The Lsm1-7-Pat1 complex enhances mRNA decapping in vivo. This study examines the RNA binding activity of Lsm1-7 and Pat1 separately and as a complex. It is demonstrated that both the Lsm complex and Pat1 display very low RNA binding by themselves, but in combination, the complex has robust RNA binding activity.
Keywords: decapping, Lsm1-7–Pat1 complex, Pat1, Sm-like proteins, mRNA decay
Abstract
A major mRNA decay pathway in eukaryotes is initiated by deadenylation followed by decapping of the oligoadenylated mRNAs and subsequent 5′-to-3′ exonucleolytic degradation of the capless mRNA. In this pathway, decapping is a rate-limiting step that requires the hetero-octameric Lsm1-7–Pat1 complex to occur at normal rates in vivo. This complex is made up of the seven Sm-like proteins, Lsm1 through Lsm7, and the Pat1 protein. It binds RNA and has a unique binding preference for oligoadenylated RNAs over polyadenylated RNAs. Such binding ability is crucial for its mRNA decay function in vivo. In order to determine the contribution of Pat1 to the function of the Lsm1-7–Pat1 complex, we compared the RNA binding properties of the Lsm1-7 complex purified from pat1Δ cells and purified Pat1 fragments with that of the wild-type Lsm1-7–Pat1 complex. Our studies revealed that both the Lsm1-7 complex and purified Pat1 fragments have very low RNA binding activity and are impaired in the ability to recognize the oligo(A) tail on the RNA. However, reconstitution of the Lsm1-7–Pat1 complex from these components restored these abilities. We also observed that Pat1 directly contacts RNA in the context of the Lsm1-7–Pat1 complex. These studies suggest that the unique RNA binding properties and the mRNA decay function of the Lsm1-7–Pat1 complex involve cooperation of residues from both Pat1 and the Lsm1-7 ring. Finally our studies also revealed that the middle domain of Pat1 is essential for the interaction of Pat1 with the Lsm1-7 complex in vivo.
INTRODUCTION
mRNA decay is an important control point in the regulation of gene expression. Modulation of mRNA turnover rates plays a key role in determining the response time of the gene expression changes elicited by various external stimuli and stresses (Perez-Ortin et al. 2007; Shalem et al. 2008; Elkon et al. 2010; Rabani et al. 2011). As a result, mRNA stability turns out to be as significant a determinant of global changes in mRNA abundance as transcription rate (Fan et al. 2002; Garcia-Martinez et al. 2004; Cheadle et al. 2005; Rabani et al. 2011).
mRNA decay pathways and machinery are highly conserved in all eukaryotes. Major pathways for bulk mRNA decay initiate with deadenylation; afterward, the oligoadenylated mRNA is degraded in a 3′-to-5′ exonucleolytic fashion by the exosome (3′-to-5′ pathway) or decapped and then degraded in a 5′-to-3′ exonucleolytic manner (5′-to-3′ pathway) by Xrn1 (Balagopal et al. 2012; Parker 2012; Wu and Brewer 2012). In the 5′-to-3′ pathway, decapping is dependent on prior deadenylation, and it is a tightly controlled rate-limiting step carried out by the Dcp2-Dcp1 decapping enzyme (Ling et al. 2011; Parker 2012). This step is positively and negatively influenced by multiple factors (Ling et al. 2011; Parker 2012). The cytoplasmically localized hetero-octameric Lsm1-7–Pat1 complex made up of the seven Sm-like proteins (Lsm1 through Lsm7) and the Pat1 protein conserved in all eukaryotes is a key activator of the decapping enzyme and is essential for the normal rates of mRNA decapping in vivo (Bouveret et al. 2000; Tharun et al. 2000; Tharun 2009b; Totaro et al. 2011). This complex also protects mRNA 3′-ends from trimming (Boeck et al. 1998; He and Parker 2001; Tharun et al. 2005; Tharun 2009a). Consistent with its role in activation of deadenylation-dependent decapping in the 5′-to-3′ pathway and in the protection of mRNA 3′-ends, the Lsm1-7–Pat1 complex purified from yeast has RNA binding activity, exhibits a unique binding preference for oligoadenylated RNAs over polyadenylated RNAs, and binds at the 3′-ends of RNAs (Chowdhury et al. 2007). Further, it selectively binds at the 3′-ends of deadenylated mRNAs in vivo (Tharun et al. 2000; Tharun and Parker 2001; Mitchell et al. 2013). Importantly, the ability of this complex to recognize the oligoadenylated status of RNAs is essential for its function in mRNA decay in vivo and is dependent on the RNA binding residues in the Sm domain of the Lsm1 subunit (Chowdhury and Tharun 2008). Nevertheless, after binding the mRNA, to activate decapping, this complex needs to facilitate additional post-binding events that are not yet fully understood (Chowdhury and Tharun 2009). This complex shares six of its subunits (Lsm2 through Lsm7) with the Lsm2-8 complex, which is localized in the nucleus and is not involved in cytoplasmic mRNA decay (Bouveret et al. 2000; Tharun et al. 2000; Ingelfinger et al. 2002; Tharun 2009b). Thus Lsm1 is a key distinguishing subunit of this complex. The Lsm1-7, Lsm2-8, and Sm complexes have a very similar doughnut-shaped quaternary structure wherein the individual Lsm/Sm subunits are organized relative to each other in an analogous fashion (Kambach et al. 1999; Sharif and Conti 2013; Zhou et al. 2013, 2014). A unique feature of the Lsm1-7 complex is that the extended C-terminal domain of Lsm1 forms a long α-helix that crosses the entire diameter of the doughnut and partially blocks the central hole (Sharif and Conti 2013; Zhou et al. 2014).
Apart from Lsm1, another key subunit of the Lsm1-7–Pat1 complex is Pat1. Like the Lsm1 through Lsm7 proteins, Pat1 is also well conserved in all eukaryotes. While a single Pat1 protein is present in yeast and invertebrates (Bonnerot et al. 2000; Bouveret et al. 2000; Tharun et al. 2000; Eulalio et al. 2007; Gallo et al. 2008), vertebrates have two paralogs, Pat1a and Pat1b (Rother et al. 1992; Scheller et al. 2007; Marnef and Standart 2010; Marnef et al. 2010). Pat1 has been implicated in several functions, and not all of them are related to its association with the Lsm1-7 complex. In yeast, Pat1, but not Lsm1, associates with a pool of mRNPs that is also bound to eIF4G, eIF4E, and Pab1 (Tharun and Parker 2001). Also yeast Pat1 functions in translational repression along with Dhh1, and therefore, translation rates do not decrease in dhh1Δ pat1Δ cells upon glucose starvation unlike in wild-type and lsm1Δ cells (Holmes et al. 2004; Coller and Parker 2005). Thus, while the Lsm1-7–Pat1 complex facilitates decapping of mRNPs, Pat1 by itself seems to play additional roles at earlier steps involving translational repression and mRNP rearrangement that set the stage for decapping. Translational repression activity has also been noted for Pat1 homologs of Xenopus but not humans (Marnef et al. 2010; Ozgur et al. 2010). Pat1 has an important role in p-body assembly as revealed by a drop in the number and size of p-bodies in pat1Δ yeast, while p-bodies are increased in lsm1Δ yeast (Teixeira and Parker 2007). Consistently, while Pat1 is absolutely required for Lsm1 to enter p-bodies, in lsm1Δ cells Pat1 is still present in p-bodies (Teixeira and Parker 2007). Overexpression of Pat1 induces p-body formation in both yeast and human cells (Coller and Parker 2005; Ozgur et al. 2010). Pat1 facilitates p-body assembly, probably by acting as a scaffolding protein since it interacts with numerous decay factors, including the decapping enzyme, decapping activators, the 5′-to-3′ exonuclease Xrn1, and components of the Ccr4 deadenylase complex as revealed by studies in human cells, Drosophila, and yeast (Braun et al. 2010; Haas et al. 2010; Nissan et al. 2010; Ozgur et al. 2010). Human and yeast Pat1 also have the potential to self-associate (Nissan et al. 2010; Ozgur et al. 2010), a property that may facilitate p-body assembly. Finally, yeast Pat1 is a substrate of cAMP-dependent protein kinase (PKA)–mediated phosphorylation, and such phosphorylation affects its ability to promote p-body formation (Ramachandran et al. 2011; Shah et al. 2013).
Pat1 has also been implicated in nuclear functions. Both Lsm1 and Pat1 are nucleocytoplasmic shuttling proteins that get exported by Crm1 (Marnef et al. 2012; Haimovich et al. 2013). Yeast Pat1 affects tRNA subcellular distribution dynamics in a manner that is different from that of Lsm1 (Hurto and Hopper 2011). Yeast Pat1 associates with topoisomerase-II and CEN DNA and has been implicated in rDNA locus stability and chromosome transmission fidelity (Wang et al. 1996, 1999; Mishra et al. 2013). Recent studies suggest that Pat1, Lsm1, and other decay factors associate with chromatin and influence transcription such that transcription and mRNA decay are coupled (Haimovich et al. 2013). Finally, human Pat1b localizes to splicing speckles in the nucleus (Marnef et al. 2012).
Pat1 proteins are large, with the yeast and human orthologs being 796 and 770 residues long, respectively. They do not have any readily recognizable motifs (Scheller et al. 2007; Marnef and Standart 2010; Marnef et al. 2010; Ozgur et al. 2010). The region close to the N terminus of Pat1 proteins is sufficient for interaction with Dhh1 orthologs in yeast, Drosophila, and humans (Braun et al. 2010; Haas et al. 2010; Ozgur et al. 2010; Sharif et al. 2013). The middle and C-terminal segments contain the polypeptide regions that are necessary for supporting decay, mediating interaction with other decay factors, including the Lsm1-7 complex, localization to p-bodies, etc. (Braun et al. 2010; Haas et al. 2010; Marnef and Standart 2010; Ozgur et al. 2010). Structural studies carried out using the C-terminal segment of Pat1 have revealed that this segment interacts with the Lsm2 and Lsm3 subunits in the Lsm1-7 ring (Sharif and Conti 2013; Wu et al. 2014).
Both human Pat1b and Drosophila HPat trigger deadenylation and decapping when tethered to mRNA (Haas et al. 2010; Ozgur et al. 2010). They also promote the decapping of mRNA whose decay is mediated by tethered GW182 (Braun et al. 2010; Haas et al. 2010). However, in yeast, neither LSM1 nor PAT1 seems to affect deadenylation (Hatfield et al. 1996; Boeck et al. 1998; Schwartz and Parker 2000). Nevertheless, consistent with both Pat1 and Lsm1 being part of the same complex (Lsm1-7–Pat1 complex), both pat1Δ and lsm1Δ yeast cells are defective in mRNA decapping and 3′-end protection as evident from the accumulation of deadenylated 3′-trimmed capped mRNA species in those cells (Hatfield et al. 1996; Boeck et al. 1998; Bouveret et al. 2000; Tharun et al. 2000, 2005; He and Parker 2001). Further, truncated versions of yeast Pat1 that fail to interact with Lsm1 in a two-hybrid assay are also impaired in their ability to support mRNA decay in vivo (Pilkington and Parker 2008). Collaboration of Lsm1 and Pat1 in facilitating the final steps of decapping is also evident from the fact that both of them are needed for the decay of an mRNA even when that mRNA is translationally impaired via insertion of the secondary structure in the 5′ UTR, while Dhh1 is dispensable for such decay because Dhh1 primarily functions in the earlier stages of decapping (Coller and Parker 2005). In any case, while these observations and the physical association of Pat1 with the Lsm1-7 complex indicate that Pat1 and Lsm1-7 complex function together in decapping, the nature of Pat1's contribution to the functioning of the Lsm1-7–Pat1 complex is not known. To gain insight into this issue, we have addressed how Pat1 affects the RNA binding properties of the Lsm1-7–Pat1 complex.
RESULTS
Lsm1-7 complex assembles in pat1Δ cells
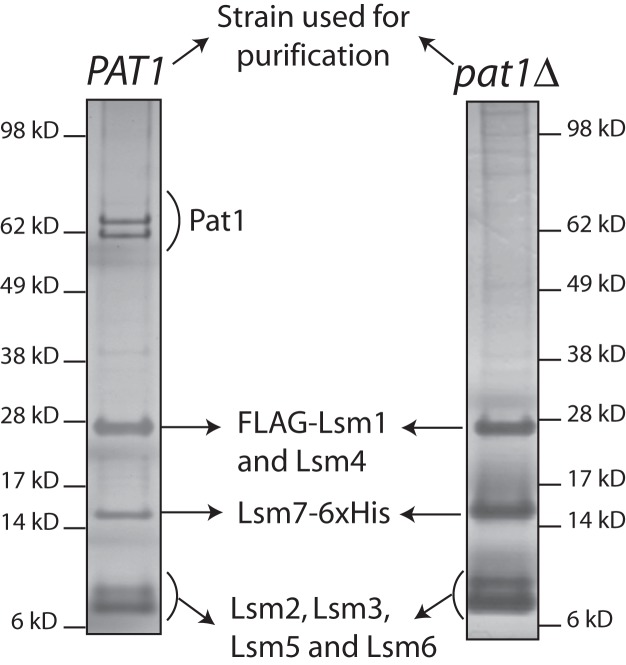
In order to determine the contribution of Pat1 to the RNA binding activity of the Lsm1-7–Pat1 complex, we purified the Lsm1-7 heptamer from a pat1Δ; FLAG-LSM1; LSM7-6xHis yeast strain using the tandem affinity chromatographic strategy we described earlier (Chowdhury et al. 2007; Tharun 2008), which employs the anti-Flag antibody matrix and the Ni-NTA matrix in the first and second affinity purification steps, respectively (targeting Flag-LSM1 and LSM7-6xHis, respectively). Comparison of the SDS–polyacrylamide gel electrophoretic (PAGE) band patterns of the complexes purified from wild-type and pat1Δ cells (Fig. 1) and determination of the tryptic peptide sequences (data not shown) by mass spectrometry analysis of the proteins present in the complex purified from pat1Δ cells revealed that all the seven Lsm proteins (Lsm1 through Lsm7) are present in the complex purified from pat1Δ cells, indicating that Pat1 is not required for the assembly of the Lsm1-7 heptamer. These results are consistent with the observation that the human and yeast Lsm1-7 complexes can be reconstituted in vitro in the absence of Pat1 (Zaric et al. 2005; Sharif and Conti 2013; Zhou et al. 2014). The complex purified from pat1Δ cells will be referred to as Lsm1-7 complex hereafter.
FIGURE 1.
Lsm1-7 complex assembles in a pat1Δ mutant. Lsm1-7–Pat1 complex purified from wild-type cells (left) and the Lsm1-7 complex purified from pat1Δ cells (right) were separated by SDS-PAGE and visualized by silver staining.
Lsm1-7 complex is severely impaired in its RNA binding activity and fails to exhibit a detectable binding preference for oligoadenylated RNA over unadenylated RNA
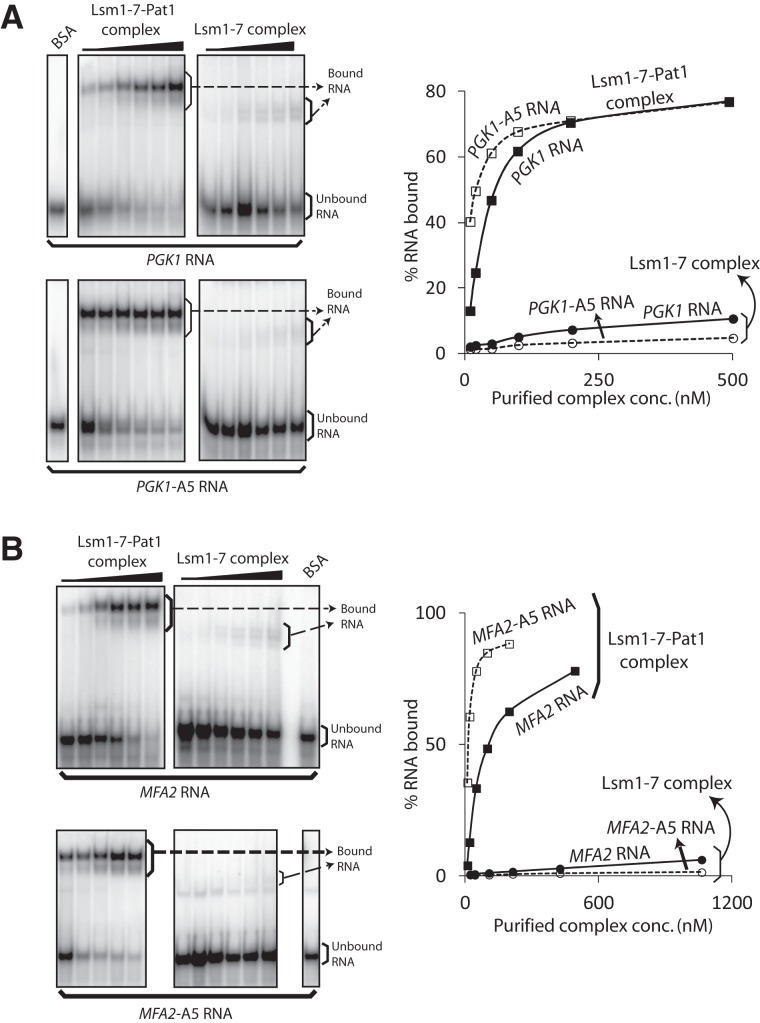
A unique RNA binding property of the Lsm1-7–Pat1 complex purified from wild-type cells is that it has significantly higher affinity for oligoadenylated RNAs over polyadenylated and unadenylated RNAs (Chowdhury et al. 2007). Such ability to recognize the presence of oligo(A) tail on the RNA is essential for mRNA decay in vivo and is readily revealed in gel shift assays (using our standard 42-mer in vitro transcripts as substrates) as a higher binding affinity toward the 3′-penta-adenylated RNA substrate over the unadenylated RNA substrate (Chowdhury et al. 2007; Chowdhury and Tharun 2008, 2009). Therefore, we studied the RNA binding ability of the purified Lsm1-7–Pat1 and Lsm1-7 complexes via gel mobility shift assays using uniformly labeled in vitro transcribed PGK1 RNA (42-mer RNA derived from the 3′ UTR of PGK1) and PGK1-A5 RNA (PGK1 RNA carrying a 3′-A5 tail) (Chowdhury et al. 2007; Chowdhury and Tharun 2008). As seen in Figure 2A, two important observations can be made from these experiments. First, overall, the RNA binding ability of the Lsm1-7 complex is very much lower than that of the wild-type Lsm1-7–Pat1 complex. The gel retarded complex formed by Lsm1-7 complex is of higher mobility than that formed by the Lsm1-7–Pat1 complex. This may partly be to do with the smaller size of the Lsm1-7 complex compared with the wild-type Lsm1-7–Pat1 complex (∼97 kD vs. ∼185 kD). Second, while the Lsm1-7–Pat1 complex clearly exhibited a higher affinity for the PGK1-A5 RNA than the PGK1 RNA as expected and as observed earlier (Chowdhury and Tharun 2009; Chowdhury et al. 2012), such binding preference could not be observed with the Lsm1-7 complex. Very similar results were also observed when the experiments were carried out using the MFA2 and MFA2-A5 RNAs (42-mer RNA derived from the 3′ UTR of MFA2 and the 3′-penta-adenylated version of such RNA) as substrates for gel shift assays (Fig. 2B). Thus, the Pat1 subunit seems to contribute significantly to both the overall RNA binding activity and the ability to recognize the presence of 3′-oligo(A) tail of the Lsm1-7–Pat1 complex.
FIGURE 2.
Lsm1-7 complex is severely impaired in its ability to bind RNA and does not exhibit a binding preference for oligoadenylated RNA. BSA or increasing concentrations of Lsm1-7–Pat1 complex purified from wild-type cells or the Lsm1-7 complex purified from pat1Δ cells (indicated on top in A and B) were subjected to gel shift assays using uniformly radiolabeled PGK1 and PGK1-A5 RNAs (A) or MFA2 and MFA2-A5 RNAs (B). Plots of the percentage of RNA bound vs. the concentration of the complex used are shown on the right of the phosphorimages of the gels. Bands of bound and unbound RNA are labeled.
Pat1 subunit contacts RNA directly
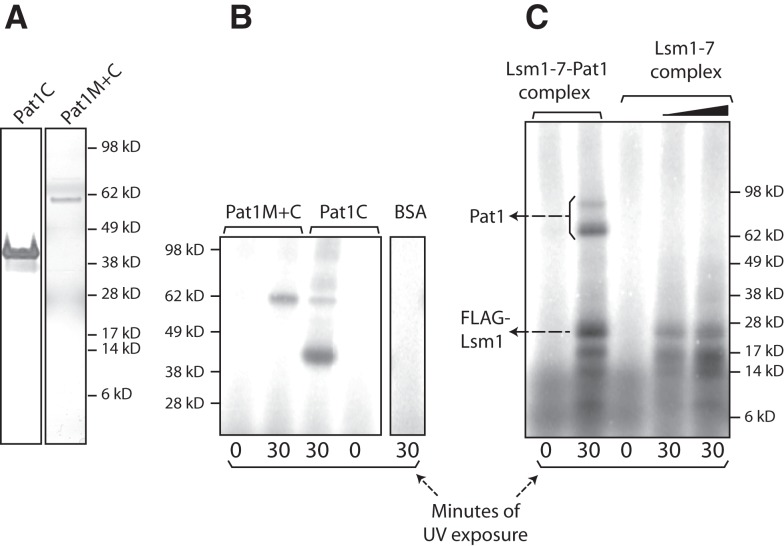
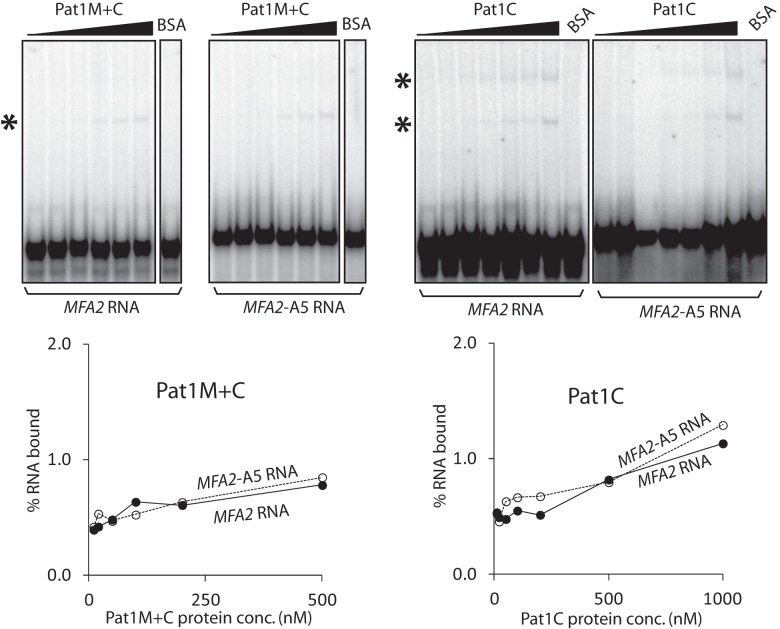
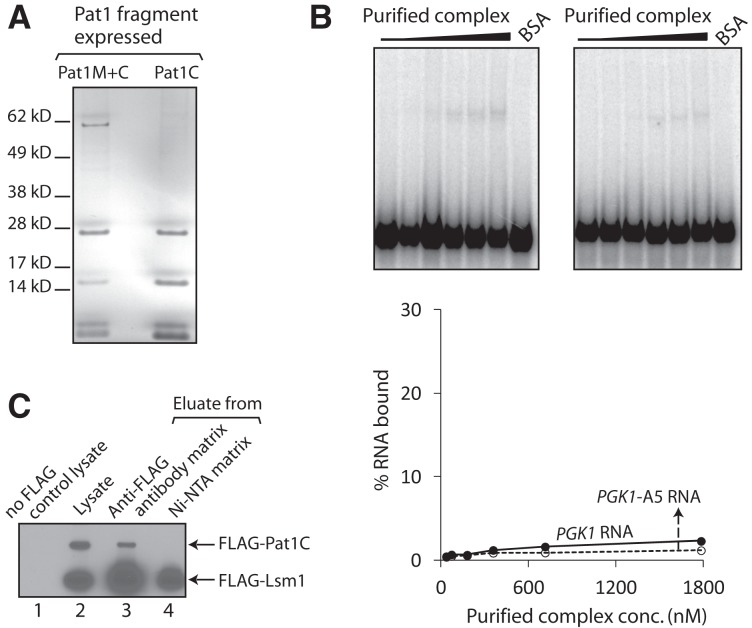
The Pat1 subunit could be enhancing the RNA binding activity of the Lsm1-7–Pat1 complex either by directly contacting RNA and thereby providing additional RNA binding surface(s) or simply by strengthening the RNA contacts of the Lsm1 through Lsm7 subunits via its influence on the conformation of those subunits. It is known that pull-down of Pat1 from yeast cell lysate coprecipitates mRNA (Tharun and Parker 2001; Mitchell et al. 2013) and that yeast Pat1 translated in vitro using reticulocyte lysate can be pulled down from such lysate using poly(U) Sepharose (Pilkington and Parker 2008). Therefore, in order to address the above issue, we first asked if Pat1 by itself is able to bind RNA directly. Given that efforts to purify full-length Pat1 have not been successful so far (Nissan et al. 2010), we purified two previously characterized N-terminally truncated fragments of yeast Pat1 carrying the middle and C-terminal domains (residues 254–796) or only the C-terminal domain (residues 422–796) of Pat1, referred to as Pat1M+C (predicted size 62 kD) and Pat1C (predicted size 43 kD), respectively (Nissan et al. 2010), after expressing them in Escherichia coli (Fig. 4A, below). These regions of yeast Pat1 mediate the mRNA decay function and the interactions with most of the decay factors (Pilkington and Parker 2008; Nissan et al. 2010). To determine if Pat1C and Pat1M+C bind RNA directly, we carried out gel shift assays using uniformly radiolabeled MFA2 and MFA2-A5 RNAs. As seen in Figure 3, Pat1C and Pat1M+C bound RNA with extremely low affinity. Interestingly, these results also showed that as for the Lsm1-7 complex, a clear binding preference for oligoadenylated RNA over unadenylated RNA was not detectable for the Pat1C and Pat1M+C fragments also. Similar results were obtained in gel shift assays carried out using radiolabeled PGK1 and PGK1-A5 RNAs (Supplemental Fig. S1; data not shown). In order to further confirm that the Pat1 fragments are able to bind RNA, we carried out UV crosslinking assays. Purified Pat1C and Pat1M+C fragments and bovine serum albumin (BSA; control) were incubated with the uniformly radiolabeled PGK1 RNA and then UV irradiated. After ribonuclease treatment, the crosslinked proteins were analyzed by SDS-PAGE followed by autoradiography. As seen in Figure 4B, both Pat1C and Pat1M+C, but not BSA, crosslinked to the RNA in a UV irradiation–dependent manner. These results suggest that yeast Pat1 by itself is capable of binding RNA directly, and are consistent with the observation that the purified C-terminal fragment of human Pat1b is able to directly bind poly (U) in vitro (Braun et al. 2010).
FIGURE 4.
Pat1 directly contacts RNA. (A) Silver-stained SDS-PAGE of Pat1C and Pat1M+C purified from bacteria. (B,C) Lsm1-7–Pat1 complex purified from wild-type yeast (2 pmol), Lsm1-7 complex purified from pat1Δ yeast (3 and 5 pmol), Pat1C (45 pmol), or Pat1M+C (24 pmol) expressed and purified from E. coli or BSA was subjected to UV crosslinking in the presence of uniformly radiolabeled PGK1 RNA for varying lengths of time (indicated at the bottom). After ribonuclease treatment, the crosslinked proteins were visualized by denaturing PAGE and autoradiography.
FIGURE 3.
The Pat1 fragments are severely impaired in their ability to bind RNA and do not exhibit a binding preference for oligoadenylated RNA. BSA or increasing concentrations of Pat1M+C or Pat1C expressed and purified from E. coli (indicated on top) were subjected to gel shift assays using uniformly radiolabeled MFA2 and MFA2-A5 RNAs. Plots of the percentage of RNA bound (quantitated using phosphorimager) vs. the concentration of the protein used are shown directly below the phosphorimages of the corresponding gels. Bands corresponding to the bound RNA are marked with asterisks on the left of the phosphorimages. At least two gel retarded bands are observed with Pat1C. The lower of the two bands may be the result of disassembly during the gel run of the RNP complex(s) representing the upper band.
The above results support the idea that Pat1 contacts RNA as a part of the Lsm1-7–Pat1 complex. To test this directly, we carried out UV crosslinking assays using uniformly radiolabeled PGK1 RNA with purified wild-type complex and the Lsm1-7 complex purified from pat1Δ cells. The following observations can be made from these studies (Fig. 4C). First, as expected and as we had observed previously with the wild-type complex (Chowdhury et al. 2007; Chowdhury and Tharun 2009), a ∼23-kD band radiolabeled in a UV irradiation–dependent manner was clearly visible in crosslinking reactions carried out using both the complexes. This band has the expected mobility of Flag-Lsm1, and we have shown earlier via immunoprecipitation analyses that it does contain Flag-Lsm1 (Chowdhury et al. 2007; Chowdhury and Tharun 2009). Second, in crosslinking reactions carried out using the wild-type complex, at least two more bands of lower mobility (∼65 kD and ∼97 kD) were also detectable in longer exposures, although one of them (∼97 kD) was weak. Two observations strongly suggest that these bands correspond to Pat1. First, although the expected size of yeast Pat1 is 88 kD, it is known to run in SDS-PAGE as multiple bands of anomalous electrophoretic mobility, including two or three bands with mobility ranging from ∼62 kD to ∼75 kD and a ∼97-kD band (Bouveret et al. 2000; Chowdhury et al. 2007, 2012; Chowdhury and Tharun 2008, 2009). In our purified Lsm1-7–Pat1 complex preparations, we always find the ∼62-kD to ∼75-kD Pat1 bands to be much stronger than the ∼97-kD band (Chowdhury et al. 2007, 2012; Chowdhury and Tharun 2008, 2009). Thus, the ∼65-kD and ∼97-kD bands (wherein the latter band is weaker than the former) observed in UV crosslinking match the typical mobility of yeast Pat1 bands. Second, both the ∼65-kD and ∼97-kD bands are undetectable in the crosslinking reactions carried out using the Lsm1-7 complex purified from pat1Δ cells (Supplemental Fig. S3A shows longer exposure of the autoradiograph shown in Fig. 4C). These observations support the idea that RNA binding by the Lsm1-7–Pat1 complex involves direct contacts of not only the Lsm proteins but also Pat1 with the RNA. The ∼65-kD and ∼97-kD crosslinked bands are often weaker than the ∼23-kD crosslinked band corresponding to Lsm1 (Supplemental Fig. S2), and this is probably why they were missed in our earlier studies (Chowdhury et al. 2007). Finally, crosslinking reactions carried out with both the wild-type complex and the Lsm1-7 complex revealed a set of additional bands of mobility higher than that of the ∼23-kD Flag-Lsm1 band, possibly corresponding to other Lsm proteins, as we had observed earlier with the wild-type complex (Chowdhury et al. 2007; Chowdhury and Tharun 2009). Additional experiments suggested that among these, the ∼14-kD crosslinked band corresponds to Lsm7 because, if crosslinking reactions were carried out using the wild-type complex purified from a strain that carries the C-terminal 6xHis tag on LSM5 instead of LSM7, this ∼14-kD crosslinked band showed a small but reproducible increase in mobility (Supplemental Fig. S2).
Association of Lsm1-7 complex with Pat1 fragments in vitro results in restoration of RNA binding activity
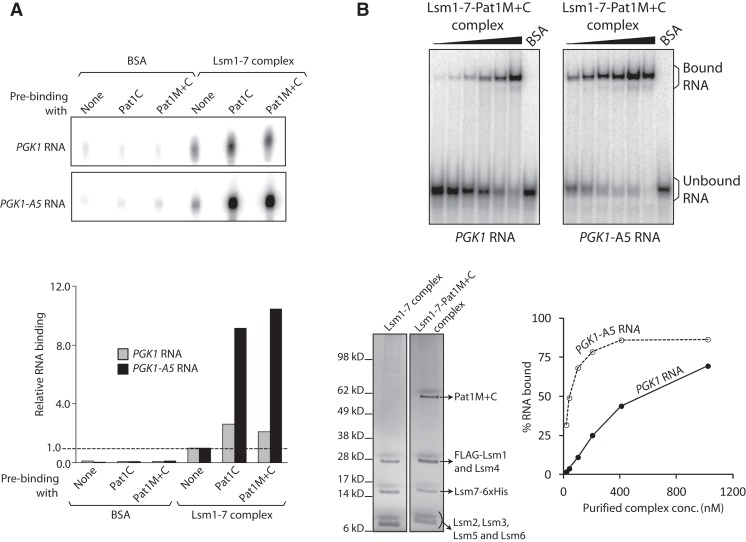
As mentioned above, the Lsm1-7 complex not only has an RNA binding activity that is much lower than that of the wild-type Lsm1-7–Pat1 complex but also lacks any detectable ability to recognize the oligo(A) tail on the RNA. While in principle this could be because the RNA binding properties of the Lsm1-7–Pat1 complex are completely an attribute of Pat1, such a scenario is less likely given the observation that the Pat1 fragments Pat1C and Pat1M+C also have very poor RNA binding activity and fail to exhibit a detectable binding preference for oligoadenylated RNA. Although the impaired RNA binding activity of the Pat1 fragments could be because of their N-terminal truncation, an alternate possibility is that neither Pat1 nor the Lsm1-7 complex has robust RNA binding activity in isolation, and the enhanced RNA binding ability and unique RNA binding preference of the Lsm1-7–Pat1 complex are generated only when Pat1 associates with the Lsm1-7 complex. Since both Pat1M+C and Pat1C associate with the Lsm1-7 complex in vitro as observed by us (Fig. 7C, below) and others (Nissan et al. 2010), in order to test this idea, we asked if a complex reconstituted in vitro by combining purified Lsm1-7 complex with Pat1M+C or Pat1C will have enhanced RNA binding activity. To this end, we immobilized the Lsm1-7 complex onto the anti-Flag antibody matrix (using the Flag tag on Lsm1) after incubating it with Pat1M+C, Pat1C, or just buffer. After washing to remove unbound proteins, the matrix was incubated with uniformly labeled PGK1 RNA and washed again before extracting the bound RNA, which was then analyzed by denaturing PAGE and autoradiography. As shown in Figure 5A, the amount of RNA retained on the matrix was increased about twofold if the Lsm1-7 complex was immobilized on it after incubation with Pat1C or Pat1M+C. Importantly, when the experiment was carried out using uniformly labeled PGK1-A5 RNA instead of PGK1 RNA, the enhancement of RNA retention caused by preincubation of the Lsm1-7 complex with the Pat1 fragments was about eightfold. The retention of RNA on the matrix was dependent on the Lsm1-7 complex immobilized onto the matrix and not due to nonspecific association because the matrix incubated with BSA did not bind RNA at significant levels. Further, preincubation of the Lsm1-7 complex with BSA instead of Pat1 fragments before its immobilization onto the matrix did not affect the amount of RNA retained on the matrix (data not shown). These observations suggest that the association of Pat1 with the Lsm1-7 complex results in the significantly enhanced RNA binding ability and the restoration of the binding preference for oligoadenylated RNA over unadenylated RNA of the Lsm1-7–Pat1 complex.
FIGURE 7.
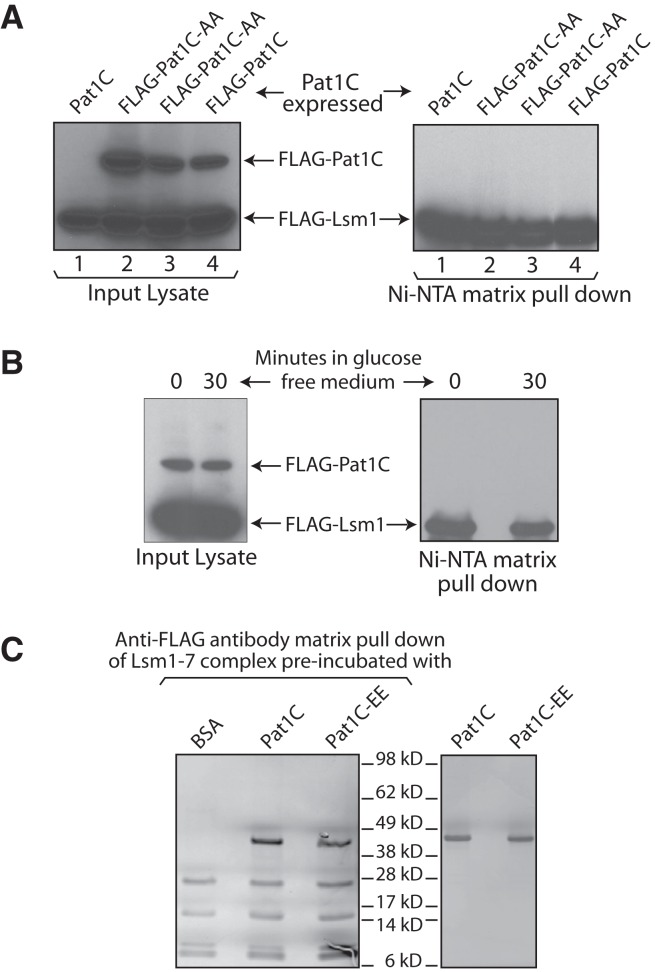
Inability of Pat1C to interact with the Lsm1-7 complex is not due to phosphorylation. (A) Lsm1-7 complex was pulled down using Ni-NTA matrix from the lysates of pat1Δ cells that express Pat1C, Flag-Pat1C, or Flag-Pat1C-AA (indicated on top) from a CEN plasmid followed by Western analysis using anti-Flag antibodies of the lysate and pull-down samples. Two independent transformants expressing Flag-Pat1C-AA were used for the experiment. (B) pat1Δ cells expressing Pat1C were grown in glucose containing medium to log phase and then transferred to fresh medium that contains or lacks glucose for 30 min before they were pelleted and lysed. The Lsm1-7 complex was pulled down from the lysates using Ni-NTA matrix followed by Western analysis using anti-Flag antibodies of the lysate and pull-down samples. (C) Lsm1-7 complex purified from pat1Δ yeast was incubated with Pat1C or Pat1C-EE purified from E. coli or BSA and then bound to the anti-Flag antibody matrix. After washing to remove unbound proteins, the bound proteins were eluted, separated by SDS-PAGE, and visualized by silver staining (left panel). (Right) Silver-stained SDS-PAGE of purified Pat1C and Pat1C-EE.
FIGURE 5.
Association of Lsm1-7 complex with Pat1 fragments results in restoration of RNA binding activity. (A) BSA or purified Lsm1-7 complex was incubated with purified Pat1C or Pat1M+C or just buffer (as indicated on top) and then bound to anti-Flag antibody matrix. After washing followed by incubation of the matrix with uniformly radiolabeled PGK1 RNA and washing again, the RNA retained on the matrix was extracted and visualized by denaturing PAGE and autoradiography (top). (Bottom) A bar diagram of the amount of RNA retained on the matrix after each reaction (quantitated using phosphorimager) normalized to the value obtained with Lsm1-7 complex pre-incubated with just buffer. (B) BSA or increasing concentrations of Lsm1-7–Pat1M+C complex purified from pat1Δ cells expressing Pat1M+C fragment were subjected to gel shift assays using uniformly radiolabeled PGK1 and PGK1-A5 RNAs. A plot of the percentage of RNA bound (quantitated using phosphorimager) vs. the concentration of the complex used and phosphorimages of the gels are shown on the lower right and upper panels, respectively. Lower left panel shows the silver-stained SDS-PAGE of the purified Lsm1-7–Pat1M+C complex.
Lsm1-7–Pat1M+C complex purified from yeast has considerable RNA binding activity and exhibits binding preference for oligoadenylated RNA
In order to further confirm the above observation, we expressed (from CEN vector using native promoter) Pat1M+C in pat1Δ yeast and purified the Lsm1-7 complex from those cells. SDS-PAGE analysis revealed that, as expected, the Pat1M+C fragment (∼62-kD band) copurifies with the Lsm1 through Lsm7 proteins (Fig. 5B). Analysis of the RNA binding activity of the purified Lsm1-7–Pat1M+C complex via gel shift assays using uniformly radiolabeled PGK1 and PGK1-A5 RNAs revealed that it has considerable RNA binding activity (Fig. 5B) that is much higher than that of the Lsm1-7 complex (Fig. 2) or the Pat1 fragments (Fig. 3) although lower than that of the wild-type complex (Fig. 2). The KD values for the interaction of PGK1 and PGK1-A5 RNAs with the purified Lsm1-7–Pat1M+C complex are ∼550 nM and ∼40 nM, respectively, while the corresponding KD values obtained with the wild-type complex for the same RNAs (Fig. 2) are ∼56 nM and ∼20 nM. Thus, although the Lsm1-7–Pat1M+C complex has lower RNA binding activity compared with the wild-type complex, it exhibits a clear binding preference for oligoadenylated RNA over unadenylated RNA.
M-domain of Pat1 is essential for interaction of Lsm1-7 complex with Pat1 in vivo
In order to gain more insight into the enhancement of RNA binding activity of the Lsm1-7 complex upon its association with the Pat1 fragments, we also expressed Pat1C (from CEN vector using native promoter) in pat1Δ yeast and purified the Lsm1-7 complex from those cells. Surprisingly, SDS-PAGE analysis of the purified material failed to detect the Pat1C fragment (∼43 kD), suggesting that Pat1C does not copurify with the Lsm1-7 complex (Fig. 6A), although under the same conditions, Lsm1-7–Pat1M+C complex could be purified from the same pat1Δ strain expressing Pat1M+C from a similar construct (Figs. 5B, 6A). Consistently, the RNA binding activity of the purified complex was very low, like that of the Lsm1-7 complex purified from pat1Δ cells (Figs. 2, 6B). In order to rule out the possibility that the Pat1C polypeptide fails to accumulate at sufficient levels in vivo (due to instability or poor expression), we expressed Flag-tagged Pat1C using a similar construct in the pat1Δ strain and purified the Lsm1-7 complex from such strain. We then subjected the eluate of the first affinity purification step (anti-Flag antibody matrix), the final eluate (after the Ni-NTA matrix step), and the input cell lysate to Western analysis using anti-Flag antibody. Such analysis revealed that while Pat1C is clearly detectable in the cell lysate and the anti-Flag antibody matrix eluate, it is undetectable in the final eluate (Fig. 6C). Thus, although Pat1C is expressed in the cells, it fails to copurify with the Lsm1-7 complex. These results therefore suggest that the C-terminal domain of Pat1 is not sufficient to support the interaction with the Lsm1-7 complex in vivo and that the middle domain is also needed additionally.
FIGURE 6.
Pat1C does not copurify with the Lsm1-7 complex from yeast. (A) Lsm1-7 complex was purified from pat1Δ cells that express Pat1C or Pat1M+C, and the purified material was separated by SDS-PAGE and visualized by silver staining. (B) BSA or increasing concentrations of the complex purified from pat1Δ cells expressing Pat1C were subjected to gel shift assays using uniformly labeled PGK1 or PGK1-A5 RNAs. Plots of the percentage of RNA bound (quantitated using phosphorimager) vs. the concentration of the protein used are shown directly below the phosphorimages of the gels. (C) The Lsm1-7 complex was purified from a pat1Δ strain expressing Flag-Pat1C and the input lysate, eluates of the first (anti-Flag antibody matrix pull-down), and second (Ni-NTA matrix pull-down) steps of purification and cell lysate from a control strain in which neither Lsm1 nor Pat1C is Flag tagged were subjected to Western analysis using anti-Flag antibodies.
Inability of Pat1C to interact with the Lsm1-7 complex is not due to phosphorylation
The observation that the C-terminal domain of Pat1 is not sufficient for the interaction with the Lsm1-7 complex is surprising because past studies have shown that Pat1C is able to interact with the Lsm1-7 complex in vitro (Nissan et al. 2010; Sharif and Conti 2013; Wu et al. 2014). Since these studies were carried out using recombinant Pat1C protein purified from E. coli, our observations suggest that the inability of Pat1C to associate with the Lsm1-7 complex in vivo in yeast could be due to post-translational modification. Indeed, recent studies have shown that in glucose-replete growth conditions, Pat1 is phosphorylated by PKA at two serine residues in its C-terminal domain, S456 and S457 (Budovskaya et al. 2005; Ramachandran et al. 2011; Shah et al. 2013). Therefore we asked if the inability of Pat1C to copurify with the Lsm1-7 complex from yeast is due to such phosphorylation since we grew the cells in glucose containing growth medium for the pull-down experiments (Fig. 6). To test this idea, we mutagenized S456 and S457 to alanines in the CEN plasmid that was used to express Flag-Pat1C in yeast for the experiment shown in Figure 6. The Lsm1-7 complex was then pulled down using Ni-NTA matrix from pat1Δ cells expressing the wild-type Pat1C or the mutant version, Pat1C-AA, from analogous plasmids. The lysate and pull-down fractions were analyzed by anti-Flag antibody Westerns. As seen in Figure 7A, Pat1C-AA, just like Pat1C, also fails to coprecipitate with the Lsm1-7 complex, although it is expressed.
It has been shown that when yeast cells grown in glucose-replete medium were shifted to a medium lacking glucose, the PKA-mediated phosphorylation of Pat1 is lost rapidly within 10–30 min (Ramachandran et al. 2011; Shah et al. 2013). Therefore, in order to test the effect of such natural loss of phosphorylation in vivo on the interaction of Pat1C with the Lsm1-7 complex, we grew the pat1Δ cells expressing Flag-Pat1C under glucose-replete conditions to log phase and then transferred them to medium lacking glucose for 30 min before lysing them and pulling down the Lsm1-7 complex from them using Ni-NTA matrix. As seen in Figure 7B, Pat1C still fails to coprecipitate with the Lsm1-7 complex.
Mutation of S456 and S457 of Pat1 to glutamic acid residues has been shown to mimic phosphorylation (Ramachandran et al. 2011; Shah et al. 2013). Therefore, we asked if Pat1C carrying such phosphomimetic mutation expressed and purified from bacteria will fail to interact with the purified Lsm1-7 complex in vitro. To this end, we introduced such mutation in the plasmid used for expressing GST-tagged Pat1C in E. coli. The wild-type Pat1C and the mutant version, Pat1C-EE, expressed using the analogous constructs in E. coli were then purified using glutathione-Sepharose matrix. After cleaving off their GST tags, equal amounts of the two proteins were incubated with the purified Lsm1-7 complex. The Lsm1-7 complex was pulled down from these reactions using an anti-Flag antibody matrix. After washing to remove unbound proteins, the bound proteins were separated by SDS-PAGE and then visualized by silver staining. As seen in Figure 7C, Pat1C-EE interacts with the Lsm1-7 complex as efficiently as Pat1C. Together, these results suggest that PKA-mediated phosphorylation is not the likely reason for the inability of the C-domain of Pat1 to interact with the Lsm1-7 complex in vivo.
Finally, Northern analysis to determine the accumulation of the poly(G) decay intermediate of the MFA2pG mRNA at steady state (which is decreased in decapping defective cells) (Decker and Parker 1993; Hatfield et al. 1996; Tharun et al. 2005) revealed that changing S456 and S457 to Alanines in full-length Pat1 does not significantly affect mRNA decay in vivo (Supplemental Fig. S3B). As seen in Supplemental Figure S3B, Pat1-AA was able to support poly(G) fragment accumulation as well as Pat1 in pat1Δ cells. This suggests that phosphorylation of S456 and S457 of Pat1 is dispensable for mRNA decay at least under normal growth conditions.
DISCUSSION
The ability of the Lsm1-7–Pat1 complex to bind RNA and to recognize the presence of the 3′ oligo(A) tail of the RNA is essential for mRNA decay in vivo as revealed by our past analysis of various lsm1 mutants (Tharun et al. 2005; Chowdhury and Tharun 2008, 2009; Chowdhury et al. 2012). However, the contribution of Pat1 to the RNA binding properties of the Lsm1-7–Pat1 complex is not known. The main finding that we report here is that the high RNA binding affinity and the ability to recognize the 3′-oligo(A) tail of the RNA are features of the Lsm1-7–Pat1 complex that are generated only when the two key components of the complex, namely, Lsm1-7 ring and Pat1, associate to form the full Lsm1-7–Pat1 complex, and hence, neither of those two components possesses such features in isolation. Multiple observations support this idea. First, comparison of the RNA binding properties of the Lsm1-7–Pat1 and Lsm1-7 complexes purified from yeast reveals that the Lsm1-7 complex is severely impaired in its RNA binding activity and ability to recognize the 3′-oligo(A) tail. Second, Pat1 fragments purified from bacteria are also severely defective in RNA binding and fail to exhibit a detectable binding preference for oligoadenylated RNA. Third, small point mutations (changing five or fewer residues) in the Sm domain of Lsm1 can abolish the ability to recognize the 3′-oligo(A) tail with or without the concomitant (moderate or almost complete) loss of the RNA binding activity of the Lsm1-7–Pat1 complex in the absence of any significant impact on the complex integrity (Tharun et al. 2005; Chowdhury and Tharun 2008, 2009). Fourth, in vitro reconstitution of the whole complex from the RNA binding–defective components (purified Lsm1-7 complex and Pat1 fragment) results in significant enhancement of RNA binding activity and restoration of the ability to recognize 3′-oligo(A) tail of the RNA. Results obtained with similar complex purified from pat1 mutant yeast (in vivo reconstitution) are consistent with that in vitro reconstitution experiments. Finally, crosslinking analyses show that both Lsm1 and Pat1 contact RNA as part of the entire Lsm1-7–Pat1 complex. Thus, the characteristic RNA binding properties of the Lsm1-7–Pat1 complex can be severely impaired by lesions in either Lsm1 or Pat1, indicating that both the lsm1-7 ring and Pat1 play crucial roles in RNA binding.
Our earlier studies support the idea that the lack of binding preference for oligoadenylated RNA of the Lsm1-7 complex and the Pat1 fragments observed here is not because the unadenylated RNA binding activities measured are very low. For example, although the mutant complexes isolated from the lsm1-27 and lsm1-28 strains (that carry large deletions in the C-terminal domain of Lsm1) are severely impaired in binding unadenylated RNA, their binding preference for oligoadenylated RNA over unadenylated RNA can be very clearly observed (Chowdhury et al. 2012). Further, very low binding activity is also observed with the wild-type complex when a poor unadenylated RNA substrate like RPP2B, TEF1(s) or MFA26xUΔ RNA is used (Chowdhury et al. 2007). However, here again, the binding preference for the oligoadenylated forms of these RNAs over the corresponding unadenylated forms can be very clearly observed (Chowdhury et al. 2007). Therefore low binding activity toward unadenylated RNA per se does not preclude the ability to detect the binding preference for oligoadenylated RNA. Finally and conversely, although the mutant complex isolated from the lsm1-14 strain is as efficient as (or slightly better than) the wild-type complex in binding unadenylated RNA, a binding preference of this mutant complex for oligoadenylated RNA over unadenylated RNA is not detectable (Chowdhury and Tharun 2008). Thus, the binding activity toward unadenylated RNA does not always impact the ability to detect or correlate with the binding preference for oligoadenylated RNA.
Our observations are consistent with the model that the Lsm1-7–Pat1 complex possesses a composite RNA binding surface made up of residues from both the Lsm1-7 ring and Pat1. The poor binding activity of the Lsm1-7 ring and Pat1 fragments could be simply either because the fewer RNA contacts (compared with the Lsm1-7–Pat1 complex) made by their individual RNA binding surfaces are not sufficient to support significantly stable RNA binding or because their binding surfaces attain optimal conformation only when they associate to form the entire Lsm1-7–Pat1 complex. Our past studies have shown that both the Sm domain and the C-terminal domain of Lsm1 are essential for the normal RNA binding activity of the Lsm1-7–Pat1 complex, suggesting that residues from both of these regions of Lsm1 contribute to the RNA binding surface of the Lsm1-7 ring (Chowdhury and Tharun 2008, 2009; Chowdhury et al. 2012). The other Lsm subunits (Lsm2 through Lsm7) also contact RNA in the Lsm1-7–Pat1 complex, although the manner in which they affect the overall binding is not clear.
A unique feature of the Lsm1-7–Pat1 complex is its strong binding preference for oligoadenylated RNA over unadenylated RNA. While the mechanism by which this binding preference is attained is not clear, it is possible that in addition to a binding surface for the RNA body, the Lsm1-7–Pat1 complex also possesses an oligo(A) tail–specific binding surface such that when the RNA has a 3′-oligo(A) tail, substrate binding is further strengthened due to additional RNA contacts and possible conformational changes caused by such contacts. Since our studies show that both the Lsm1-7 ring and Pat1 contribute not only to the binding affinity for unadenylated RNA but also to the binding preference for oligoadenylated RNA of the Lsm1-7–Pat1 complex, it is likely that both of these binding surfaces are composites involving residues from Pat1 and the Lsm1-7 ring. However, it is likely that the oligo(A) tail–specific binding surface involves residues from the Sm domain but not the C-terminal domain of Lsm1 since the latter domain is not needed for the ability to recognize the oligo(A) tail (Chowdhury et al. 2012). In any case, the mechanism by which this complex distinguishes between oligoadenylated and polyadenylated RNA is not clear.
The Lsm1-7–Pat1 complex facilitates both mRNA decay and 3′-end protection in vivo. We showed earlier that while the RNA binding ability of this complex is sufficient to support 3′-end protection, the mRNA decay function additionally needs the ability of this complex to recognize the oligo(A) tail (Chowdhury and Tharun 2008, 2009). Therefore the impairment of both of these abilities in the Lsm1-7 complex is consistent with the observation that both 3′-end protection and mRNA decay are impaired in pat1Δ cells (Hatfield et al. 1996; Bouveret et al. 2000; Tharun et al. 2000; He and Parker 2001).
An important observation we have made is that while a larger fragment of Pat1 carrying both the middle and C-terminal domains (Pat1M+C) copurifies with the Lsm1-7 complex from yeast, a smaller fragment of Pat1 carrying only the C-terminal domain (Pat1C) fails to do so. This is surprising since Pat1C purified from E. coli was shown to be able to interact with the Lsm1-7 ring in vitro (Nissan et al. 2010; Sharif and Conti 2013; Wu et al. 2014). In vitro studies on Pat1 fragments support the notion that the middle and C-terminal domains of Pat1 interact in the context of full-length Pat1 (Nissan et al. 2010). Further, Pat1C also self-associates in vitro (Nissan et al. 2010). Given this, one possibility is that Pat1C interacts with the Lsm1-7 complex as a dimer (the dimer may mimic interacting middle and C-terminal domains) in vitro and that such interaction fails to occur in vivo because Pat1C does not accumulate to sufficient levels in vivo to drive dimer formation. However, this possibility is ruled out by the observation that Pat1C and the Lsm1-7 ring bind with 1:1 stoichiometry in vitro (Sharif and Conti 2013). An alternate possibility is that PKA-mediated phosphorylation of S456 and S457 disrupts the contacts between the C-terminal domain of Pat1 and the Lsm1-7 complex such that Pat1C is unable to interact with Lsm1-7 complex, while Pat1M+C is able to associate due to additional contacts made by the middle domain of Pat1. Our results presented here do not support this possibility because neither mutating these serines to alanines nor allowing their dephosphorylation in vivo results in copurification of Pat1C with the Lsm1-7 complex. Further, conversely, phosphomimetic mutation of these serines to glutamic acid residues does not abolish the interaction between Pat1C and the Lsm1-7 complex observed in vitro. Thus, together these results suggest that, in addition to the C-terminal domain, the middle domain of Pat1 is also needed for the interaction of Pat1 with the Lsm1-7 ring in vivo and that the inability of Pat1C to associate with the Lsm1-7 complex is not due to phosphorylation of S456 and S457 of Pat1.
Our observation that the C-terminal domain of Pat1 is not sufficient for the interaction with the Lsm1-7 ring is consistent with earlier studies showing that only Pat1M+C, but not Pat1C, is able to suppress the mRNA decay defect of pat1Δ cells and to interact with Lsm1 in yeast two-hybrid assay (Pilkington and Parker 2008). Nevertheless, in vitro, just like Pat1M+C, Pat1C is able to not only interact with the Lsm1-7 ring but also facilitate the enhancement of the RNA binding activity and restore of the binding preference for oligoadenylated RNA of the resulting complex upon such interaction. Further, like Pat1M+C, Pat1C is also able to activate the decapping enzyme in vitro (Nissan et al. 2010). Therefore these observations suggest that the interaction of Pat1C with the Lsm1-7 complex observed in vitro is not biologically irrelevant but may rather represent a conformational state of the C-terminal segment of Pat1 that is transient in vivo but for some reason is stabilized in vitro.
MATERIALS AND METHODS
Lsm1-7 complex and the wild-type Lsm1-7–Pat1 complex were purified from pST17 (Chowdhury et al. 2007) transformants of yST330 (Matα, ade2, his3, leu2, ura3, pat1Δ::NEOr, lsm1Δ::TRP1, LSM7-6xHis-NEOr) and yST253 (MatA, trp1, his3, ura3, leu2, ade2, can1, lsm1Δ::TRP1, LSM7-6xHis-NEOr), respectively. C-terminal His-tagging of LSM7 was carried out using the PCR-based method of gene modification (Longtine et al. 1998) to generate yST253. Pat1C-EE, Pat1C, and Pat1M+C were purified from E. coli Rosetta-2 cells carrying pST446, pRP1837, and pRP1838, respectively, as described before (Nissan et al. 2010). pST446 was made by using pRP1837 (Nissan et al. 2010) as the template for QuikChange mutagenesis. Lsm1-7–Pat1M+C complex was purified from yST330 transformed with pRP1476 (Pilkington and Parker 2008) and pST123. pST123 was made by cloning the NotI fragment of pST17 into NotI-digested pRS413 (Sikorski and Hieter 1989). Pat1C, Flag-Pat1C, and Flag-Pat1C-AA were expressed in yeast using plasmids pRP1475, pRP1487 (Pilkington and Parker 2008), and pST444, respectively. pST444 was made by using pRP1487 as template for QuikChange mutagenesis. S456A and S457A mutations were introduced into full-length Pat1 by using pRP1424 (Pilkington and Parker 2008) as the template for QuikChange mutagenesis. pat1Δ strain used in Supplemental Figure S3B is yRP1372 (Tharun et al. 2000).
Purification of the Lsm1-7, Lsm1-7–Pat1, and Lsm1-7–Pat1M+C complexes; preparation of radiolabeled RNAs; gel shift assays; pull-down assays; Western analyses; and UV crosslinking analyses were done as described before (Chowdhury et al. 2007, 2012; Chowdhury and Tharun 2008, 2009).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Roy Parker and Dr. Tracy Nissan for Pat1 expression plasmids. This work was supported by funds from National Institute of General Medical Sciences (NIH) RO1 grant (GM072718) and Uniformed Services University of the Health Sciences (USUHS) exploratory grant (RO71JX) to S.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045252.114.
REFERENCES
- Balagopal V, Fluch L, Nissan T 2012. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta 1819: 593–603 [DOI] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol 18: 5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B 2000. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol 20: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J 19: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Tritschler F, Haas G, Igreja C, Truffault V, Weichenrieder O, Izaurralde E 2010. The C-terminal α-α superhelix of Pat is required for mRNA decapping in metazoa. EMBO J 29: 2368–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci 102: 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG 2005. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics 6: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Tharun S 2008. lsm1 mutations impairing the ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo. RNA 14: 2149–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Tharun S 2009. Activation of decapping involves binding of the mRNA and facilitation of the post-binding steps by the Lsm1-7–Pat1 complex. RNA 15: 1837–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S 2007. The decapping activator Lsm1p-7p–Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13: 998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Raju KK, Kalurupalle S, Tharun S 2012. Both Sm-domain and C-terminal extension of Lsm1 are important for the RNA-binding activity of the Lsm1-7–Pat1 complex. RNA 18: 936–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Elkon R, Zlotorynski E, Zeller KI, Agami R 2010. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics 11: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yang X, Wang W, Wood WH III, Becker KG, Gorospe M 2002. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci 99: 10611–10616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G 2008. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol 323: 76–87 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J, Aranda A, Perez-Ortin JE 2004. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell 15: 303–313 [DOI] [PubMed] [Google Scholar]
- Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E 2010. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol 189: 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Medina DA, Causse SZ, Garber M, Millan-Zambrano G, Barkai O, Chavez S, Perez-Ortin JE, Darzacq X, Choder M 2013. Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell 153: 1000–1011 [DOI] [PubMed] [Google Scholar]
- Hatfield L, Beelman CA, Stevens A, Parker R 1996. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol 16: 5830–5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Parker R 2001. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158: 1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes LE, Campbell SG, De Long SK, Sachs AB, Ashe MP 2004. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol Cell Biol 24: 2998–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurto RL, Hopper AK 2011. P-body components, Dhh1 and Pat1, are involved in tRNA nuclear-cytoplasmic dynamics. RNA 17: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375–387 [DOI] [PubMed] [Google Scholar]
- Ling SH, Qamra R, Song H 2011. Structural and functional insights into eukaryotic mRNA decapping. Wiley Interdiscip Rev RNA 2: 193–208 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Marnef A, Standart N 2010. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem Soc Trans 38: 1602–1607 [DOI] [PubMed] [Google Scholar]
- Marnef A, Maldonado M, Bugaut A, Balasubramanian S, Kress M, Weil D, Standart N 2010. Distinct functions of maternal and somatic Pat1 protein paralogs. RNA 16: 2094–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Weil D, Standart N 2012. RNA-related nuclear functions of human Pat1b, the P-body mRNA decay factor. Mol Biol Cell 23: 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Ottmann AR, Basrai MA 2013. Structural integrity of centromeric chromatin and faithful chromosome segregation requires Pat1. Genetics 195: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Jain S, She M, Parker R 2013. Global analysis of yeast mRNPs. Nat Struct Mol Biol 20: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, Parker R 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell 39: 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Chekulaeva M, Stoecklin G 2010. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol 30: 4308–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R 2012. RNA degradation in Saccharomyces cerevisae. Genetics 191: 671–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortin JE, Alepuz PM, Moreno J 2007. Genomics and gene transcription kinetics in yeast. Trends Genet 23: 250–257 [DOI] [PubMed] [Google Scholar]
- Pilkington GR, Parker R 2008. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol Cell Biol 28: 1298–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. 2011. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29: 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Shah KH, Herman PK 2011. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol Cell 43: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother RP, Frank MB, Thomas PS 1992. Purification, primary structure, bacterial expression and subcellular distribution of an oocyte-specific protein in Xenopus. Eur J Biochem 206: 673–683 [DOI] [PubMed] [Google Scholar]
- Scheller N, Resa-Infante P, de la Luna S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, Diez J 2007. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim Biophys Acta 1773: 1786–1792 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Parker R 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20: 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah KH, Zhang B, Ramachandran V, Herman PK 2013. Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics 193: 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, Pilpel Y 2008. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif H, Conti E 2013. Architecture of the lsm1-7-Pat1 complex: a conserved assembly in eukaryotic mRNA turnover. Cell Rep 5: 283–291 [DOI] [PubMed] [Google Scholar]
- Sharif H, Ozgur S, Sharma K, Basquin C, Urlaub H, Conti E 2013. Structural analysis of the yeast Dhh1–Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic Acids Res 41: 8377–8390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Parker R 2007. Analysis of P-Body assembly in Saccharomyces cerevisiae. Mol Biol Cell 18: 2274–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S 2008. Purification and analysis of the decapping activator Lsm1p-7p-Pat1p complex from yeast. Methods Enzymol 448: 41–55 [DOI] [PubMed] [Google Scholar]
- Tharun S 2009a. Lsm1-7-Pat1 complex: a link between 3′ and 5′-ends in mRNA decay? RNA Biol 6: 228–232 [DOI] [PubMed] [Google Scholar]
- Tharun S 2009b. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int Rev Cell Mol Biol 272: 149–189 [DOI] [PubMed] [Google Scholar]
- Tharun S, Parker R 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell 8: 1075–1083 [DOI] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518 [DOI] [PubMed] [Google Scholar]
- Tharun S, Muhlrad D, Chowdhury A, Parker R 2005. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics 170: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A, Renzi F, La Fata G, Mattioli C, Raabe M, Urlaub H, Achsel T 2011. The human Pat1b protein: a novel mRNA deadenylation factor identified by a new immunoprecipitation technique. Nucleic Acids Res 39: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Watt PM, Louis EJ, Borts RH, Hickson ID 1996. Pat1: a topoisomerase II-associated protein required for faithful chromosome transmission in Saccharomyces cerevisiae. Nucleic Acids Res 24: 4791–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Watt PM, Borts RH, Louis EJ, Hickson ID 1999. The topoisomerase II-associated protein, Pat1p, is required for maintenance of rDNA locus stability in Saccharomyces cerevisiae. Mol Gen Genet 261: 831–840 [DOI] [PubMed] [Google Scholar]
- Wu X, Brewer G 2012. The regulation of mRNA stability in mammalian cells: 2.0. Gene 500: 10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Muhlrad D, Bowler MW, Jiang S, Liu Z, Parker R, Song H 2014. Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation. Cell Res 24: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaric B, Chami M, Remigy H, Engel A, Ballmer-Hofer K, Winkler FK, Kambach C 2005. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem 280: 16066–16075 [DOI] [PubMed] [Google Scholar]
- Zhou L, Hang J, Zhou Y, Wan R, Lu G, Yin P, Yan C, Shi Y 2013. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature 506: 116–120 [DOI] [PubMed] [Google Scholar]
- Zhou L, Zhou Y, Hang J, Wan R, Lu G, Yan C, Shi Y 2014. Crystal structure and biochemical analysis of the heptameric Lsm1-7 complex. Cell Res 24: 497–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.