Abstract
Background
CDK11p58, a Ser/Thr kinase that belongs to the cell division cycle 2-like 1 (CDC2L1) subfamily, is associated with cell cycle progression, tumorigenesis and apoptotic signaling. CDK11p58 is also involved in the regulation of steroid receptors, such as androgen and estrogen receptors. We previously found that CDK11p58 was abnormally expressed in prostate cancer. However, its role in breast cancer remains unclear.
Methods
CDK11p58 expression was evaluated by immunohistochemical staining in a tissue array. A Transwell assay was used to detect invasion and metastasis in breast cancer cells. The TaqMan® Metastasis Gene Expression Assay was used to search for potential downstream factors in the CDK11p58 signaling pathway. qRT-PCR was used to evaluate mRNA levels, and the dual luciferase array was used to analyze promoter activity. Western blotting was used to detect the protein level.
Results
CDK11p58 expression was negatively correlated with node status (P = 0.012), relapse status (P = 0.002) and metastasis status (P = 0.023). Kaplan-Meier survival curves indicated that the disease-free survival (DFS) was significantly poor in breast cancer patients with low CDK11 expression. Interestingly, using the breast cancer cell lines ZR-75-30 and MDA-MB-231, we found that CDK11p58 was capable of repressing the migration and invasion of ERα-positive breast cancer cells, but not ERα-negative breast cancer cells, in a kinase-dependent manner. Gene expression assays demonstrated that integrin β3 mRNA was dramatically repressed by CDK11p58, and luciferase results confirmed that the integrin β3 promoter was inhibited by CDK11p58 through ERα repression. The expression of integrin β3 was highly related to ERα signaling; ERα overexpression stimulated integrin β3 expression, whereas siRNA-mediated knockdown of ERα attenuated integrin β3 expression.
Conclusions
These data indicate that CDK11p58 is an anti-metastatic gene in ERα-positive breast cancer and that the regulation of integrin β3 by CDK11p58 via the repression of ERα signaling may constitute part of a signaling pathway underlying breast cancer invasion.
Keywords: CDK11p58, Metastasis, Integrinβ3, ERα, Tissue array, TaqMan assay
Background
Breast cancer is the most common malignancy among women and represents an important public health issue [1]. It is the major cause of cancer-related death in women, and treatment is particularly difficult in patients with tumor metastasis [2]. Despite recent improvements in survival rates, many patients relapse, and the majority of these patients die from disseminated metastatic disease [3].
The chromosome 1p36 locus is frequently mutated in many types of human tumors, especially in breast tumors. This lesion is associated with progression and lymph node metastasis [4], poor prognosis [5], higher rate of recurrence [6], large tumor size and DNA aneuploidy [7]. Deletion of this chromosome region occurs late in oncogenesis and is correlated with aggressive tumor growth, suggesting that one or more tumor-related genes may reside in this region [8]. However, no specific tumor suppressor gene at 1p36 has yet been identified. Previous studies have identified CHD5 [9] and KIF1B [10] as candidate tumor suppressor genes in this region, and more recently it was reported that disruption of PER3 function may indicate the likelihood of tumor recurrence in patients with ERα-positive tumors [11]. However, to date, no specific roles for these genes in breast cancer development have been demonstrated.
CDK11p58 belongs to the large family of p34cdc2-related kinases [12] and is located on human chromosome 1p36.33, a region frequently mutated in numerous human tumors, including breast cancer. CDK11p58 is specifically expressed in the G2/M phase, and expression is closely associated with cell cycle arrest, oncogenesis and apoptosis [13, 14]. Centrosome abnormalities are a common feature of breast cancer [15, 16]. The overexpression of centrosome-related genes has been detected in premalignant lesions, in situ breast tumors and over 70% of invasive breast tumors, and is associated with aneuploidy and tumor development [17]. CDK11p58 is a centrosome-associated mitotic kinase involved in centrosome maturation and bipolar spindle formation [18] and is required for centriole duplication and Plk4 recruitment to mitotic centrosomes [19]. Furthermore, we found that Thr-370 in CDK11p58 is required for its autophosphorylation, dimerization and kinase activity [13].
Previous studies have shown that CDK11p58 is involved in the negative regulation of steroid receptors in a kinase activity-dependent manner, including androgen [20], vitamin D [21] and estrogen receptors (ERs) [22]. We previously demonstrated that CDK11p58 promotes the ubiquitin/proteasome-mediated degradation of ERα to repress its transcriptional activity [22]. The sex steroid estrogen plays a major role in the development and progression of breast cancer and promotes breast cancer proliferation through a number of established pathways [23]. Estrogen promotes breast cancer cell motility and invasion in ERα-positive breast cancer cells, largely through ERα via FAK and N-WASP [24]. Additionally, the estrogen-ERα complex stimulates the transcriptional activity of MMP-26 and contributes to the survival of ERα-positive breast cancer patients [25]. Because CDK11p58 is involved in the negative regulation of the ERα signaling pathway, we speculated whether CDK11p58 may function as a tumor suppressor in breast cancer via the inhibition of ERα.
In this study, we demonstrate that CDK11p58 inhibited the invasion of ERα-positive breast cancer cells by downregulating integrin β3 expression via ERα signaling. Moreover, the functions of CDK11p58 were highly dependent on its kinase activity. These data indicate that CDK11p58 plays an important role in the negative regulation of breast cancer invasion.
Methods
Patient samples
RNALater-preserved solid tissues, formalin-fixed, paraffin-embedded (FFPE) breast cancer tissues and adjacent normal tissue (ANCT) were obtained from Fudan University Shanghai Cancer Center between 2002 and 2004. The tumors were assessed according to the WHO classification by two academic pathologists. This study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center for Clinical Research. Written informed consent was obtained from all patients.
Immunohistochemical (IHC) analysis
A total of 250 FFPE blocks of breast cancer tissues and ANCT were collected for tissue microarrays. Two breast cancer tissue cores and two ANCT cores from the same patient’s FFPE blocks were arranged on a recipient paraffin block (with a 1-mm core per specimen). Paraffin sections (3-μm thick) were deparaffinized in xylene and rehydrated in a graded alcohol series, boiled with 10 mmol/L of citrate buffer (pH 6) for 15 min and pre-incubated in blocking solution (10% normal goat sera) for 1 h at room temperature. The steps were performed using the Envision two-step method. The Envision and DAB Color Kit was purchased from Gene Tech Company Limited (Shanghai, China). A rabbit anti-human polyclonal antibody against CDK11 was used at a 1:100 dilution. PBS (phosphate buffered saline) was used as a negative control. The tissue microarray slides were concurrently evaluated by two of the authors. Granular nuclear staining was assessed as positive. The staining index (SI, range 0–9) was considered as the product of the intensity score (0, no staining; 1+, faint/equivocal; 2+, moderate; 3+, strong) and the distribution score (0, no staining; 1+, staining of <10% of cells; 2+, between 10% and 50% of cells; and 3+, >50% of cells). For CDK11 protein in this study, a moderate/strong cytoplasm staining (SI = 3–9) was defined as positive staining, and a weak or negative staining (SI = 0–2) was defined as negative staining.
Cell culture and transfection
MCF-7, MDA-MB-231, ZR-75-30 and 293 T cells were grown using RPMI1640 supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin (Cat. 10378-016, Invitrogen, USA) at 37°C and 5% CO2. The constructions of CDK11p58 mutant plasmids T370A and T370D were mentioned in our previous report [13]. Cell transfection was performed with Lipofectamine 2000 transfection reagent (Cat. 11668-019, Invitrogen) according to the manufacturer’s instructions.
RNA interference
Small interfering RNAs (siRNA) designed for CDK11 (three siRNAs, siRNA1–3) and ERα (siERα) were ordered from Shanghai GenePharma Co., Ltd. The sequences of the three siRNAs for siCDK11 were as follows: siRNA1, 5′-GAAGCAUGCUAGAGUGAAATT-3′, siRNA2, 5′-GGGAAUGGGAAAGACAGAATT-3′, and siRNA3, 5′-GCAGCAACAUGGACAAGAUTT-3′. The siERα sequence was 5′-GGAGAAUGUUGAAGCACAATT-3′.
Transwell invasion
Cell invasion was assayed using BD BioCoat Growth Factor Reduced (GFR) Matrigel Invasion Chambers (BD, CA). Transfected ZR-75-30 cells (0.5 ml; 2.5 × 104 cells/ml) were added to the inside of the inserts and incubated for 3 h. After incubation, non-invading cells were removed from the upper surface of the membrane using cotton-tipped swabs. The cells on the lower surface of the membrane were stained with Crystal violet and counted in the central field of triplicate membranes.
TaqMan® Array human metastasis gene expression assays and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
TaqMan® Array Metastasis 96-Well Plates were obtained from Life Technologies Corporation (California, USA) and contained lyophilized TaqMan® Gene Expression Assays. After converting total RNA to cDNA via reverse transcription, TaqMan® Array Plates and the associated reagents were used to quantitate mRNA expression levels. The results were analyzed using the RQ manager software. Genes altered in the Taqman Array gene expression assays were subsequently verified using a qRT-PCR assay system. Total RNA was extracted using TRIzol reagent (Invitrogen). After converting total RNA to cDNA in a reverse transcription (RT) reaction by Cdna Synthesis Kit (Cat. 6110A, TaKaRa, Dalian, China), qPCR were used to quantitate the mRNA expression levels by SYBR® Premix Ex Taq™(Cat. RR420Q, TaKaRa). The cycling conditions were as follows: 95°C for 5 minutes; 40 cycles of 95°C for 15 seconds and 60°C for 32 seconds. GAPDH was used as an endogenous control. Each qRT-PCR cycle was repeated three times to confirm the results.
Dual luciferase reporter gene assays
Dual luciferase assays were performed as previously described [21]. Luciferase activity was measured using the dual luciferase reporter gene assay (Promega, USA), as previously reported [20], and a SynergyHT Multi-Mode Microplate Reader (BioTek, USA).
Statistical analysis
The experimental data are expressed as the mean ± standard deviation, and the statistical significance between the different groups was determined using t-tests. The relationship between CDK11 expression and the clinicopathological features of breast cancer patients was analyzed using χ2 and Fisher’s exact tests. All statistical tests were two sided, and P values less than 0.05 were considered significant.
Results
IHC analysis of CDK11 protein expression
We first evaluated CDK11 protein expression in 250 FFPE breast cancer tissues and ANCTs. CDK11 was clearly expressed in the nucleus in breast cancer tissues, and CDK11 positive expression was found to be both at a higher staining intensity and in a higher proportion of positively stained cells (Figure 1A). Table 1 summarizes the correlation of CDK11 expression with the clinicopathological features of breast cancer patients. CDK11 expression was not correlated with the breast cancer patients’ menopausal status, tumor size, HER-2 status, differentiation or TNM status, but was negatively correlated with age, node status, ER, relapse and metastasis status (P < 0.05). The patients with lower CDK11 expression tended to have a higher risk of relapse and metastasis. All patients were followed up for at least 8 years. Disease free survival (DFS) was significantly worse in breast cancer patients with low CDK11 expression (P = 0.028; Figure 1B). These results indicate that low CDK11 expression in breast cancer is related to a worse prognosis.
Figure 1.
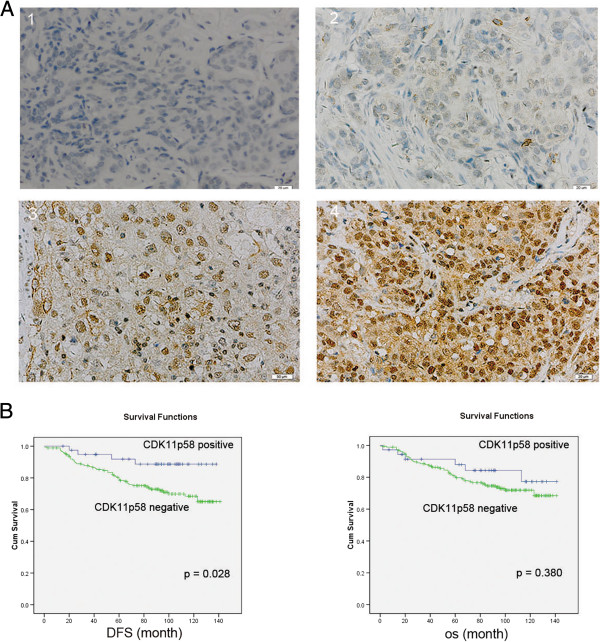
Immunohistochemistry of CDK11 in tissue microarrays. (A) CDK11 immunostaining was determined in breast cancer and scored as (1) (2) low expression, (3) (4) high expression. All immunohistochemical photomicrographs are magnified 400×. (B) Relationship between CDK11 expression and disease free survival (DFS)/overall survival (OS). P values were calculated using the unadjusted log-rank test.
Table 1.
Relationship between CDK11 p58 expression and clinicopathological features in breast cancer patients
| Characteristics | CDK11p58 | ||||
|---|---|---|---|---|---|
| Low | High | n | x 2 | p | |
| Age(years) | 7.024 | 0.008 | |||
| <50 | 27(8.1%) | 78(31.5%) | 105 | ||
| ≥50 | 18(7.3%) | 125(50.4%) | 143 | ||
| Menopausal status | 0.375 | 0.540 | |||
| Pre | 29(11.6%) | 122(48.8%) | 151 | ||
| Post | 16(6.4%) | 83(33.2%) | 99 | ||
| Tumor size (cm) | 0.939 | 0.333 | |||
| ≦2 cm | 18(7.4%) | 95(39.1%) | 113 | ||
| >2 cm | 27(11.1%) | 103(42.4%) | 130 | ||
| Node status | 6.312 | 0.012 | |||
| Negative | 18(7.2%) | 124(49.6%) | 142 | ||
| Positive | 27(10.8%) | 81(32.4%) | 108 | ||
| ER status | 4.065 | 0.044 | |||
| Negative | 33(13.2) | 117(46.8%) | 150 | ||
| Positive | 12(4.8%) | 88(35.2) | 100 | ||
| HER-2 status | 2.823 | 0.093 | |||
| Negative | 22(8.8%) | 128(51.2%) | 150 | ||
| Positive | 23(9.2%) | 77(30.8) | 100 | ||
| Differentiation | 1.456 | 0.228 | |||
| III | 26(13.7%) | 142(74.7%) | 130 | ||
| III | 14(7.4%) | 49(25.8%) | 60 | ||
| TNM stage | 1.567 | 0.211 | |||
| III | 23(11%) | 129(61.4%) | 152 | ||
| III | 13(6.2%) | 45(21.4%) | 58 | ||
| Relapse | 9.323 | 0.002 | |||
| No | 29(11.8%) | 179(72.8%) | 208 | ||
| Yes | 13(5.3%) | 25(10.2%) | 38 | ||
| Metastasis | 4.803 | 0.023 | |||
| No | 26(10.6%) | 159(64.6%) | 185 | ||
| Yes | 16(6.5%) | 45(18.3%) | 61 | ||
Statistically significant p < 0.05 are indicated in bold. ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2.
CDK11p58 inhibits ERα-positive breast cancer metastasis
As CDK11 expression was negatively correlated with node status, relapse and metastasis status in breast cancer patients, we further investigated the role of CDK11p58 in the migration and invasion of breast cancer by Transwell assays. Our previous study showed that the function of CDK11p58 was kinase related, and we identified T370D as a constantly activated CDK11p58 kinase mutant and T370A as a kinase-dead mutant [13]. The invasion of ZR-75-30 cells transfected with wild-type CDK11p58 and the T370D mutant was significantly lower compared with the control cells (P < 0.05; Figure 2A). In contrast, the kinase-dead T370A mutant failed to suppress the metastasis of ZR-75-30 cells. Notably, CDK11p58 failed to inhibit the migration and invasion of ER-negative MDA-MB-231 breast cancer cells (Figure 2B). Together these data suggest that CDK11p58 inhibits ER-positive breast cancer metastasis in a kinase-dependent manner.
Figure 2.
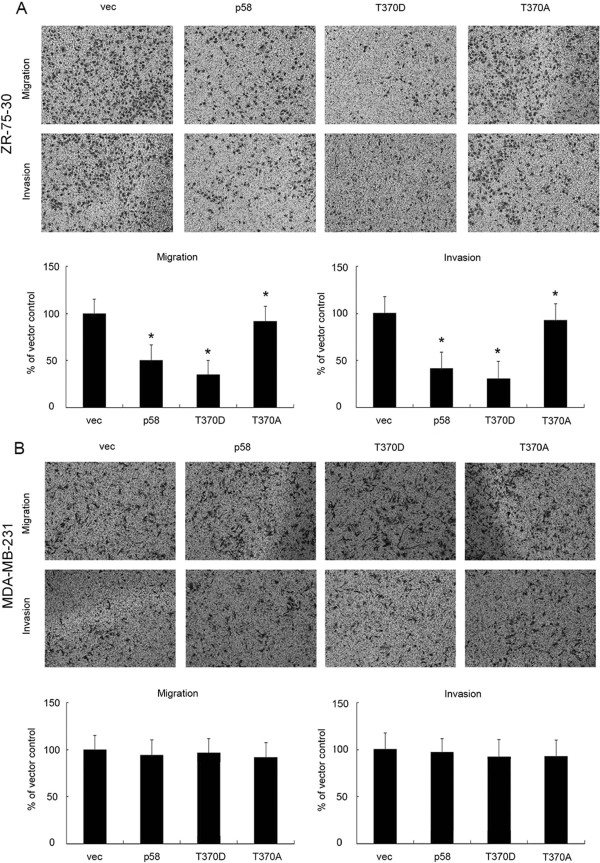
CDK11 p58 inhibits breast cancer cell metastasis in an ERα-dependent manner. (A) ZR-75-30 cells were transfected with wild-type CDK11p58 (p58), kinase activated mutant (T370D), kinase dead mutant (T370A) or control vectors. After 6 h, Transwell assays were performed as described. Crystal violet staining of invading cells is shown. Data are expressed as the mean ± SEM of the number of invading cells in more than five separate areas. *P < 0.05 versus vector controls (n = 3 experiments). (B) MDA-MB-231 cells were transfected with CDK11p58 or various mutants. After 6 h, Transwell assays were performed as described. Crystal violet staining of invading cells is shown. Data are expressed as the mean ± SEM of the number of invading cells in more than five separate areas.
Inhibition of metastasis by CDK11p58 is regulated through integrin β3
To investigate the mechanism by which CDK11p58 inhibits metastasis, we used TaqMan Array human metastasis expression plates to examine the expression of key genes involved in cancer metastasis. MCF-7 cells were transfected with CDK11p58, and the overexpression of CDK11p58 mRNA and protein was confirmed by qRT-PCR and western blotting, respectively (Figure 3A). We observed significant changes in the expression of numerous metastasis-related genes in the CDK11p58-transfected cells compared with control MCF-7 cells (Figure 3B and C). Notably, integrin β3 was significantly repressed in CDK11p58-transfected MCF-7 cells (Figure 3B), and these results were further confirmed by qRT-PCR (Figure 3D). Thus, we speculated that CDK11p58 might inhibit the metastasis of breast cancer cells via the repression of integrin β3.
Figure 3.
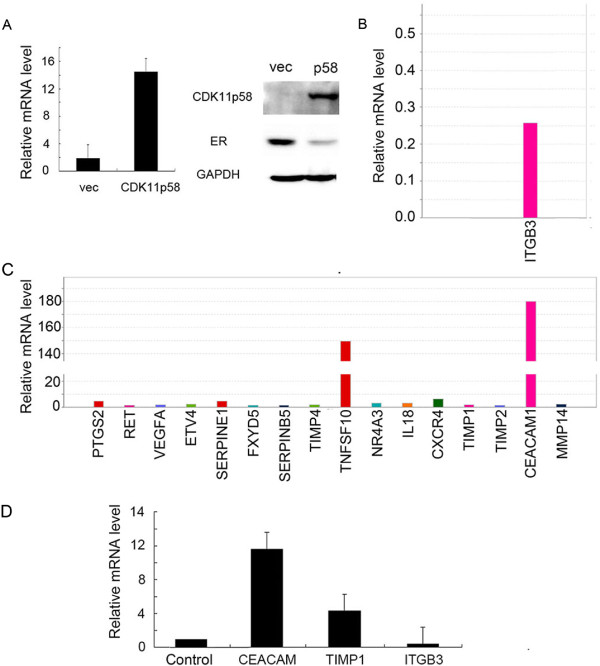
CDK11 p58 downregulates integrin β3 in ERα-positive breast cancer cells. (A) Overexpression of CDK11p58 in MCF-7 ER-positive breast cancer cells. The expression of CDK11p58 was detected by qRT-PCR and western blotting. (B) Genes downregulated in the metastasis pathway in the TaqMan Array human metastasis gene expression assay. (C) Genes upregulated in the metastasis pathway in the TaqMan Array human metastasis gene expression assay. (D) Validation of TaqMan Array human gene expression assays by qRT-PCR.
CDK11p58 represses integrin β3 by inhibiting ERα signaling
As CDK11p58 inhibited the mRNA level of integrin β3, we speculated that integrin β3 promoter activity was regulated by CDK11p58. We cloned the ITGB3 promoter (~2.5 kb) spanning -2000 to +50 from the transcriptional start site into the pGL3-Basic (pGL3B) luciferase vector, and transfected the vector into MCF-7 and ZR-75-30 cells for dual luciferase assays (Figure 4A and B). Cotransfection of wild-type or T370D CDK11p58 vectors significantly repressed ITGB3 promoter activity compared with controls, whereas the T370A variant had no effect. These data suggested that CDK11p58 inhibited the promoter activity of integrin β3 in a kinase-dependent manner.
Figure 4.
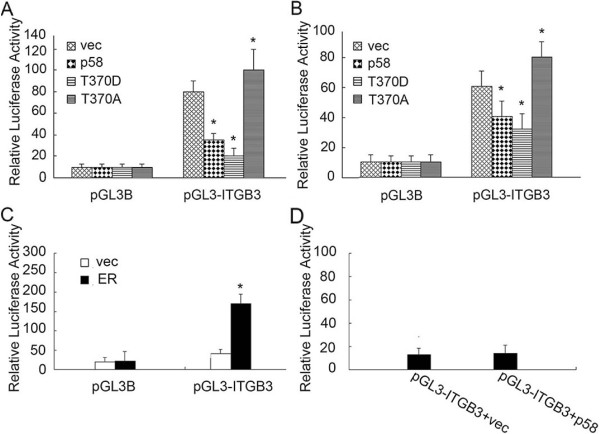
CDK11 p58 inhibits ITGB3 promoter activity in MCF-7 and ZR-75-30 cells. (A and B) CDK11p58 inhibits ITGB3 promoter activity in a kinase-dependent manner. Dual luciferase assays were performed in ZR-75-30 and MCF-7 cells as described in Methods. P < 0.05 vs. controls (n = 3 experiments). (C) 293 T cells were transfected with the indicated plasmids, and luciferase assays were performed 48 h later. (D) MDA-MB-231 cells were transfected with the indicated plasmids and luciferase assays were performed 48 h later.
We then mapped the promoter region of integrin β3 to identify potential transcription factor binding sites and found three potential estrogen response element (ERE) consensus sites in the -1000 to -500 region. Thus, we speculated that integrin β3 was regulated by ERα. To test this hypothesis, pGL3-ITGB3 and ERα were co-transfected into 293 T cells, and dual luciferase assay results showed that ERα could increase the integrin β3 promoter activity (Figure 4C). We previously demonstrated that CDK11p58 represses the transcription activity of ERα. Therefore, we considered that CDK11p58 may repress integrin β3 promoter activity through ERα signaling. To further determine whether the inhibition of integrin β3 by CDK11p58 is ERα dependent, we performed dual luciferase assays in the MDA-MB-231 ERα-negative cell line. As expected, CDK11p58 showed no effect on the integrin β3 promoter in MDA-MB-231 cells (Figure 4D). Together these data suggest that CDK11p58 inhibits integrin β3 through ERα signaling.
We then examined this inhibition at the protein level. We first performed CDK11p58 knockdown in ZR-75-30 cells using three siRNAs. The third siRNA was the most efficient in knocking down CDK11p58 and was used for subsequent experiments (Figure 5A). Western blotting revealed that the overexpression of CDK11p58 attenuated integrin β3 expression, whereas siRNA-mediated knockdown of CDK11p58 had no effect to attenuate integrin β3 expression (Figure 5B). Targeted knockdown of ERα by siRNA led to the inhibition of integrin β3 expression, whereas activation of ERα signaling by 17b-estradiol (E2) stimulated integrin β3 expression (Figure 5C and D). The T370D mutant significantly repressed ERα and integrin β3 protein levels, while the T370A mutant failed to repress ERα and integrin β3 protein levels (Figure 5E). Metastasis-related matrix metalloproteinases (MMPs), including MMP2, MMP3 and MMP9, were also repressed by CDK11p58 in a kinase-dependent manner. Together these data indicated that CDK11p58 represses integrin β3 expression to inhibit the metastasis of breast cancer via the regulation of ERα signaling.
Figure 5.

CDK11 p58 inhibits cell motility and invasion in ERα-positive breast cancer cells through integrin β3. (A) ZR-75-30 cells were transfected with specific siRNAs targeting CDK11p58. After 48 h, the cells were harvested and subject to immunoblotting analysis as indicated. Protein levels were normalized to GAPDH. (B) ZR-75-30 cells were transfected with the CDK11p58 plasmid or siRNA3 targeting CDK11p58 as indicated, and lysates were subjected to an immunoblotting analysis. (C) ZR-75-30 cells were transfected with siRNA targeting ERα, and after 48 h, cell lysates were subjected to immunoblotting analysis. (D) ZR-75-30 cells were transfected with ERα, and after 36 h, the cells were treated with ethanol or 10 nM E2 for an additional 12 h. The cells were harvested and subjected to immunoblotting analysis as indicated. (E) ZR-75-30 cells were transfected with CDK11p58 wild-type or mutants. After 48 h, the cells were harvested and the lysates subjected to immunoblotting analysis as indicated.
Discussion
CDK11p58 is involved in a variety of important regulatory pathways in eukaryotic cells, including cell cycle control, apoptosis, neuronal physiology, differentiation and transcription [26–28]. CDK11p58 is a Ser/Thr kinase, and the majority of its functions are dependent on its kinase activity. In our previous study, we found that CDK11p58 inhibited ERα transcriptional activity by promoting its degradation via the ubiquitin-proteasome pathway. In the present study, we demonstrate, for the first time, that CDK11p58 expression is involved in the negative regulation of breast cancer invasion in a kinase-dependent manner.
Recent studies have reported the existence of various genomic alterations of 1p36 in ERα-positive breast cancers. CDK11p58 is located on chromosome 1p36 and plays a role in the negative regulation of ERα signaling. Approximately 70–75% of breast cancers express ER and/or progesterone receptor. In recent years, we have witnessed tremendous advances in the understanding of ERα biology, revealing a complex process of ERα signaling that includes interactions with other signaling pathways [29]. The ERα signaling pathway plays an important role in the development and progression of breast cancer, and we previously showed that CDK11p58 interacts with ERα in breast cancer cells. Estrogens and ERα act as promoters of cell movement in different tissues, including breast tissue [30]. Previous studies showed that ERα induced breast cancer cell migration and invasion via the phosphorylation of FAK and N-WASP [24]. Another report revealed that within the broader range of actions of ERα, rapid extra-nuclear signaling to the actin cytoskeleton through the Gα13/RhoA/ROCK/moesin cascade is relevant for the determination of estrogen-dependent breast cancer cell movement and invasion, which are related to cancer metastasis [31]. Previous studies also demonstrated that the MAPK-integrated signaling crosstalk between Ras and ERα leads to increased breast cancer invasion [32]. Because CDK11p58 plays a negative role in the regulation of ERα, we speculated that CDK11p58 may participate in the progression of breast cancer via the regulation of the ERα pathway.
First we examined the expression of CDK11 in 250 breast cancer patients and found that high expression of CDK11 indicated a better prognosis, while low expression of CDK11 was related with a worse breast cancer node status, relapse and metastasis. These data suggest that CDK11p58 is related to migration and invasion. As expected, CDK11p58 inhibited the migration and invasion of ZR-75-30 ERα-positive breast cancer cells in a kinase-dependent manner. To examine the mechanism by which CDK11p58 inhibited breast cancer cell invasion, we specifically examined CDK11p58-dependent gene expression changes in invasion signaling pathways using TaqMan array human metastasis plates. We observed that the expression of integrin β3 was significantly attenuated in CDK11p58-transfected cells. Integrins are cell surface glycoproteins that control cell attachment to the extracellular matrix [33]. Integrins play a role in mammary gland biology, with expression in all cell types within the gland [34], and activate intracellular signaling pathways that control proliferation, differentiation, apoptosis, cell motility, migration and survival [35, 36]. Integrin β3 is expressed on the surface of endothelial cells, smooth muscle cells, monocytes and platelets [37]. Expression is predominantly associated with tumor metastasis and has been reported to increase the metastatic potential of melanoma, breast cancer and lymphoma cells. Interestingly, the opposite effect is observed in ovarian cancer [38].
In this study, we found that integrin β3 promoter activity was significantly repressed by CDK11p58 and that this repression was dependent on CDK11p58 kinase activity. ERα belongs to the superfamily of nuclear receptors, comprising structurally conserved transcription factors that enhance the transcription of specific genes upon hormone binding [39]. Upon E2 binding, ERα specifically interacts with ERE sequences in target promoters and stimulates the transcription of a variety of E2-responsive genes [40]. As previously published, CDK11p58 is capable of repressing the transcriptional activity of ERα and promotes its degradation. In the present study, we further demonstrate that the promoter activity of integrin β3 is stimulated by ERα and that the inhibition of integrin β3 by CDK11p58 is dependent on ERα.
Decreased expression of ERα correlated with a decrease in integrin β3 protein. Furthermore, the induction of ERα expression following transfection with ERα plasmids or the activation of ERα signaling with E2 was correlated with the enhanced expression of integrin β3 protein. Overall, CDK11p58 and T370D were capable of repressing the expression and transactivation of ERα and ultimately inhibiting the expression of integrin β3, thereby inhibiting the invasion of breast cancer cells. The kinase-dead mutants T370A failed to inhibit the expression of ERα and integrin β3, and were incapable of inhibiting cancer invasion. Taken together, we conclude that CDK11p58 may inhibit breast cancer cell invasion via the inhibition of the ERα signaling pathway, leading to the inhibition of integrin β3. Furthermore, metastasis-promoting MMP2, MMP3 and MMP9 proteins were subsequently inhibited. Thus, CDK11p58 inhibits the invasion and metastasis of breast cancer via the inhibition of integrin β3.
Conclusions
Our data show that CDK11p58 inhibits the invasion of ERα-positive breast cancer cells by repressing integrin β3. Thus, we demonstrate, for the first time, that CDK11p58 is involved in the negative regulation of breast cancer invasion in a kinase activity-dependent manner.
Acknowledgment
This work was supported by the National Natural Scientific Foundation of China (81102002).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JW and XZ conceived and designed the study. YC and SH performed the experiments. LW and RZ analyzed the data. LS, XX, DL and YC contributed reagents, materials and analysis tools. YC wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Yayun Chi, Email: yychi@126.com.
Sheng Huang, Email: sammer312@126.com.
Lei Wang, Email: wanglei56713@yahoo.com.cn.
Ruoji Zhou, Email: zrj1204@gmail.com.
Lisha Wang, Email: rabbitstanely@yahoo.com.cn.
Xiuying Xiao, Email: xiaoxiuying2002@163.com.
Dali Li, Email: lidalipopeye@yahoo.com.cn.
Ying Cai, Email: cy3163@163.com.
Xiaoyan Zhou, Email: xyzhou100@yahoo.com.
Jiong Wu, Email: wujiong1122@vip.sina.com.
References
- 1.Snyder RD. New concepts in breast cancer therapy. Intern Med J. 2004;34(5):266–269. doi: 10.1111/j.1444-0903.2004.00586.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Zhou BP. Epithelial-mesenchymal Transition-A Hallmark of Breast Cancer Metastasis. Cancer Hallm. 2013;1(1):38–49. doi: 10.1166/ch.2013.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almendro V, Kim HJ, Cheng YK, Gonen M, Itzkovitz S, Argani P, Van Oudenaarden A, Sukumar S, Michor F, Polyak K. Genetic and phenotypic diversity in breast tumor metastases. Cancer Res. 2014;74(5):1338–1348. doi: 10.1158/0008-5472.CAN-13-2357-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukamoto K, Ito N, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G, Nakamura Y, Emi M. Allelic loss on chromosome 1p is associated with progression and lymph node metastasis of primary breast carcinoma. Cancer. 1998;82(2):317–322. doi: 10.1002/(SICI)1097-0142(19980115)82:2<323::AID-CNCR11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 5.Ragnarsson G, Eiriksdottir G, Johannsdottir JT, Jonasson JG, Egilsson V, Ingvarsson S. Loss of heterozygosity at chromosome 1p in different solid human tumours: association with survival. Br J Cancer. 1999;79(9–10):1468–1474. doi: 10.1038/sj.bjc.6690234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han W, Han MR, Kang JJ, Bae JY, Lee JH, Bae YJ, Lee JE, Shin HJ, Hwang KT, Hwang SE, Kim SW, Noh DY. Genomic alterations identified by array comparative genomic hybridization as prognostic markers in tamoxifen-treated estrogen receptor-positive breast cancer. BMC Cancer. 2006;6:92. doi: 10.1186/1471-2407-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borg A, Zhang QX, Olsson H, Wenngren E. Chromosome 1 alterations in breast cancer: allelic loss on 1p and 1q is related to lymphogenic metastases and poor prognosis. Genes Chromosomes Cancer. 1992;5(4):311–320. doi: 10.1002/gcc.2870050406. [DOI] [PubMed] [Google Scholar]
- 8.Lahti JM, Xiang J, Heath LS, Campana D, Kidd VJ. PITSLRE protein kinase activity is associated with apoptosis. Mol Cell Biol. 1995;15(1):1–11. doi: 10.1128/mcb.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128(3):459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 10.Schlisio S, Kenchappa RS, Vredeveld LC, George RE, Stewart R, Greulich H, Shahriari K, Nguyen NV, Pigny P, Dahia PL, Pomeroy SL, Maris JM, Look AT, Meyerson M, Peeper DS, Carter BD, Kaelin WG., Jr The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22(7):884–893. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Climent J, Perez-Losada J, Quigley DA, Kim IJ, Delrosario R, Jen KY, Bosch A, Lluch A, Mao JH, Balmain A. Deletion of the PER3 gene on chromosome 1p36 in recurrent ER-positive breast cancer. J Clin Oncol. 2010;28(23):3770–3778. doi: 10.1200/JCO.2009.27.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, Morgan DO, Tsai LH, Wolgemuth DJ. Cyclin-dependent kinases: a family portrait. Nat Cell Biol. 2009;11(11):1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi Y, Zhang C, Zong H, Hong Y, Kong X, Liu H, Zou W, Wang Y, Yun X, Gu J. Identification of CDK11p58 Thr-370 responsible for its autophosphorylation, dimerization and kinase activity. J Biol Chem. 2010;286(3):1748–1757. doi: 10.1074/jbc.M110.107367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Wang H, Zong H, Sun Q, Kong X, Jiang J, Gu J. Downregulation of beta1,4-galactosyltransferase 1 inhibits CDK11(p58)-mediated apoptosis induced by cycloheximide. Biochem Biophys Res Commun. 2005;327(2):628–636. doi: 10.1016/j.bbrc.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 15.Guo HQ, Gao M, Ma J, Xiao T, Zhao LL, Gao Y, Pan QJ. Analysis of the cellular centrosome in fine-needle aspirations of the breast. Breast Cancer Res. 2007;9(4):R48. doi: 10.1186/bcr1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneeweiss A, Sinn HP, Ehemann V, Khbeis T, Neben K, Krause U, Ho AD, Bastert G, Kramer A. Centrosomal aberrations in primary invasive breast cancer are associated with nodal status and hormone receptor expression. Int J Cancer. 2003;107(3):346–352. doi: 10.1002/ijc.11408. [DOI] [PubMed] [Google Scholar]
- 17.Olson JE, Wang X, Pankratz VS, Fredericksen ZS, Vachon CM, Vierkant RA, Cerhan JR, Couch FJ. Centrosome-related genes, genetic variation, and risk of breast cancer. Breast Cancer Res Treat. 2011;125(1):221–228. doi: 10.1007/s10549-010-0950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petretti C, Savoian M, Montembault E, Glover DM, Prigent C, Giet R. The PITSLRE/CDK11p58 protein kinase promotes centrosome maturation and bipolar spindle formation. Embo Rep. 2006;7(4):418–424. doi: 10.1038/sj.embor.7400639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franck N, Montembault E, Rome P, Pascal A, Cremet JY, Giet R. CDK11(p58) is required for centriole duplication and Plk4 recruitment to mitotic centrosomes. PLoS One. 2011;6(1):e14600. doi: 10.1371/journal.pone.0014600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zong H, Chi Y, Wang Y, Yang Y, Zhang L, Chen H, Jiang J, Li Z, Hong Y, Wang H, Yun X, Gu J. Cyclin D3/CDK11p58 complex is involved in the repression of androgen receptor. Mol Cell Biol. 2007;27(20):7125–7142. doi: 10.1128/MCB.01753-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi Y, Hong Y, Zong H, Wang Y, Zou W, Yang J, Kong X, Yun X, Gu J. CDK11p58 represses vitamin D receptor-mediated transcriptional activation through promoting its ubiquitin-proteasome degradation. Biochem Biophys Res Commun. 2009;386(3):493–498. doi: 10.1016/j.bbrc.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Zong H, Chi Y, Hong Y, Yang Y, Zou W, Yun X, Gu J. Repression of estrogen receptor alpha by CDK11p58 through promoting its ubiquitin-proteasome degradation. J Biochem. 2009;145(3):331–343. doi: 10.1093/jb/mvn177. [DOI] [PubMed] [Google Scholar]
- 23.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354(3):270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez AM, Flamini MI, Baldacci C, Goglia L, Genazzani AR, Simoncini T. Estrogen receptor-{alpha} promotes breast cancer cell motility and invasion via focal adhesion kinase and N-WASP. Mol Endocrinol. 2010;24(11):2114–2125. doi: 10.1210/me.2010-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savinov AY, Remacle AG, Golubkov VS, Krajewska M, Kennedy S, Duffy MJ, Rozanov DV, Krajewski S, Strongin AY. Matrix metalloproteinase 26 proteolysis of the NH2-terminal domain of the estrogen receptor beta correlates with the survival of breast cancer patients. Cancer Res. 2006;66(5):2716–2724. doi: 10.1158/0008-5472.CAN-05-3592. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Yan J, Sun Q, Yao L, Jian Y, Lu J, Gu J. Induction of apoptosis by p110C requires mitochondrial translocation of the proapoptotic BCL-2 family member BAD. Febs Lett. 2006;580(3):813–821. doi: 10.1016/j.febslet.2005.12.097. [DOI] [PubMed] [Google Scholar]
- 27.Harper JW, Adams PD. Cyclin-dependent kinases. Chem Rev. 2001;101(8):2511–2526. doi: 10.1021/cr0001030. [DOI] [PubMed] [Google Scholar]
- 28.Trembley JH, Loyer P, Hu D, Li T, Grenet J, Lahti JM, Kidd VJ. Cyclin dependent kinase 11 in RNA transcription and splicing. Prog Nucleic Acid Res Mol Biol. 2004;77:263–288. doi: 10.1016/S0079-6603(04)77007-5. [DOI] [PubMed] [Google Scholar]
- 29.Jung HH, Park YH, Jun HJ, Kong J, Kim JH, Kim JA, Yun J, Sun JM, Won YW, Lee S, Kim ST, Ahn JS, Im YH. Matrix metalloproteinase-1 expression can be upregulated through mitogen-activated protein kinase pathway under the influence of human epidermal growth factor receptor 2 synergized with estrogen receptor. Mol Cancer Res. 2010;8(7):1037–1047. doi: 10.1158/1541-7786.MCR-09-0469. [DOI] [PubMed] [Google Scholar]
- 30.Ricketts D, Turnbull L, Ryall G, Bakhshi R, Rawson NS, Gazet JC, Nolan C, Coombes RC. Estrogen and progesterone receptors in the normal female breast. Cancer Res. 1991;51(7):1817–1822. [PubMed] [Google Scholar]
- 31.Giretti MS, Fu XD, De Rosa G, Sarotto I, Baldacci C, Garibaldi S, Mannella P, Biglia N, Sismondi P, Genazzani AR, Simoncini T. Extra-nuclear signalling of estrogen receptor to breast cancer cytoskeletal remodelling, migration and invasion. PLoS One. 2008;3(5):e2238. doi: 10.1371/journal.pone.0002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi X, Tang J, Loesch M, Pohl N, Alkan S, Chen G. p38gamma mitogen-activated protein kinase integrates signaling crosstalk between Ras and estrogen receptor to increase breast cancer invasion. Cancer Res. 2006;66(15):7540–7547. doi: 10.1158/0008-5472.CAN-05-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakubowska A, Rozkrut D, Antoniou A, Hamann U, Lubinski J. The Leu33Pro polymorphism in the ITGB3 gene does not modify BRCA1/2-associated breast or ovarian cancer risks: results from a multicenter study among 15,542 BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2010;121(3):639–649. doi: 10.1007/s10549-009-0595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw LM. Integrin function in breast carcinoma progression. J Mammary Gland Biol Neoplasia. 1999;4(4):367–376. doi: 10.1023/A:1018766317055. [DOI] [PubMed] [Google Scholar]
- 35.Taddei I, Faraldo MM, Teuliere J, Deugnier MA, Thiery JP, Glukhova MA. Integrins in mammary gland development and differentiation of mammary epithelium. J Mammary Gland Biol Neoplasia. 2003;8(4):383–394. doi: 10.1023/B:JOMG.0000017426.74915.b9. [DOI] [PubMed] [Google Scholar]
- 36.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2(2):91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 37.Lei Y, Huang K, Gao C, Lau QC, Pan H, Xie K, Li J, Liu R, Zhang T, Xie N, Nai HS, Wu H, Dong Q, Zhao X, Nice EC, Huang C, Wei Y. Proteomics identification of ITGB3 as a key regulator in reactive oxygen species-induced migration and invasion of colorectal cancer cells. Mol Cell Proteomics. 2011;10(10):M110–M5397. doi: 10.1074/mcp.M110.005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desgrosellier JS, Barnes LA, Shields DJ, Huang M, Lau SK, Prevost N, Tarin D, Shattil SJ, Cheresh DA. An integrin alpha(v)beta(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat Med. 2009;15(10):1163–1169. doi: 10.1038/nm.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83(6):851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/14/577/prepub


