untranslated regions interact to inhibit protein translation. Mol Carcinog 49: 190–199 [DOI] [PubMed] [Google Scholar]
- Dewing AS, Rueli RH, Robles MJ, Nguyen-Wu ED, Zeyda T, Berry MJ, Bellinger FP 2012. Expression and regulation of mouse selenoprotein P transcript variants differing in non-coding RNA. RNA Biol 9: 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R 2008. miR-148 targets human DNMT3b protein coding region. RNA 14: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA 2010a. Emerging roles for natural microRNA sponges. Curr Biol 20: R858–R861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA 2010b. MicroRNA sponges: progress and possibilities. RNA 16: 2043–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT 2010. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol 402: 491–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA 2008. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci 105: 14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner JJ, Parker SCJ, Prickett TD, Dutton-Regester K, Stitzel ML, Lin JC, Davis S, Simhadri VL, Jha S, Katagiri N, et al. 2013. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci 110: 13481–13486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA 2010. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog 6: e1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hao J, Zhang S, Zhou Y, Liu C, Hu X, Shao C 2011. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem Biophys Res Commun 406: 552–557 [DOI] [PubMed] [Google Scholar]
- Hausser J, Syed AP, Bilen B, Zavolan M 2013. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res 23: 604–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27: 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. 2011. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, et al. 2008. p16INK4a translation suppressed by miR-24. PLoS One 3: e1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. 2009. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell 35: 610–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD 2009. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 19: 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Lonnerdal B 2010. Global microRNA characterization reveals that miR-103 is involved in IGF-1 stimulated mouse intestinal cell proliferation. PLoS One 5: e12976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SP, Fu RH, Yu HH, Li KW, Tsai CH, Shyu WC, Lin SZ 2009. MicroRNAs regulation modulated self-renewal and lineage differentiation of stem cells. Cell Transplant 18: 1039–1045 [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci 104: 9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Muller C, Li D, Bharadwaj U, Li M, Chen C, Hodges SE, Fisher WE, Mo Q, Hung M-C, Yao Q 2013. A tumorigenic factor interactome connected through tumor suppressor microRNA-198 in human pancreatic cancer. Clin Cancer Res 19: 5901–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. 2010. A MicroRNA targeting dicer for metastasis control. Cell 141: 1195–1207 [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I 2006. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Moretti F, Thermann R, Hentze MW 2010. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA 16: 2493–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Mao G, Wilfred BR, Xie K, Jennings MH, Gao Z, Wang X 2011. Specific sequence determinants of miR-15/107 microRNA gene group targets. Nucleic Acids Res 39: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polster BJ, Westaway SK, Nguyen TM, Yoon MY, Hayflick SJ 2010. Discordant expression of miR-103/7 and pantothenate kinase host genes in mouse. Mol Genet Metab 101: 292–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan P, Mohr AM, Grandgenett PM, Steele MM, Batra SK, Hollingsworth MA 2013. MicroRNA-200c modulates the expression of MUC4 and MUC16 by directly targeting their coding sequences in human pancreatic cancer. PLoS One 8: e73356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Rigoutsos I 2009. New tricks for animal microRNAs: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res 69: 3245–3248 [DOI] [PubMed] [Google Scholar]
- Rigoutsos I, Tsirigos A 2010. MicroRNA target prediction. In MicroRNAs in development and cancer (ed. Slack F), pp. 237–259 Imperial College Press, London, UK [Google Scholar]
- Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, et al. 2006. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol 24: 4677–4684 [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C 2011. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12: 683–691 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Spiridonov NA, Kashina A 2013. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res 41: 2073–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C 2008. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol 28: 4609–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Fujimoto J, Men T, Ye X, Deng J, Lacroix L, Clifford JL, Mao L, Van Pelt CS, Lee JJ, et al. 2007. Identification of the retinoic acid–inducible Gprc5a as a new lung tumor suppressor gene. J Natl Cancer Inst 99: 1668–1682 [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. 2011. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M 2011. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474: 649–653 [DOI] [PubMed] [Google Scholar]
- Tsai NP, Lin YL, Wei LN 2009. MicroRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem J 424: 411–418 [DOI] [PubMed] [Google Scholar]
- Vora M, Shah M, Ostafi S, Onken B, Xue J, Ni JZ, Gu S, Driscoll M 2013. Deletion of microRNA-80 activates dietary restriction to extend C. elegans healthspan and lifespan. PLoS Genet 9: e1003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862 [DOI] [PubMed] [Google Scholar]
- Xia Z, Clark P, Huynh T, Loher P, Zhao Y, Chen H-W, Rigoutsos I, Zhou R 2012. Molecular dynamics simulations of Ago silencing complexes reveal a large repertoire of admissible ‘seed-less’ targets. Sci Rep 2: 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Lim B, Lodish HF 2009. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 58: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GH, Wang F, Yu J, Wang XS, Yuan JY, Zhang JW 2009. MicroRNAs are involved in erythroid differentiation control. J Cell Biochem 107: 548–556 [DOI] [PubMed] [Google Scholar]
- Yao J, Hennessey T, Flynt A, Lai E, Beal MF, Lin MT 2010. MicroRNA-related cofilin abnormality in Alzheimer's disease. PLoS One 5: e15546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.