The Neurospora VS ribozyme contains a stem–loop substrate domain and a stem–loop catalytic domain that form a kissing-loop interaction. This article uses a variety of thermodynamic techniques to demonstrate that this interaction is 7–8 kcal/mol more stable than a comparable duplex of Watson–Crick base pairs.
Keywords: Neurospora VS ribozyme, substrate recognition, U-turn, kissing-loop interaction, isothermal titration calorimetry
Abstract
Kissing loops are tertiary structure elements that often play key roles in functional RNAs. In the Neurospora VS ribozyme, a kissing-loop interaction between the stem–loop I (SLI) substrate and stem–loop V (SLV) of the catalytic domain is known to play an important role in substrate recognition. In addition, this I/V kissing-loop interaction is associated with a helix shift in SLI that activates the substrate for catalysis. To better understand the role of this kissing-loop interaction in substrate recognition and activation by the VS ribozyme, we performed a thermodynamic characterization by isothermal titration calorimetry using isolated SLI and SLV stem–loops. We demonstrate that preshifted SLI variants have higher affinity for SLV than shiftable SLI variants, with an energetic cost of 1.8–3 kcal/mol for the helix shift in SLI. The affinity of the preshifted SLI for SLV is remarkably high, the interaction being more stable by 7–8 kcal/mol than predicted for a comparable duplex containing three Watson–Crick base pairs. The structural basis of this remarkable stability is discussed in light of previous NMR studies. Comparative thermodynamic studies reveal that kissing-loop complexes containing 6–7 Watson–Crick base pairs are as stable as predicted from comparable RNA duplexes; however, those with 2–3 Watson–Crick base pairs are more stable than predicted. Interestingly, the stability of SLI/ribozyme complexes is similar to that of SLI/SLV complexes. Thus, the I/V kissing loop interaction represents the predominant energetic contribution to substrate recognition by the trans-cleaving VS ribozyme.
INTRODUCTION
Kissing-loop interactions are widespread elements of RNA tertiary structures that result from base-pairing between the terminal loops of two hairpins. Their formation is central to many RNA-mediated processes, such as antisense recognition, plasmid replication control, retroviral dimerization, translation initiation, and ribosomal frameshifting (Wagner and Simons 1994; Guo et al. 2001; Brunel et al. 2002; Baranov et al. 2005; Brantl 2007). One of the best-studied kissing-loop interactions occurs during retroviral genome dimerization in HIV-1, where it forms between palindromic base sequences of the dimer initiation site (Skripkin et al. 1994; Ennifar et al. 2001; Brunel et al. 2002). Although kissing loops form long-range interactions between different RNAs or remote domains within the same RNA, they also play important roles in stabilizing the complex architecture of functional RNA elements, such as tRNAs (Quigley and Rich 1976) riboswitch aptamers (Batey et al. 2004; Serganov et al. 2004, 2008; Lemay et al. 2006; Blouin and Lafontaine 2007; Garst et al. 2008), and ribozymes (De la Peña et al. 2003; Khvorova et al. 2003; Chi et al. 2008; Dufour et al. 2009). Thus, kissing-loop motifs play crucial roles in RNA structure and function; however, only a few of these have been thermodynamically characterized.
Like several other functional RNAs, the Neurospora VS ribozyme depends on the formation of a kissing-loop interaction. Early work on the self-cleaving VS ribozyme revealed that the catalytic domain (VS Rz) recognizes its substrate (stem–loop I) through a kissing-loop interaction that is stabilized by magnesium ions (Fig. 1A; Beattie et al. 1995; Rastogi et al. 1996). This interaction involves three Watson–Crick (W-C) base pairs between residues 630, 631, and 632 of stem–loop I (SLI) and residues 697, 698, and 699 of stem–loop V (SLV). It is required for proper folding (Andersen and Collins 2001; Hiley and Collins 2001) as well as for the efficient phosphodiester bond cleavage between G620 and A621 and the reverse ligation reaction (Rastogi et al. 1996; Andersen and Collins 2000). Interestingly, formation of the I/V kissing-loop interaction involves a well-characterized conformational change in SLI from an unshifted to a shifted conformation (Fig. 1B; Andersen and Collins 2000, 2001; Michiels et al. 2000; Flinders and Dieckmann 2001; Hoffmann et al. 2003), and either the catalytic domain or an isolated SLV can produce this helix shift within the SLI substrate (Andersen and Collins 2000). Through the SLI helix shift, the I/V kissing-loop interaction activates the substrate for catalysis, since ribozymes containing SLI variants that stabilized the unshifted conformation are inactive, whereas those that allow or favor the shifted conformation (preshifted substrates) are active (Andersen and Collins 2000). Formation of the I/V kissing-loop interaction also facilitates docking of the cleavage site internal loop of SLI with the A730 loop of SLVI (Andersen and Collins 2001; Hiley and Collins 2001; Hiley et al. 2002). Our current understanding is that the internal loops of SLI and SLVI must precisely interact to create the active site, where the nucleobases of G638 and A756 play key roles in the general acid–base cleavage mechanism (Lafontaine et al. 2001b, 2002b; Sood and Collins 2002; Jones and Strobel 2003; Zhao et al. 2005; Smith and Collins 2007; Wilson et al. 2007, 2010; Jaikaran et al. 2008). Thus, the I/V kissing-loop interaction is important for several steps of the VS ribozyme cleavage reaction, including substrate recognition, activation, and docking for catalysis.
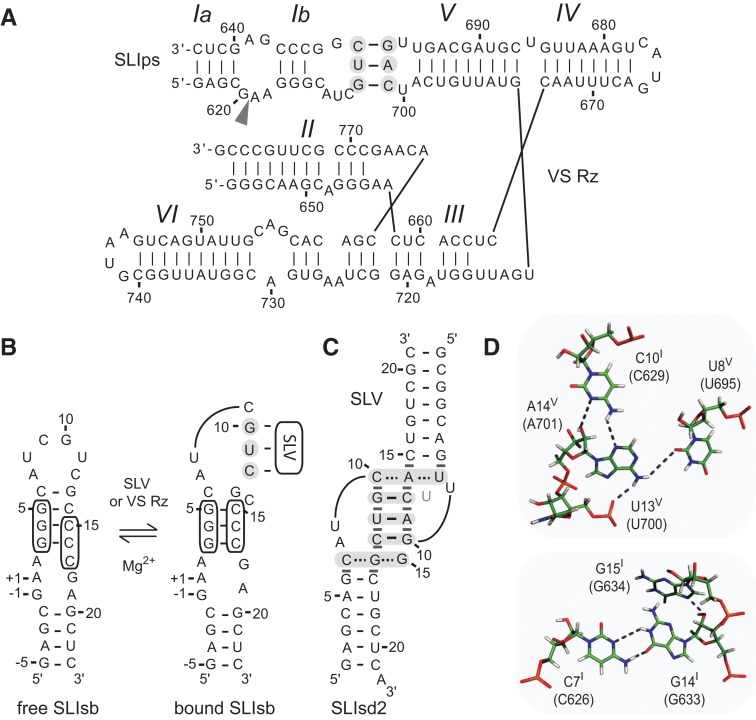
FIGURE 1.
Primary and secondary structures of the Neurospora VS ribozyme. (A) The catalytic domain of the VS ribozyme containing helical domains II-VI (VS Rz) and an SLI substrate (SLIps) containing stems Ia and Ib. The cleavage site is indicated by an arrowhead. The I/V kissing-loop interaction involves W-C base pairs (black lines) between shaded residues of SLI and SLV (Beattie et al. 1995; Rastogi et al. 1996). (B) Formation of the I/V kissing-loop is accompanied by a structural rearrangement of the SLI substrate from an unshifted (free) to a shifted (bound) conformation. The cleavage site is between residues −1 and +1 of SLIsb. (C,D) Structural characteristics of the I/V kissing-loop based on the NMR structure of the SLIds2/SLV complex (Bouchard and Legault 2014). In C, W-C and noncanonical base pairs are represented by solid and dashed lines, respectively, on the secondary structure of the complex, whereas stable base stacking at the kissing-loop junction is illustrated by gray rectangles. U13 is in gray to illustrate its extrusion from the loop V fold. In D, hydrogen bonds within the base triples of the minimized averaged structure are represented by dotted lines.
Until recently, structural insights into the I/V kissing-loop interaction were provided by three-dimensional models of the full ribozyme (Hiley and Collins 2001; Lafontaine et al. 2001a, 2002a; Lipfert et al. 2008) as well as high-resolution NMR structures of free SLV (Campbell and Legault 2005; Campbell et al. 2006) and of the unshifted and shifted conformations of SLI (Michiels et al. 2000; Flinders and Dieckmann 2001; Hoffmann et al. 2003). As part of our efforts to characterize the I/V kissing-loop interaction, we previously reported a structure-function study that addressed the role of loop V residues in SLI substrate recognition (Bouchard et al. 2008). One intriguing observation is that, although any base can replace the extruded U700 loop residue, deletion of this nucleotide substantially reduces ribozyme cleavage activity (Bouchard et al. 2008), possibly by affecting substrate binding. More recently, we performed NMR studies of several SLI/SLV complexes formed with RNA hairpins derived from the VS ribozyme and determined the NMR structure of an SLI/SLV kissing-loop complex (Bouchard and Legault 2014). These NMR studies provide clear structural evidence for the helix shift in SLI upon SLV binding. In addition, the NMR structure of an SLI/SLV complex confirms that both the SLI and SLV loops adopt U-turn structures, which facilitate formation of the three expected W-C base pairs between SLI and SLV, and additional hydrogen bonding and base stacking interactions at the kissing-loop junction (Fig. 1C,D). Complementary thermodynamic investigations of the I/V kissing-loop interaction are needed to evaluate the affinity of the SLI/SLV interaction in light of the NMR structure and to help understand the effect of SLI and SLV sequence variations on substrate recognition and catalysis.
Binding studies have been previously carried out using various SLI substrates and a trans-cleaving VS ribozyme (Zamel and Collins 2002). Although these studies revealed the importance of the shifted SLI conformation in facilitating binding to the ribozyme (Zamel and Collins 2002), it used a ribozyme system stabilized by an artificial intermolecular stem and thus yielded KD values that do not directly reflect the stability of the I/V kissing loop (Zamel and Collins 2002). In the present work, we use isothermal titration calorimetry (ITC) (Reymond et al. 2009; Salim and Feig 2009) to thermodynamically characterize the I/V kissing-loop interaction formed between isolated SLI and SLV hairpins. As a result, our study provides KD values as well as energetic contributions directly associated with formation of the I/V kissing-loop interaction. In addition, since most of the SLI/SLV complexes investigated here have been structurally characterized by NMR spectroscopy (Bouchard et al. 2008; Bouchard and Legault 2014), the thermodynamic parameters can be directly related to structural features of SLI/SLV complexes. In particular, we examine the effect of kinetically characterized SLV sequence variations on the thermodynamic parameters associated with the I/V kissing-loop interaction. We also investigate the thermodynamic contribution of the helix shift in SLI on the stability of SLI/SLV complexes using several shiftable, preshifted, and double-stranded SLI variants. Finally, using noncleavable SLI substrates, we compare the affinity of SLI/SLV complexes with that of SLI/ribozyme complexes to evaluate the relative contribution of the I/V kissing-loop interaction to substrate recognition in the VS ribozyme.
RESULTS
Thermodynamic characterization of the VS ribozyme I/V kissing-loop interaction
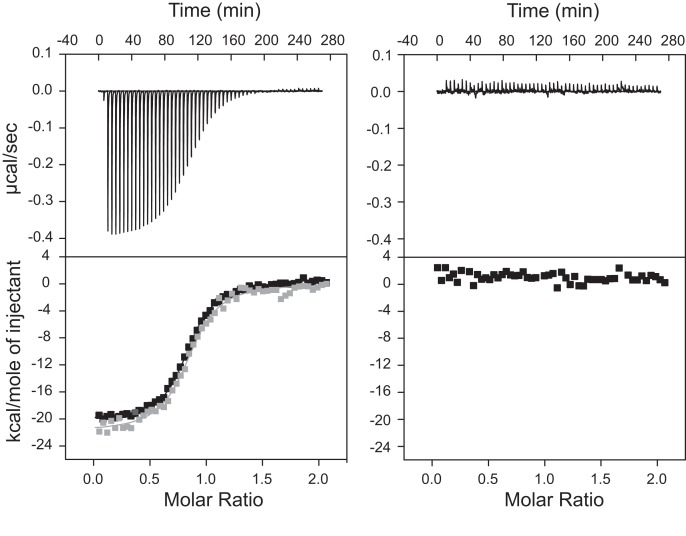
To better understand substrate recognition by the VS ribozyme, we determined the thermodynamic parameters associated with the I/V kissing-loop interaction by ITC using two isolated stem–loops derived from the VS ribozyme: the SLI substrate and the SLV receptor (Fig. 2). Biophysical characterization of the I/V kissing-loop interaction using isolated RNA stem–loops is justified by the fact that an isolated SLV receptor hairpin is sufficient to bind an isolated SLI substrate and cause the same helix shift in SLI that is observed with the full VS ribozyme catalytic domain (Andersen and Collins 2001; Bouchard et al. 2008; Bouchard and Legault 2014). To further simplify the thermodynamic characterization of the I/V kissing-loop interaction, the initial SLI substrate was designed to adopt only an active preshifted conformation (SLIps) (Fig. 2A; Andersen and Collins 2000; Zamel and Collins 2002). The same SLIps sequence was previously used for kinetic characterization of VS ribozyme variants (Bouchard et al. 2008). Using NMR spectroscopy and native gel electrophoresis, we previously verified that both SLIps and SLV RNAs form stable hairpins under conditions used for ITC studies (Bouchard et al. 2008; Bouchard and Legault 2014). NMR spectroscopy studies also indicate that SLIps and SLV form a stable 1:1 kissing-loop complex in the presence of MgCl2 (Bouchard and Legault 2014). ITC experiments were first carried out at 25°C in a buffer containing 20 mM MgCl2 (Fig. 3). Analysis of duplicate ITC experiments for formation of this 1:1 SLIps/SLV complex yields the following average thermodynamic values: KD = 0.24 ± 0.01 μM; ΔH = −22.0 ± 0.1 kcal/mol; −TΔS= 12.9 ± 0.1 kcal/mol; and ΔGITC= −9.04 ± 0.03 kcal/mol (Table 1).
FIGURE 2.
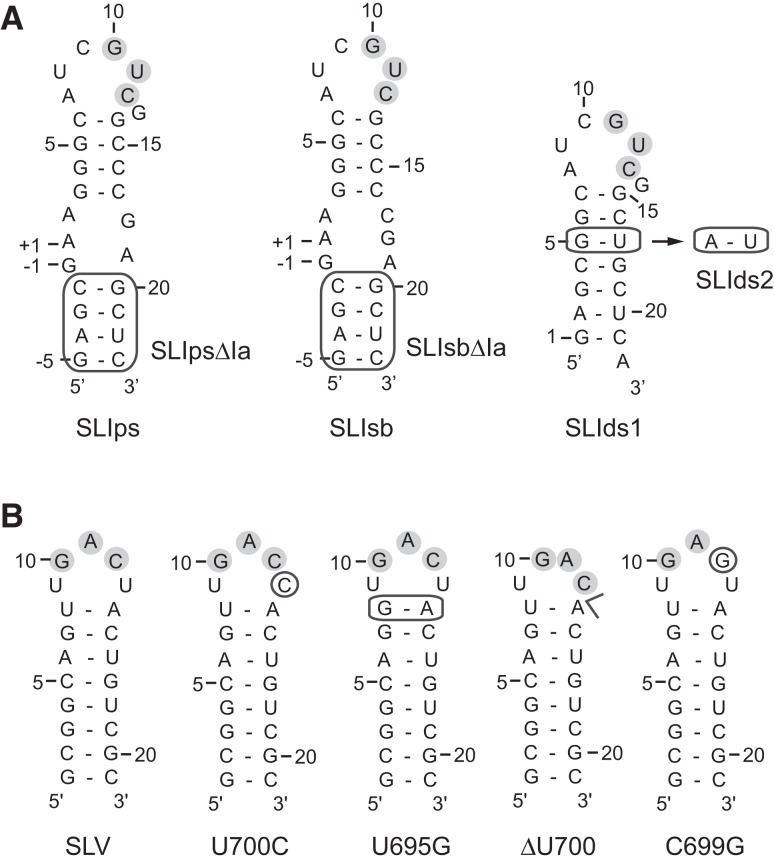
Primary and secondary structures of SLI and SLV RNAs used in this study. (A) The SLIps substrate RNA and related variants. SLIpsΔIa and SLIsbΔIa are derivatives of SLIps and SLIsb, respectively, in which stem Ia is absent (boxed region). SLIsd2 is a G5A variant of SLIds1. The VS ribozyme cleavage site for SLIps, SLIpsΔIa, SLIsb, and SLIsbΔIa is between residues −1 and +1 of SLI. (B) The wild-type SLV RNA and related SLV variants. In A and B, residues that participate in the three W-C base pairs at the I/V kissing-loop junction are shaded in gray.
FIGURE 3.
ITC data analysis of wild-type SLV binding to SLIps at 20 mM MgCl2. The raw data of a representative titration experiment are shown on the left top panel and the corresponding control experiment (only buffer in the cell) is shown on the right top panel. The isothermal curves, which represent the integrated data of the above titration experiments, are shown on the bottom panels. On the left bottom panel, two isothermal curves of the titration experiment are shown, before (black) and after (gray) subtraction of the control experiment. They both yield the same KD value (0.24 μM). In the titration experiment, 100 μM of wild-type SLV was used in the syringe and 10 μM of SLIps was used in the cell.
TABLE 1.
ITC data for binding of SLV variants to a preshifted SLI substrate (SLIps)
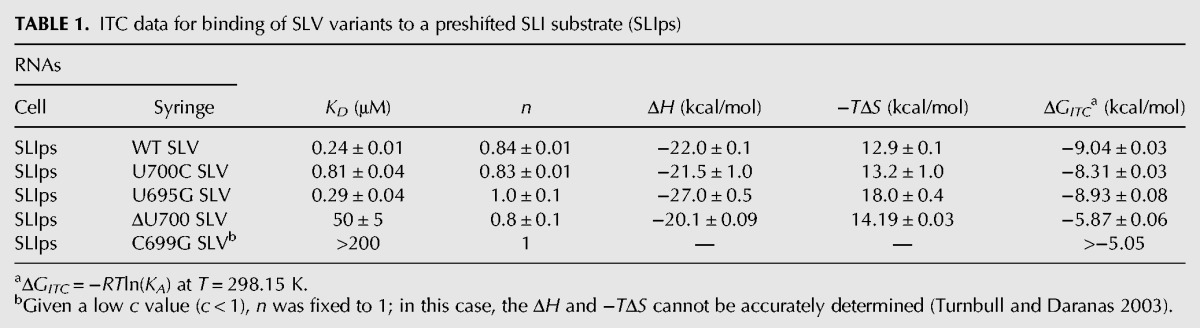
Magnesium dependence of the I/V kissing-loop interaction
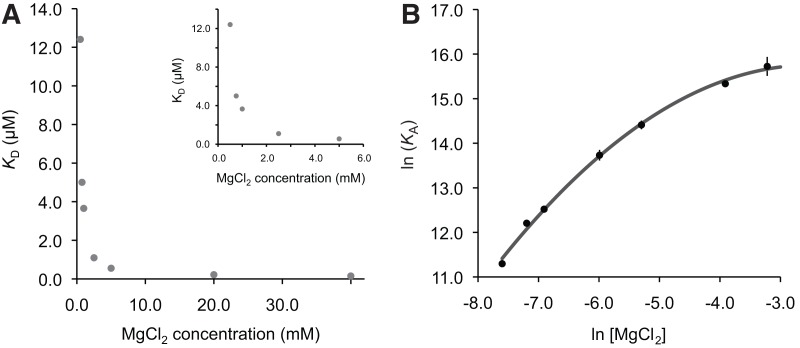
Since Mg2+ ions are essential for formation of the I/V kissing-loop interaction, ITC experiments were performed at 25°C under varying concentrations of MgCl2 to examine the Mg2+ ion dependence of this interaction and determine conditions at which the Mg2+ ion concentration is saturated. A plot of the dissociation constants (KD) against the MgCl2 concentration indicates that the affinity of SLV for the SLIps substrate increases (decrease of KD) with increasing Mg2+ ion concentration and reaches a plateau around 20 mM MgCl2 (Fig. 4). Thus, subsequent ITC experiments were carried out in a buffer containing 20 mM MgCl2.
FIGURE 4.
Magnesium ion dependence of the I/V kissing-loop interaction. (A) Dissociation constant (KD) for the SLIps/SLV complex with respect to MgCl2 concentration. The inset shows the same data on a different scale. (B) Dependence of ln(KA) on ln[MgCl2] for the SLIps/SLV complex. These data are best fit by a second-order polynomial equation: y = 0.1651x2 − 0.8162x + 14.748. In A and B, error bars were calculated for each point from the standard deviations of two experiments, and are, in some cases, smaller than the data points.
The Mg2+ dependence of SLIps/SLV binding can also be analyzed as
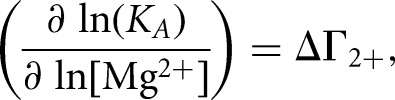 |
where ΔΓ2+ is the apparent interaction coefficient that represents the net uptake or release of divalent cations associated with the interaction (Draper 2008; Leipply et al. 2009). Although the plot of ln(KA) versus ln[Mg2+] is generally linear within a certain range of Mg2+ concentration, it is not expected over the full range of Mg2+ concentration (Weixlbaumer et al. 2004; Draper 2008; Leipply et al. 2009), as observed here for the SLIps/SLV complex (Fig. 4B). Thus, our data are best fit by a second-order polynomial equation:
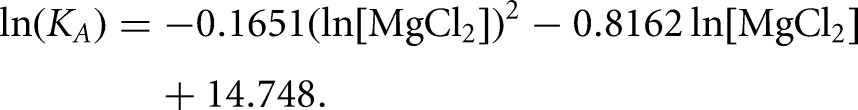 |
The slope of this plot evaluated at the midpoint of the curve ([MgCl2] = 4.5 mM) was used to estimate an apparent ΔΓ2+ value of 0.97 (Leipply et al. 2009). Maximum and minimum values of 1.46 and 0.71, respectively, for ΔΓ2+ were derived by linear regression analysis using either the first four points or the last four points of the ln(KA) versus ln[MgCl2] plot (Fig. 4B). The derived ΔΓ2+ values are termed “apparent” ΔΓ2+ because the experimental conditions, particularly the use of Tris buffer and the absence of monovalent salts, were selected to be compatible with those used for NMR structural characterization (Campbell et al. 2006; Bouchard et al. 2008; Bouchard and Legault 2014) but do not allow determination of accurate ΔΓ2+ values (Leipply et al. 2009). Nevertheless, this analysis indicates that there is an approximate net uptake of one Mg2+ ion associated with formation of the SLI/SLV complex in the concentration range between 0.5 mM and 40 mM MgCl2.
Effect of SLV sequence variations on the stability of the SLI/SLV complex
As part of a previous study, VS ribozyme variants were kinetically characterized to evaluate the contribution of U700 and other SLV loop residues to SLI recognition (Bouchard et al. 2008). This study revealed that replacing U700 by A, C, or G does not significantly affect the ribozyme cleavage activity, whereas U700 deletion dramatically impairs this activity (Bouchard et al. 2008). In order to address the role of U700 in SLI substrate recognition, we investigated the effect of U700 deletion and other SLV sequence modifications on the thermodynamic stability of the SLI/SLV complex. ITC experiments were performed at 25°C in a buffer containing 20 mM MgCl2 with SLIps and different SLV variants (Fig. 2B). It was previously verified by native gel electrophoresis and NMR spectroscopy that under these conditions, these SLV variants adopt a stable hairpin conformation (Bouchard et al. 2008). As expected, all SLV variants bound to the SLIps substrate with an approximate stoichiometry (n) of 1 (Table 1), except for the C699G variant for which no binding could be detected under our ITC conditions.
Changing the U700 base to a C (U700C) or changing the U-A closing base pair to a G-A (U695G) yields KD values of 0.81 μM and 0.29 μM, respectively, which are on the same order of magnitude as that obtained with the wild-type SLV (KD = 0.24 μM) (Table 1). In contrast, an SLV variant containing a U700 deletion (ΔU700) displays a substantial decrease in affinity for SLIps with a KD value of 50 μM (Table 1). For the C699G variant, thermodynamic parameters could not be derived even at high concentrations of RNA (1 mM C699G SLV in the syringe and 100 μM of SLIps in the cell), and a KD > 200 μM was estimated, assuming that the poor quality of the titration curve reflects a situation in which c < 1 [c = (concentration of RNA in the cell)/KD] (Wiseman et al. 1989; Turnbull and Daranas 2003). Accordingly, this SLV variant was not expected to stably bind to the SLIps substrate, since the C699G variation disrupts one of the three W-C base pairs at the kissing-loop junction (Rastogi et al. 1996; Bouchard and Legault 2014).
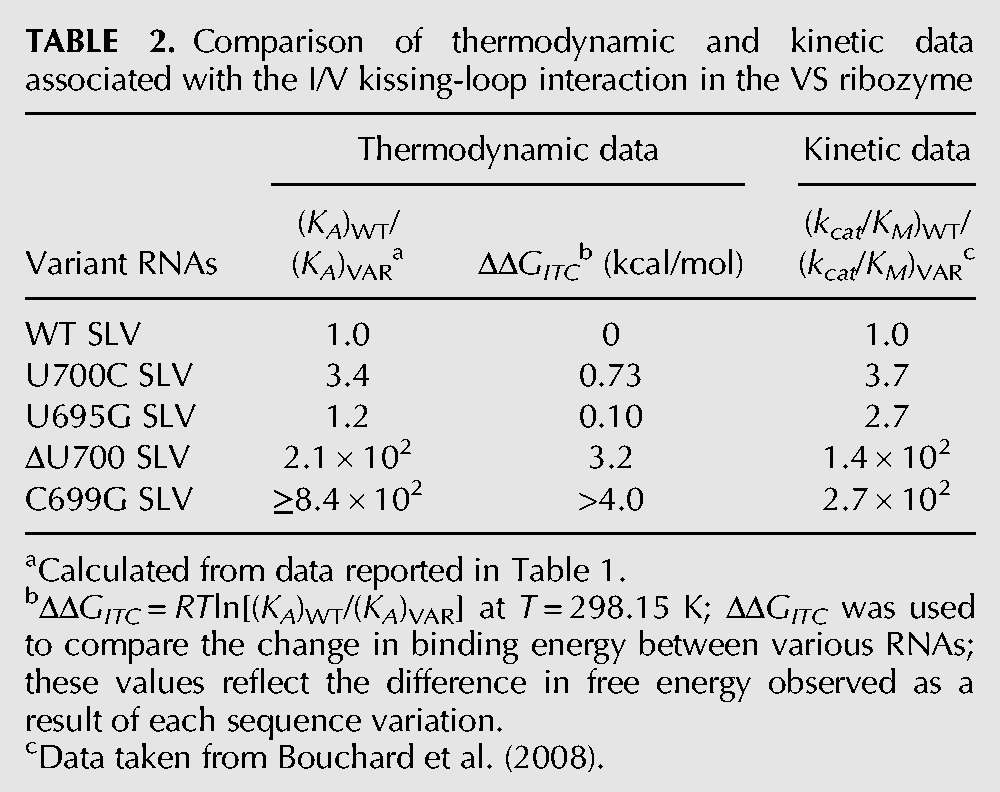
To compare the effect of SLV sequence variations on SLI binding with their effect on SLI cleavage by the VS ribozyme, we tabulated the relative KA values [(KA)WT/(KA)VAR = (KD)VAR/(KD)WT] obtained in the present study with the relative kcat/KM values [(kcat/KM)WT/(kcat/KM)VAR] obtained previously (Table 2; Bouchard et al. 2008). In addition, the contributions of modified residues to the free energy of the I/V kissing-loop interaction were derived from the relative KA values {ΔΔGITC = RTln[(KA)WT/(KA)VAR]} (Table 2). For the U700C and U695G variants, the (KA)WT/(KA)VAR ratios (3.4 and 1.2, respectively) are similar to the (kcat/KM)WT/(kcat/KM)VAR ratios (3.7 and 2.7, respectively). In both cases, the small ratios (<4) indicate that neither the affinity of the I/V kissing-loop interaction nor ribozyme activity is significantly affected by these modifications. In addition, the ΔΔGITC (≤0.7 kcal/mol) indicates that no substantial loss of free energy of binding resulted from these SLV modifications (Table 2). For the ΔU700 variant, the large decrease in ribozyme activity corresponds to a (kcat/KM)WT/(kcat/KM)VAR ratio of 140, which is on the same order of magnitude as the respective (KA)WT/(KA)VAR ratio of 210 (Table 2). The ΔΔGITC value of this variant (3.2 kcal/mol) indicates that the deletion of U700 results in an energetic cost of ∼3 kcal/mol for SLI binding (Table 2).
TABLE 2.
Comparison of thermodynamic and kinetic data associated with the I/V kissing-loop interaction in the VS ribozyme
Effect of SLI sequence variations on the stability of the SLI/SLV complex
In the VS ribozyme, the kissing-loop interaction between SLI and SLV is accompanied by a structural rearrangement in SLI (Andersen and Collins 2000, 2001), in which SLI changes from an inactive (unshifted) to an active (shifted) conformation. The cytosines C635 to C637 (C15–17) (Fig. 1B) in helix Ib shift their position in the stem, excluding C634 (C14) (Fig. 1B) from the helix and rearranging the cleavage site internal loop (Andersen and Collins 2000). To better understand the importance of this helix shift in substrate recognition, we evaluated its energetic contribution by ITC using six different SLI variants (Fig. 2A). We selected shiftable variants that convert from an inactive to an active conformation upon SLV binding in the presence of Mg2+ ions (SLIsb and SLIsbΔIa) (Fig. 2A). We also selected preshifted variants that adopt a single active shifted conformation in their free form (SLIps and SLIpsΔIa) (Fig. 2A). Similar shiftable and preshifted variants that lack stem Ia (ΔIa) were also investigated to evaluate the energetic cost of stem Ia in the formation of the SLI/SLV complex, since it was previously shown that removal of stem Ia increases ribozyme cleavage activity (Beattie and Collins 1997; Rastogi and Collins 1998; Zamel et al. 2004; Poon et al. 2006). Finally, we also investigated SLI variants containing a noncleavable double-stranded stem (SLIds1 and SLIds2) (Fig. 2A). The secondary structure of these six SLI variants in their free form was confirmed by native gel electrophoresis and NMR spectroscopy (Bouchard and Legault 2014). NMR spectroscopy was also used to demonstrate formation of stable kissing-loop complexes between several SLI variants and SLV (SLIsbΔIa/SLV, SLIps/SLV, SLIpsΔIa/SLV, SLIds1/SLV, and SLIds2/SLV) (Bouchard and Legault 2014), which include the three proposed W-C base pairs. Moreover, these studies provide direct evidence that shiftable SLI variants, but not the preshifted or double-stranded variants, undergo the proposed helix shift upon SLV binding (Bouchard and Legault 2014).
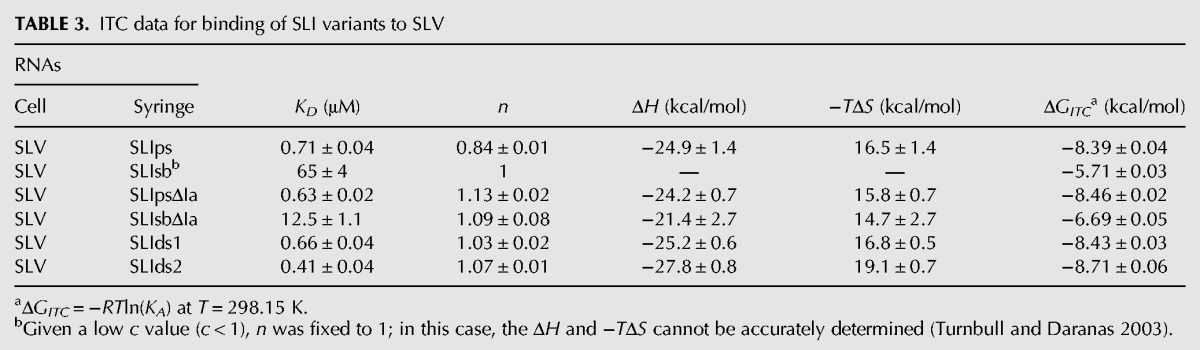
The ITC results reveal higher affinities (lower KD) of SLV for preshifted SLI variants compared to shiftable SLI variants (SLIps versus SLIsb and SLIpsΔIa versus SLIsbΔIa) (Table 3). Among the shiftable SLI variants, a lower affinity for SLV is observed in the presence of stem Ia (SLIsb) than in its absence (SLIsbΔIa) (Table 3). In contrast, the presence of stem Ia does not significantly decrease the affinity of SLV to a preshifted SLI variant (SLIps versus SLIpsΔIa) (Table 3). Not surprisingly, SLI variants containing a double-stranded stem bind SLV with affinities that closely match those of the preshifted SLI variants (SLIds1 and SLIds2 versus SLIps and SLIpsΔIa) (Table 3). In terms of free energy (Table 3), these results show that SLV forms more stable complexes with preshifted SLI variants than with shiftable SLI variants. An energetic cost of 1.8 kcal/mol is associated with complex formation of shiftable versus preshifted SLI variants in the absence of stem Ia, and this can be directly attributed to the structural rearrangement of the shiftable substrate upon complex formation. The presence of stem Ia further reduces by 1 kcal/mol the thermodynamic stability of the SLI/SLV complex formed with a shiftable substrate but has essentially no effect on the stability of complexes formed with preshifted substrates. These results also indicate that the stability of SLI/SLV complexes formed with several nonshiftable SLI variants (SLIps, SLIpsΔIa, SLIds1, SLIds2) is influenced neither by the sequence nor the structure of the helical domain adjoining the terminal loop.
TABLE 3.
ITC data for binding of SLI variants to SLV
The VS ribozyme I/V kissing-loop interaction is more stable than predicted
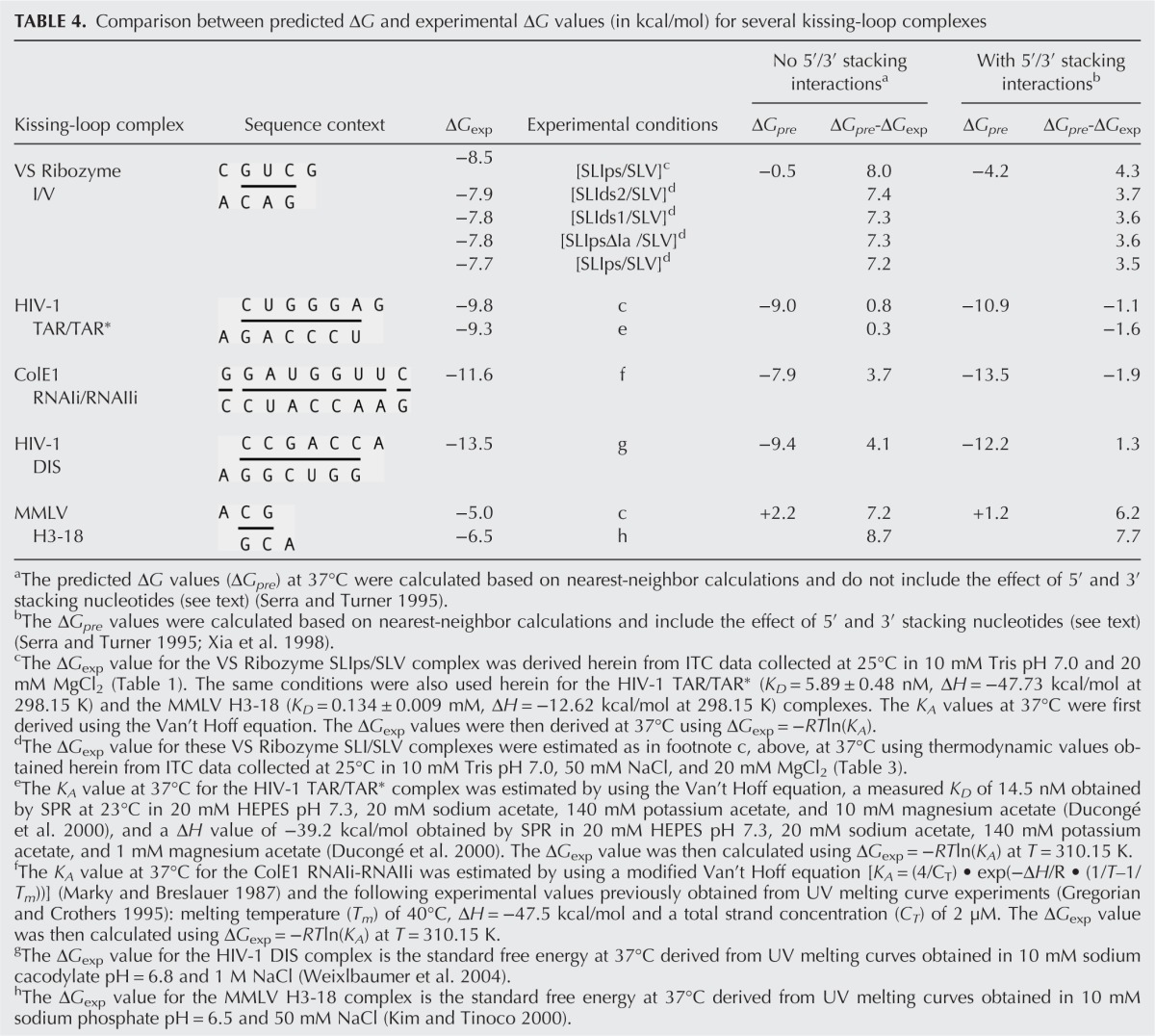
In this study, we obtained KD values ranging from 0.24 to 0.71 μM for binding of nonshiftable SLI variants to SLV (Tables 1, 3), indicating that small variations in experimental conditions (e.g., switching the titrant, addition of 50 mM NaCl) do not significantly affect the measured KD value (less than threefold). The observed KD values reflect the thermodynamic stability of the I/V kissing-loop interaction, which appears to be remarkably high given that only three W-C base pairs are formed at the kissing-loop junction (Rastogi et al. 1996; Bouchard and Legault 2014). We compared the free energy values estimated at 37°C obtained for formation of SLI/SLV complexes with these nonshiftable SLI variants (ΔGITC of −7.7 to −8.5 kcal/mol) (Table 4) to the predicted ΔG value for formation of a comparable RNA duplex at 37°C according to nearest-neighbor calculations (Xia et al. 1998; Lorenz et al. 2006). The SLI/SLV kissing-loop complex was investigated under high-salt conditions (20 mM MgCl2) that are comparable to those used for establishing nearest-neighbor calculations. The three proposed W-C base pairs at the I/V kissing-loop junction form a short A-form helix, as shown in the NMR structure (Bouchard and Legault 2014). According to the nearest-neighbor thermodynamic calculation (Xia et al. 1998), the predicted ΔG (ΔGpre) for the short RNA duplex
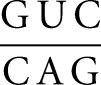 |
composed of the same sequence as the I/V kissing-loop interaction
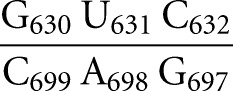 |
is −0.5 kcal/mol under standard conditions (Table 4). The NMR structure of the SLIds2/SLV complex also reveals 5′- and 3′-stacking interactions, which are known to contribute to helix stability (Fig. 1C). They include a 5′-stacking interaction between C629 and G630 as well as 3′-stacking interactions between C632 and G633 in SLI and between C699 and A701 in SLV (Bouchard and Legault 2014). If we incorporate these stacking interactions to our thermodynamic calculations using nearest-neighbor interactions involving 3′ and 5′ unpaired nucleotides (Serra and Turner 1995), a ΔGpre value of −4.2 kcal/mol is obtained for the resulting
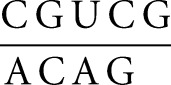 |
duplex (Table 4). Nonetheless, this ΔGpre value still differs by 3.5–4.3 kcal/mol from the range of ΔGITC values associated with formation of the I/V kissing-loop interaction. Thus, the I/V kissing-loop interaction is significantly more stable than a three-base-pair RNA duplex of the same sequence, even when stacking interactions with 5′ and 3′ nucleotides are included in the free energy calculation of the RNA duplex.
TABLE 4.
Comparison between predicted ΔG and experimental ΔG values (in kcal/mol) for several kissing-loop complexes
Comparison with thermodynamic parameters of other kissing-loop interactions
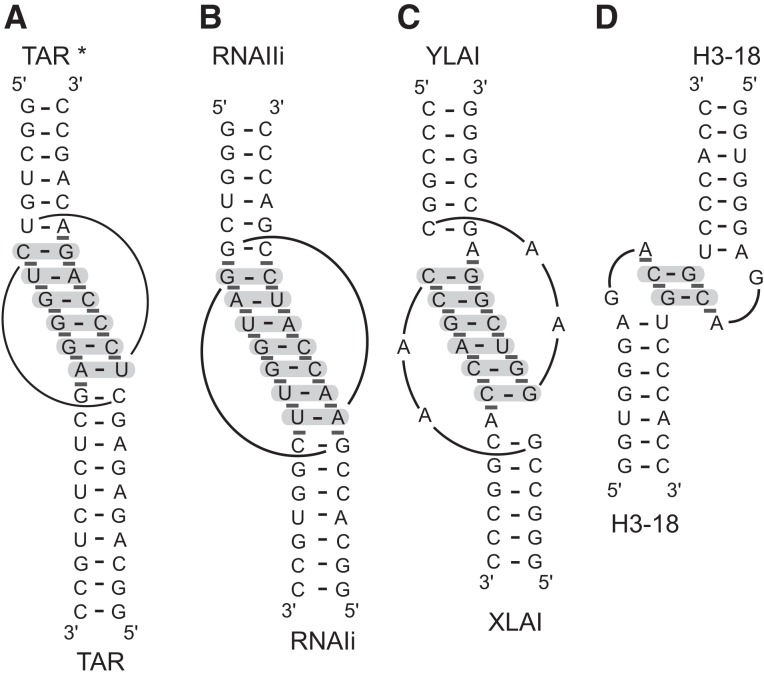
A few other known kissing-loop complexes have been thoroughly investigated both structurally and thermodynamically (Fig. 5), and we examined their stability by comparing their experimental ΔG values obtained from previous studies (ΔGexp) with their ΔG value predicted according to nearest-neighbor calculations (ΔGpre). With the exception of the Moloney murine leukemia virus (MMLV) H3-18 complex, the kissing-loop complexes that we considered have been thermodynamically investigated under high-salt conditions, either at 1 M NaCl or high Mg2+ concentrations (5–10 mM MgCl2) (Table 4), that are comparable to those used for establishing nearest-neighbor calculations (Xia et al. 1998; Lorenz et al. 2006). The ΔGpre values were calculated as for the SLI/SLV complex based on available high-resolution structures (Fig. 5). For the HIV-1 TAR/TAR*, ColE1 RNAIi/RNAIIi, and HIV-1 DIS complexes, the experimental ΔG values (ΔGexp) and the ΔGpre can differ considerably (0.3–4.1 kcal/mol) when only base-pair stacking interactions are taken into account for calculation of the ΔGpre value but generally differ less (<2 kcal/mol) when 5′/3′ stacking interactions are taken into account (Table 4). In contrast, for the MMLV H3-18 complex, which contains only two W-C base pairs at the kissing-loop junction (Fig. 5), ΔGpre-ΔGexp = 7.7 kcal/mol when 5′/3′ stacking interactions are taken into account (Table 4), indicating a more stable interaction than predicted by the thermodynamic model. In summary, except for MMLV H3-18 kissing-loop interaction and the VS ribozyme SLI/SLV interaction, the values of ΔGexp and ΔGpre obtained for the other kissing-loop interactions investigated are similar.
FIGURE 5.
Primary and secondary structures of other kissing-loop interactions considered in this study. (A) The HIV-1 TAR/TAR* kissing-loop complex used for the present study. Similar complexes were used in previous thermodynamic studies of the TAR/TAR* complex (Ducongé et al. 2000; Nair et al. 2000). (B) The ColE1 RNAIi/RNAIIi complex previously thermodynamically characterized by UV melting curves (Gregorian and Crothers 1995). (C) The HIV-1 DIS complex (XLAI/YLAI) previously investigated by UV melting studies (Weixlbaumer et al. 2004). (D) The MMLV H3-18 kissing-loop complex used here and in previous UV melting studies (Kim and Tinoco 2000). In A–D, interactions at the kissing-loop junction are highlighted by pale gray shading for W-C base pair and dark gray boxes for base stacking, according to previously determined three-dimensional structures of the HIV-1 TAR/TAR* (pdb code 1KIS.pdb) (Chang and Tinoco 1997), ColE1 RNAIi/RNAIIi (pdb code 1BJ2) (Lee and Crothers 1998), HIV-1 DIS (pdb code 2B8R) (Ennifar et al. 2001), and MMLV H3-18 kissing-loop complexes (pdb code 1F5U) (Kim and Tinoco 2000).
In order to verify that our ITC conditions were not responsible for the unexpectedly large ΔGpre-ΔGexp value obtained for the I/V kissing-loop interaction, we performed control ITC experiments with the HIV-1 TAR/TAR* and MMLV H3-18 complexes (Fig. 5) under the same conditions as those used for the SLIps/SLV complex (Table 1). For the HIV-1 TAR/TAR* complex, we derived a ΔGITC of −9.8 kcal/mol (Table 4), which is similar to the value of ΔGexp of −9.3 kcal/mol derived from a previous study (Table 4). For the MMLV H3-18 complexes, the use of our ITC conditions yielded a ΔGITC of −5.0 kcal/mol (Table 4), which indicates a weaker interaction than previously established under lower salt conditions (ΔGexp of −6.5 kcal/mol) (Kim and Tinoco 2000). However, with this new experimental ΔGexp value (ΔGITC), the difference with ΔGpre is still fairly high (6.2 kcal/mol). Thus, both the MMLV H3-18 and I/V kissing-loop interactions are substantially more stable than predicted from thermodynamic calculations, and their remarkable stability is not due to exceptional experimental conditions.
Thermodynamic characterization of SLI/ribozyme complexes
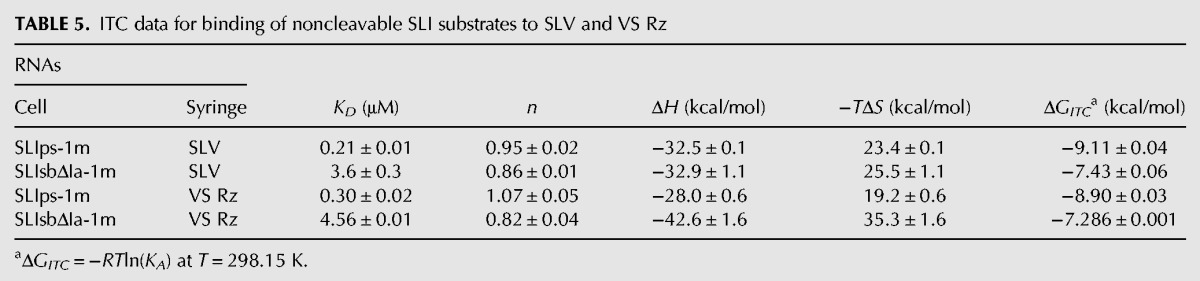
To characterize the thermodynamic parameters for SLI substrate recognition in the context of the trans-cleaving VS ribozyme (VS Rz) (Fig. 1), we selected two SLI substrate variants (Fig. 2A) in which a 2′-O-methyl was introduced at nucleotide G-1 to prevent cleavage by the ribozyme, either a shiftable substrate with no stem Ia (SLIsbΔIa-1m) or a preshifted substrate with stem Ia (SLIps-1m). ITC experiments were performed at 25°C and 20 mM MgCl2 using each of these noncleavable SLI analogs with either SLV or the VS Rz to extract the thermodynamic parameters associated with the resulting complexes (Table 5). The KD values associated with SLV binding to the noncleavable SLI analogs are within threefold of those obtained for SLV binding to the corresponding unmodified SLI variants (Tables 3, 4) (SLIsbΔIa-1m/SLV versus SLIsbΔIa/SLV and SLIps-1m/SLV versus SLIps/SLV), indicating that the 2′-O-methyl modification does not significantly affect the binding affinity. Such small changes in KD values could be simply due to differences in experimental conditions (the titrant is an SLI derivative for Table 3, but SLV for Table 4) as described above for the SLIps/SLV interaction. More importantly, the KD values for binding of the noncleavable SLI analogs to the VS Rz are within 1.5-fold of those obtained for binding of the same SLI analogs to the free SLV (Table 5) (SLIsbΔIa-1m/VS Rz versus SLIsbΔIa-1m/SLV and SLIps-1m/VS Rz versus SLIps-1m/SLV). These results indicate that binding of these SLI variants to either SLV or VS Rz involves similar free energies. Although the 2′-O-methyl modification used for our studies prevents interaction(s) with the 2′-OH at the cleavage site, SLI docking interactions with the VS ribozyme does not contribute significantly to the ground state of the SLI/ribozyme complexes tested here. As a result, the I/V kissing-loop interaction is the key determinant of SLI substrate recognition in the VS ribozyme.
TABLE 5.
ITC data for binding of noncleavable SLI substrates to SLV and VS Rz
DISCUSSION
The I/V kissing-loop interaction is remarkably stable
In the present study, ITC experiments were performed to determine the thermodynamic parameters associated with the I/V kissing-loop interaction of the VS ribozyme. Interestingly, the ITC data revealed that formation of the SLI/SLV complex with several SLI variants is remarkably high, more stable by 7–8 kcal/mol than predicted for a comparable RNA duplex with three Watson–Crick base pairs. As discussed below, this is likely due to extensive stacking at the kissing-loop junction (≥4 kcal/mol), the stabilizing effect of Mg2+ ions, the U-turn loop structures of SLI and SLV, and the ribose-phosphate of the extruded U700 in SLV.
The high stability of the SLI/SLV complex with three W-C base pairs at the kissing-loop junction cannot be predicted from thermodynamic calculations even when both base-pair stacking and 5′/3′ stacking interactions are considered in estimating ΔGpre (ΔGpre-ΔGexp = ∼4 kcal/mol), and this is not a result of exceptional experimental conditions. For other kissing-loop interactions, namely the HIV-1 TAR/TAR*, the ColE1 RNAIi/RNAIIi, and the HIV-1 DIS complexes, the ΔGexp and ΔGpre values differ by <2 kcal/mol when both base-pair stacking and 5′/3′ stacking interactions are considered in estimating ΔGpre. Thus, the rules governing RNA duplex stability can be useful for estimating the stability of these kissing-loop complexes containing 6–7 W-C base pairs when both base-pair stacking and 5′/3′ stacking interactions are taken into account. In contrast, similar thermodynamic calculations were not successful in predicting the high stability of the MMLV H3-18 complex (ΔGpre-ΔGexp = 7–8 kcal/mol), which contains only two W-C base pairs at the kissing-loop junction (Kim and Tinoco 2000; Li et al. 2006; Chen and Garcia 2012). Thus, it appears that kissing-loop complexes with 2–3 Watson–Crick base pairs are generally more stable than predicted. Clearly, other factors than those computed by the thermodynamic model must contribute to the high stability of both the MMLV H3-18 complex and the VS ribozyme I/V kissing-loop complex.
Extensive stacking at the I/V junction
The compact network of hydrogen-bonding and stacking interactions observed at the kissing-loop junction in the NMR structure of the SLIds2/SLV complex (Bouchard and Legault 2014) may not be justly accounted for by the thermodynamic model. In this structure, the kissing-loop junction contains the three predicted W-C base pairs forming a short helix and also two base triples, one at each end of the short helix. For the two base triples, the thermodynamic model takes into account stacking interactions as if they were from unpaired nucleotides at the end of a helix but does not consider the base triples as a whole. Both base triples are part of an intricate structure that extends from the kissing-loop junction toward the stems of SLI and SLV to create a continuously stacked structure. This continuous stacking and several hydrogen bonds not computed by the thermodynamic model likely contribute to the remarkable stability of the SLI/SLV complex.
Role of Mg2+ in the I/V kissing-loop interaction
Our study confirms that Mg2+ is required for formation of a stable I/V kissing-loop interaction, as previously inferred from chemical probing data (Rastogi et al. 1996; Rastogi and Collins 1998; Sood et al. 1998; Hiley and Collins 2001). The KD of the SLIps/SLV interaction could not be measured by ITC at MgCl2 concentrations below 0.5 mM because of the lower affinity of the complex at lower MgCl2 concentrations. Starting at 0.5 mM MgCl2, we observed a decrease in KD with increasing Mg2+ ion concentration to reach a plateau around 20 mM MgCl2. In the interval between 0.5 mM and 40 mM MgCl2, a net uptake of ∼1 Mg2+ ion is associated with formation of the SLI/SLV complex.
It was previously estimated from the variation of Tm values with Mg2+ concentration that formation of kissing-loop complexes derived from the HIV-1 TAR-TAR*, the HIV-1 DIS, and the ColE1 RNAI-RNAII interactions also involves net uptakes of 1–2 Mg2+ (Gregorian and Crothers 1995; Ducongé et al. 2000; Weixlbaumer et al. 2004). Such Mg2+ uptakes are likely necessary to neutralize the high charge density created by phosphate clusters at the loop–loop junction of these complexes (Chang and Tinoco 1997; Lee and Crothers 1998; Ennifar et al. 2001; Draper 2008; Lebars et al. 2008; Van Melckebeke et al. 2008). In the I/V kissing-loop complex, Mg2+ ions likely play a similar role, since several phosphate clusters are found at the kissing-loop junction (Bouchard and Legault 2014). From detailed NMR structural characterization of the free SLI and SLV loops (Flinders and Dieckmann 2001; Campbell and Legault 2005; Campbell et al. 2006) as well as the SLI/SLV complex (Bouchard and Legault 2014), we know that formation of the SLI/SLV complex leads to a more ordered and compact SLI loop, but creates only minor changes in the SLV loop structure (Bouchard and Legault 2014). Thus, there is a net increase in electronegative charge density upon complex formation that is due to both SLI folding into a more compact loop structure and formation of intermolecular interactions at the kissing-loop junction (Bouchard and Legault 2014). Our current structural understanding of Mg2+ interactions associated with formation of the I/V kissing-loop complex is limited to the free forms of each stem–loop; whereas Mg2+ do not grossly affect the disordered loop I (Flinders and Dieckmann 2001), it induces a conformation change in the SLV loop, stabilizing a canonical U-turn fold with four specific Mg2+ binding sites (Campbell and Legault 2005; Campbell et al. 2006). Three of these four preferential sites identified by NMR in the free SLV loop interact with phosphate groups in the major groove and likely play a similar role in the SLI/SLV complex. Future studies aimed at localizing Mg2+ binding sites in the SLI/SLV complex should help better understand how specific Mg2+ binding sites at the kissing-loop junction contribute to the stability of the I/V interaction.
Importance of U turns in the I/V kissing-loop interaction
U-turn loop structures facilitate other RNA interactions, such as codon-anticodon interactions (Grosjean et al. 1976; Moras et al. 1986) and antisense RNA regulation (Franch et al. 1999; Franch and Gerdes 2000). It has been known for some time that trinucleotides bind more tightly to a complementary tRNA anticodon than would be expected for complementary RNA oligonucleotides (Grosjean et al. 1976). In pioneering binding experiments with a kissing-loop complex formed between the complementary anticodons of yeast tRNAPhe and Escherichia coli tRNAGlu, a KA of 3.6 × 105 M−1 and ΔH of −25 kcal/mol were measured at 25°C and 10 mM MgSO4 (ΔGexp at 37°C of −6.88 kcal/mol), which is more stable by 7.7 kcal/mol than predicted for the corresponding RNA duplex when only base-pair stacking interactions are taken into account (ΔGpre of +0.81 kcal/mol) (Grosjean et al. 1976). Interestingly, this ΔGpre-ΔGexp value is very similar to that of 7.2–8.0 kcal/mol obtained for the SLI/SLV complexes (Table 4). In the same study, three potential sources of affinity enhancement were identified: (1) the “loop constraint” or closure of the anticodon sequences into hairpin loops; (2) the stabilizing effect of dangling ends; and (3) the extra stability provided by modified nucleotides that contribute to 3′ stacking. This “loop constraint” that was identified is likely due to the U-turn loop conformation that may promote intermolecular binding in several ways. In the SLI/SLV complex, it promotes stacking of the interacting bases in a near A-form geometry compatible with W-C base-pairing. Such stacking is present in free SLV, but not in free SLI, and thus is increased upon formation of the SLI/SLV complex. In addition, the U turn conformation allows continuous stacking between bases of the kissing loop and adjoining stems in the SLI/SLV complex. Furthermore, the U-turn structure may promote kissing-loop formation by retracting the phosphate backbone away from the interface, thereby reducing electrostatic repulsion. Binding of Mg2+ to the U-turn loop structure, as observed for the U-turn of SLV (Campbell et al. 2006) and anticodon loops (Westhof et al. 1988; Ogle et al. 2001), likely helps further reduce electrostatic repulsion at the loop–loop junction, thereby contributing to the remarkable stability of kissing-loop interactions involving U-turn structures.
Importance of U700 in the I/V kissing-loop interaction
Although the role of loop V nucleotides involved in the U-turn (U696G697A698) and the I/V kissing-loop interaction (G697A698C699) are well understood, that of U700 has remained more elusive (Bouchard et al. 2008). NMR structural studies of SLV demonstrate that upon addition of Mg2+, the largest conformational change in loop V involves U700, which becomes completely extruded from the SLV loop fold (Campbell and Legault 2005; Campbell et al. 2006). In our previous enzymatic study of SLV loop variants (Bouchard et al. 2008), modification of U700 by a C, A, or G had negligible effects on ribozyme trans cleavage activity (kcat/KM), whereas deletion of U700 significantly reduced the kcat/KM. Similarly, ITC investigations of the I/V kissing-loop interaction using isolated stem–loops demonstrate that a U700C modification does not significantly affect the affinity of the SLI/SLV interaction, whereas deletion of U700 significantly reduces the affinity of this interaction. Thus, the role of U700 is confined to the I/V kissing-loop interaction and likely not other aspects of the cleavage reaction.
Both thermodynamic and kinetic data indicate that it is the ribose phosphate of the extruded U700 residue and not its nucleobase that contributes to the stability of the I/V kissing-loop interaction. In agreement with these results, the U700 base is disordered in the structure of the SLI/SLV complex and does not appear to form any stable interactions with SLI (Bouchard and Legault 2014). Interestingly, a comparison between the thermodynamic parameters of the wild-type SLV and the ΔU700 variant indicates that the free energy loss of ∼3 kcal/mol is due to both unfavorable entropic (1.3 kcal/mol) and enthalpic changes (1.9 kcal/mol) (Table 1). As previously suggested, U700 could provide an entropic advantage for substrate recognition by increasing the loop dynamics and/or by serving as a spacer to increase the flexibility of the loop (Bouchard et al. 2008). Such loop dynamics may contribute the necessary plasticity within the SLV loop structure to optimize the intermolecular contacts upon formation of the kissing-loop interaction (Bouchard and Legault 2014). In addition, the ribose-phosphate backbone of U700 possibly contributes to the free enthalpy of the SLI/SLV complex through hydrogen bonding between one of the two nonbridging phosphate oxygens of U700 and an amino proton of A701, as these atoms are relatively close to another (3.0 ± 0.4 Å in the 20 lowest-energy structures) in the NMR structures (Fig. 1D; Bouchard and Legault 2014). This interaction with the phosphate of U700 likely stabilizes the A14V-U8V-C10I base triple at the kissing-loop junction (Fig. 1D). The ribose-phosphate of U700 could also contribute to the enthalpic stability of the SLI/SLV complex through its interaction with a Mg2+ ion (Bouchard et al. 2008). This possibility is supported by our previous observations that the structure of the free SLV loop is stabilized by Mg2+ ions and that a Mg2+ ion directly coordinates the 5′–phosphate of U700 (Campbell et al. 2006). Although Mg2+ ion binding sites have not been mapped within the SLI/SLV complex, the SLV loop within this complex adopts a structure that resembles its free conformation in the presence of Mg2+ ions (Bouchard and Legault 2014). Taking into account these considerations, it is likely that the Mg2+-U700 5′-phosphate interaction somehow contributes to the stability of the complex. Thus, the ribose-phosphate of U700 likely contributes in several different ways to the remarkable stability of the I/V kissing-loop interaction.
Preshifting of the SLI substrate enhances its affinity for SLV
The SLI/SLV complex is highly stable when formed with preshifted substrates, but less so when formed with shiftable substrates. Using SLI substrates lacking stem Ia, we measured an increase of one order of magnitude in KD for the SLI/SLV complex when replacing a preshifted SLI (SLIpsΔIa) by a comparable shiftable SLI (SLIsbΔIa), indicating that shifting the SLI helix in this context entails an energetic cost of 1.8 kcal/mol. These results are consistent with a previous study in which KD values were measured for several VS ribozyme/SLI complexes (Zamel and Collins 2002). Two such complexes were formed using so-called “opened-loop” SLI products that are similar to SLIpsΔIa and SLIsbΔIa because they are devoid of stem Ia and contain a G627A mutation in the loop (equivalent to A7 in SLIpsΔIa and SLIsbΔIa) (Fig. 2A). Using these opened-loop SLI RNAs, an increase of one order of magnitude in KD was measured for the VS ribozyme/SLI interaction when replacing the preshifted SLI by the comparable shiftable SLI (Zamel and Collins 2002). The concordance with our results is particularly noteworthy given that these VS ribozyme/SLI complexes were artificially stabilized by an intermolecular helix (Zamel and Collins 2002). Moreover, it confirms that only SLV and not other parts of the VS ribozyme contribute to shifting the SLI helix upon SLI binding.
The energetic cost for shifting the SLI helix further increases when forming the SLI/SLV complex in the presence of stem Ia. Using shiftable SLI substrates, we observed a fivefold decrease in KD (ΔΔG = 1.0 kcal/mol) for the SLI/SLV complex when replacing a shiftable SLI containing stem Ia (SLIsb) by a comparable SLI lacking stem Ia (SLIsbΔIa). In contrast, no significant change in KD values was observed for SLI/SLV complexes formed by preshifted SLI substrates with (SLIps) or without (SLIpsΔIa) stem Ia. These results are consistent with chemical modification and mutational studies in which the stem Ia was shown to be an inhibitory element of shiftable substrates for catalysis of self-cleaving ribozymes (Beattie and Collins 1997; Rastogi and Collins 1998; McLeod and Lilley 2004; Poon et al. 2006). However, it was not possible to conclude from these studies whether the disruption of helix Ia affected formation of the SLI/SLV interaction or other aspect(s) of catalysis. Particularly, mutations that disrupt stem Ia also increase the size of the linker between SLI and the rest of the ribozyme, and longer linkers have been associated with faster cleavage of self-cleaving ribozymes (Poon et al. 2006). Our thermodynamic results clearly indicate that the presence of stem Ia destabilizes the SLI/SLV interaction of shiftable SLI substrates but has no effect on preshifted SLI substrates. With the SLI and SLV variants used here, formation of the SLI/SLV complex with a preshifted SLI substrate containing stem Ia (SLIps) is more energetically favorable by ∼3 kcal/mol than with a shiftable SLI substrate containing stem Ia. In summary, stem Ia clearly contributes to stabilizing the unshifted inactive conformation and can affect cleavage rates of VS ribozymes when formation of the I/V interaction is rate limiting, as is likely the case for the wild-type self-cleaving ribozyme.
The I/V kissing-loop interaction defines substrate recognition by the VS ribozyme
The strong correlation observed between the thermodynamic and kinetic properties of several SLV variants supports the concept that the I/V kissing-loop interaction is a functionally modular unit for substrate recognition by the VS ribozyme. Structurally, the kissing-loop interaction forms an independent unit that is distinct from residues involved in cleavage chemistry (Hiley and Collins 2001; Lafontaine et al. 2001a, 2002a; Lipfert et al. 2008). For cleavage to occur, the SLI internal loop must dock with the catalytic domain to form the active site (Lafontaine et al. 2001b, 2002b; Sood and Collins 2002; Jones and Strobel 2003; Zhao et al. 2005; Smith and Collins 2007; Wilson et al. 2007, 2010; Jaikaran et al. 2008), and we now know that such docking does not contribute significantly to ground-state substrate binding at least with the trans SLI substrate/ribozyme system investigated here. Rather, the I/V kissing-loop interaction is thermodynamically dominant for substrate recognition by the VS ribozyme. The remarkable stability of this interaction is modulated by Mg2+ concentration and SLI conformational shifting such that formation of the I/V kissing-loop can consequently directly regulate the VS ribozyme cleavage reaction.
MATERIALS AND METHODS
Preparation of RNA for ITC studies
The RNA hairpins used in this study, including the SLI and SLV variants (Fig. 2) as well as the TAR, TAR*, and Moloney Murine Leukemia virus (MMLV) H3-18 RNAs (Fig. 5), were synthesized in vitro with the T7 RNA polymerase using synthetic DNA templates (Integrated DNA Technologies) and purified as previously described (Campbell and Legault 2005). Two SLI variants (SLIps and SLIsbΔIa) (Fig. 2A) were also chemically synthesized (Integrated DNA Technologies) with a 2′-O-methyl modification at position G-1 (SLIps-1m and SLIsbΔIa-1m) and purified as previously described (Campbell and Legault 2005). The Avapl ribozyme, herein named VS Rz (Fig. 1A), was synthesized from plasmid pAvaplVS derived from plasmid pAvapl (Lacroix-Labonté et al. 2012) to allow synthesis of the ribozyme with its substrate at the 3′ end. The transcription reaction was performed in vitro at 37°C using the pAvaplVS plasmid linearized with EcoRI, the T7 RNA polymerase, unlabeled NTPs, and 25 mM MgCl2. After 3 h, 15 mM MgCl2 was added and the reaction mixture was incubated for 1 h to allow self-cleavage of the VS substrate. The cleavage reaction was stopped by addition of 40 mM EDTA, and then the VS Rz was purified as previously described (Bouchard et al. 2008). Purified RNA samples were transferred to ITC buffers by use of Amicon Ultra-4 3K ultrafiltration devices: ITC buffer A (10 mM Tris pH 7.0) for experiments with SLIps and SLV variants (Fig. 4; Table 1), TAR, TAR*, and MMLV H3-18 RNAs (Table 4) or ITC buffer B (10 mM Tris pH 7.0 and 50 mM NaCl) for experiments with SLV and SLI variants (Table 3) and VS Rz (Table 5). Prior to use, the RNAs were refolded by heating (2 min at 95°C for RNA hairpins or at 37°C for the VS Rz) and cooling in ice water. The RNA samples were subsequently transferred to ITC buffer supplemented with MgCl2, generally 20 mM MgCl2, although other MgCl2 concentrations were used to investigate the Mg2+ ion dependence of the I/V kissing-loop interaction. The pH of the Tris buffer was adjusted at room temperature. RNA concentrations were estimated from UV absorbance at 260 nm (A260) using extinction coefficients (ε260) determined by taking the sum of ε260 values for each nucleotide (Cavaluzzi and Borer 2004) within the RNA sequence and multiplying this sum by a hyperchromicity factor corresponding to the ratio of A260 values before and after Nuclease P1 digestion (Zaug et al. 1988; Legault 1995).
Isothermal titration calorimetry (ITC) studies
ITC measurements were performed at 25°C using a VP-ITC or ITC200 Microcalorimeter (GE Healthcare). All solutions used with the VP-ITC were degassed for 10–15 min at 22°C before each experiment. For ITC titrations with SLV variants (Table 1), the concentrations of the SLV titrants in the injection syringe were 100 μM (wild-type, U700C, and U695G SLV RNAs), 300 μM (ΔU700 and C699G SLV RNAs), or 1 mM (ΔU700 and C699G SLV RNAs); and the concentration of SLIps in the cell was 10 times lower than that of the titrants, except for the titration with 300 μM of ΔU700 and C699G SLV RNAs in which 20 μM of SLIps was used in the cell. For ITC titrations with SLI variants (Table 3), the concentrations of the SLI titrants in the syringe were 100 μM (SLIpsΔIa and SLIds1 RNAs), 200 μM (SLIsbΔIa, SLIps, SLIsb, and SLIds2 RNAs), or 600 μM (SLIsb RNA); and the concentration of SLV in the cell was 10 times lower than that of the titrants. For ITC titrations with the VS ribozyme catalytic domain (VS Rz) (Table 5), the concentration of VS Rz in the syringe was 100 μM, and the concentration of noncleavable SLI (SLIsbΔIa-1m and SLIps-1m) (Table 5) in the cell was 10 μM. For ITC titration studies of the TAR/TAR* kissing-loop interaction (Table 4), the concentration of TAR RNA in the syringe was 50 μM, and the concentration of TAR* RNA in the cell was 5 μM. For ITC studies of the MMLV H3-18 kissing-loop dimer (Table 4), dilution experiments were performed with 500 μM MMLV H3-18 hairpin in the syringe and buffer only in the cell.
Most ITC titrations were collected with c values ≥1 (c = [RNA in the cell]/KD; c values between 2 and 850), from which accurate thermodynamic parameters were extracted (Wiseman et al. 1989; Turnbull and Daranas 2003). For titration data collected with c values <1 (c = 0.3 and c = 0.6 for complexes SLIps/SLVC699G and SLIsb/SLV, respectively), the stoichiometry (n) was fixed to 1, such that only KA and ΔG could be accurately determined (Turnbull and Daranas 2003). For each titration experiment, a control experiment was performed under the same conditions, except that only buffer was used in the cell. The control experiment was subtracted from the titration experiment in order to eliminate the effect of diluting the titrant. Data collected for each titration experiment were then fit to a single binding site model or to a dimer-monomer dissociation model (for MMLV H3-18 only) using equations from the ORIGIN software package version 7.0 (OriginLab Corporation). The quality of the fits was obtained from the square of the correlation coefficient (R2), and in all cases R2 was ≥0.99. The values of n, KA (or KD = 1/KA), and ΔH were directly obtained from these fits. The free energy (ΔGITC) was calculated as ΔGITC = −RTlnKA, where R denotes the gas constant (R = 1.987 cal • mol−1 • K−1) and T is the temperature. The values of TΔS were derived from that of ΔG and ΔH, given that ΔG =ΔH − TΔS. At least two ITC experiments were collected for each measurement. The reported values and errors on the thermodynamic parameters are, respectively, the average and standard deviations from these multiple measurements.
ACKNOWLEDGMENTS
We thank Elyse Benoit, Dominique Chaussé, Étienne Beaulieu, and Pierre Dagenais for RNA preparation; Ao Yang for help with ITC data analysis; and James G. Omichinski for critical reading of the manuscript. This work was supported by the Canadian Institutes of Health Research (CIHR) to P.L. [MOP-86502]. P.B. was supported by a Frederick Banting and Charles Best Canada Graduate Scholarship PhD scholarship from CIHR and graduate scholarships from the Université de Montréal. The ITC instruments were purchased with funds from the Natural Sciences and Engineering Research Council of Canada. P.L. holds a Canada Research Chair in Structural Biology and Engineering of RNA.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.046144.114.
REFERENCES
- Andersen A, Collins RA 2000. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol Cell 5: 469–478 [DOI] [PubMed] [Google Scholar]
- Andersen AA, Collins RA 2001. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc Natl Acad Sci 98: 7730–7735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT 2005. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology 332: 498–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415 [DOI] [PubMed] [Google Scholar]
- Beattie TL, Collins RA 1997. Identification of functional domains in the self-cleaving Neurospora VS ribozyme using damage selection. J Mol Biol 267: 830–840 [DOI] [PubMed] [Google Scholar]
- Beattie TL, Olive JE, Collins RA 1995. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc Natl Acad Sci 92: 4686–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S, Lafontaine DA 2007. A loop–loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13: 1256–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard P, Legault P 2014. Structural insights into substrate recognition by the Neurospora Varkud satellite ribozyme: importance of U-turns at the kissing-loop junction. Biochemistry 53: 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard P, Lacroix-Labonté J, Desjardins G, Lampron P, Lisi V, Lemieux S, Major F, Legault P 2008. Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme. RNA 14: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S 2007. Regulatory mechanisms used by cis-encoded antisense RNAs. Curr Opin Microbiol 10: 102–109 [DOI] [PubMed] [Google Scholar]
- Brunel C, Marquet R, Romby P, Ehresmann C 2002. RNA loop–loop interactions as dynamic functional motifs. Biochimie 84: 925–944 [DOI] [PubMed] [Google Scholar]
- Campbell DO, Legault P 2005. NMR structure of the Varkud satellite ribozyme stem–loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry 44: 4157–4170 [DOI] [PubMed] [Google Scholar]
- Campbell DO, Bouchard P, Desjardins G, Legault P 2006. NMR structure of Varkud satellite ribozyme stem–loop V in the presence of magnesium ions and localization of metal-binding sites. Biochemistry 45: 10591–10605 [DOI] [PubMed] [Google Scholar]
- Cavaluzzi MJ, Borer PN 2004. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res 32: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KY, Tinoco I Jr 1997. The structure of an RNA “kissing” hairpin complex of the HIV TAR hairpin loop and its complement. J Mol Biol 269: 52–66 [DOI] [PubMed] [Google Scholar]
- Chen AA, Garcia AE 2012. Mechanism of enhanced mechanical stability of a minimal RNA kissing complex elucidated by nonequilibrium molecular dynamics simulations. Proc Natl Acad Sci 109: E1530–E1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YI, Martick M, Lares M, Kim R, Scott WG, Kim SH 2008. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol 6: e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña M, Gago S, Flores R 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 22: 5561–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE 2008. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys J 95: 5489–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducongé F, Di Primo C, Toulmé JJ 2000. Is a closing “GA pair” a rule for stable loop-loop RNA complexes? J Biol Chem 275: 21287–21294 [DOI] [PubMed] [Google Scholar]
- Dufour D, de la Peña M, Gago S, Flores R, Gallego J 2009. Structure–function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop–loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res 37: 368–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P 2001. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat Struct Biol 8: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Flinders J, Dieckmann T 2001. A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA. J Mol Biol 308: 665–679 [DOI] [PubMed] [Google Scholar]
- Franch T, Gerdes K 2000. U-turns and regulatory RNAs. Curr Opin Microbiol 3: 159–164 [DOI] [PubMed] [Google Scholar]
- Franch T, Petersen M, Wagner EGH, Jacobsen JP, Gerdes K 1999. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J Mol Biol 294: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT 2008. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283: 22347–22351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorian RS Jr, Crothers DM 1995. Determinants of RNA hairpin loop-loop complex stability. J Mol Biol 248: 968–984 [DOI] [PubMed] [Google Scholar]
- Grosjean H, Söll DG, Crothers DM 1976. Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J Mol Biol 103: 499–519 [DOI] [PubMed] [Google Scholar]
- Guo L, Allen EM, Miller WA 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell 7: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Hiley SL, Collins RA 2001. Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J 20: 5461–5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiley SL, Sood VD, Fan J, Collins RA 2002. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J 21: 4691–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Mitchell GT, Gendron P, Major F, Andersen AA, Collins RA, Legault P 2003. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc Natl Acad Sci 100: 7003–7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaikaran D, Smith MD, Mehdizadeh R, Olive J, Collins RA 2008. An important role of G638 in the cis-cleavage reaction of the Neurospora VS ribozyme revealed by a novel nucleotide analog incorporation method. RNA 14: 938–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FD, Strobel SA 2003. Ionization of a critical adenosine residue in the Neurospora Varkud satellite ribozyme active site. Biochemistry 42: 4265–4276 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10: 708–712 [DOI] [PubMed] [Google Scholar]
- Kim CH, Tinoco I Jr 2000. A retroviral RNA kissing complex containing only two G-C base pairs. Proc Natl Acad Sci 97: 9396–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix-Labonté J, Girard N, Lemieux S, Legault P 2012. Helix-length compensation studies reveal the adaptability of the VS ribozyme architecture. Nucleic Acids Res 40: 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Norman DG, Lilley DM 2001a. Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J 20: 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Wilson TJ, Norman DG, Lilley DM 2001b. The A730 loop is an important component of the active site of the VS ribozyme. J Mol Biol 312: 663–674 [DOI] [PubMed] [Google Scholar]
- Lafontaine DA, Norman DG, Lilley DM 2002a. The global structure of the VS ribozyme. EMBO J 21: 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Wilson TJ, Zhao ZY, Lilley DMJ 2002b. Functional group requirements in the probable active site of the VS ribozyme. J Mol Biol 323: 23–34 [DOI] [PubMed] [Google Scholar]
- Lebars I, Legrand P, Aimé A, Pinaud N, Fribourg S, Di Primo C 2008. Exploring TAR–RNA aptamer loop–loop interaction by X-ray crystallography, UV spectroscopy and surface plasmon resonance. Nucleic Acids Res 36: 7146–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJ, Crothers DM 1998. The solution structure of an RNA loop–loop complex: the ColE1 inverted loop sequence. Structure 6: 993–1005 [DOI] [PubMed] [Google Scholar]
- Legault P 1995. Structural studies of ribozymes by heteronuclear NMR spectroscopy. University of Colorado at Boulder, Boulder, CO [Google Scholar]
- Leipply D, Lambert D, Draper DE 2009. Ion-RNA interactions: thermodynamic analysis of the effects of mono- and divalent ions on RNA conformational equilibria. Methods Enzymol 469B: 433–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA 2006. Folding of the adenine riboswitch. Chem Biol 13: 857–868 [DOI] [PubMed] [Google Scholar]
- Li PT, Bustamante C, Tinoco I Jr 2006. Unusual mechanical stability of a minimal RNA kissing complex. Proc Natl Acad Sci 103: 15847–15852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DM 2008. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 16: 1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Piganeau N, Schroeder R 2006. Stabilities of HIV-1 DIS type RNA loop–loop interactions in vitro and in vivo. Nucleic Acids Res 34: 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky LA, Breslauer KJ 1987. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers 26: 1601–1620 [DOI] [PubMed] [Google Scholar]
- McLeod AC, Lilley DM 2004. Efficient, pH-dependent RNA ligation by the VS ribozyme in trans. Biochemistry 43: 1118–1125 [DOI] [PubMed] [Google Scholar]
- Michiels PJA, Schouten CHJ, Hilbers CW, Heus HA 2000. Structure of the ribozyme substrate hairpin of Neurospora VS RNA: a close look at the cleavage site. RNA 6: 1821–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D, Dock AC, Dumas P, Westhof E, Romby P, Ebel JP, Giege R 1986. Anticodon–anticodon interaction induces conformational changes in tRNA: yeast tRNAAsp, a model for tRNA–mRNA recognition. Proc Natl Acad Sci 83: 932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair TM, Myszka DG, Davis DR 2000. Surface plasmon resonance kinetic studies of the HIV TAR RNA kissing hairpin complex and its stabilization by 2-thiouridine modification. Nucleic Acids Res 28: 1935–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Poon AH, Olive JE, McLaren M, Collins RA 2006. Identification of separate structural features that affect rate and cation concentration dependence of self-cleavage by the Neurospora VS ribozyme. Biochemistry 45: 13394–13400 [DOI] [PubMed] [Google Scholar]
- Quigley GJ, Rich A 1976. Structural domains of transfer RNA molecules. Science 194: 796–806 [DOI] [PubMed] [Google Scholar]
- Rastogi T, Collins RA 1998. Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J Mol Biol 277: 215–224 [DOI] [PubMed] [Google Scholar]
- Rastogi T, Beattie TL, Olive JE, Collins RA 1996. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J 15: 2820–2825 [PMC free article] [PubMed] [Google Scholar]
- Reymond C, Bisaillon M, Perreault JP 2009. Monitoring of an RNA multistep folding pathway by isothermal titration calorimetry. Biophys J 96: 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim NN, Feig AL 2009. Isothermal titration calorimetry of RNA. Methods 47: 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ 2004. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol 11: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ 2008. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455: 1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra MJ, Turner DH 1995. Predicting thermodynamic properties of RNA. Methods Enzymol 259: 242–261 [DOI] [PubMed] [Google Scholar]
- Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci 91: 4945–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Collins RA 2007. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc Natl Acad Sci 104: 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood VD, Collins RA 2002. Identification of the catalytic subdomain of the VS ribozyme and evidence for remarkable sequence tolerance in the active site loop. J Mol Biol 320: 443–454 [DOI] [PubMed] [Google Scholar]
- Sood VD, Beattie TL, Collins RA 1998. Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J Mol Biol 282: 741–750 [DOI] [PubMed] [Google Scholar]
- Turnbull WB, Daranas AH 2003. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc 125: 14859–14866 [DOI] [PubMed] [Google Scholar]
- Van Melckebeke H, Devany M, Di Primo C, Beaurain F, Toulmé JJ, Bryce DL, Boisbouvier J 2008. Liquid-crystal NMR structure of HIV TAR RNA bound to its SELEX RNA aptamer reveals the origins of the high stability of the complex. Proc Natl Acad Sci 105: 9210–9215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EG, Simons RW 1994. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol 48: 713–742 [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Werner A, Flamm C, Westhof E, Schroeder R 2004. Determination of thermodynamic parameters for HIV DIS type loop–loop kissing complexes. Nucleic Acids Res 32: 5126–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Dumas P, Moras D 1988. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr A 44 (Pt 2):112–123 [PubMed] [Google Scholar]
- Wilson TJ, McLeod AC, Lilley DM 2007. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J 26: 2489–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Li NS, Lu J, Frederiksen JK, Piccirilli JA, Lilley DM 2010. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc Natl Acad Sci 107: 11751–11756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin LN 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem 179: 131–137 [DOI] [PubMed] [Google Scholar]
- Xia T, SantaLucia J Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry 37: 14719–14735 [DOI] [PubMed] [Google Scholar]
- Zamel R, Collins RA 2002. Rearrangement of substrate secondary structure facilitates binding to the Neurospora VS ribozyme. J Mol Biol 324: 903–915 [DOI] [PubMed] [Google Scholar]
- Zamel R, Poon A, Jaikaran D, Andersen A, Olive J, De Abreu D, Collins RA 2004. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc Natl Acad Sci 101: 1467–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Grosshans CA, Cech TR 1988. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme–substrate complexes. Biochemistry 27: 8924–8931 [DOI] [PubMed] [Google Scholar]
- Zhao ZY, McLeod A, Harusawa S, Araki L, Yamaguchi M, Kurihara T, Lilley DM 2005. Nucleobase participation in ribozyme catalysis. J Am Chem Soc 127: 5026–5027 [DOI] [PubMed] [Google Scholar]