Abstract
Oligometastasis is a cancer disease state characterized by a limited number of metastatic tumors involving single or few organs and with biological properties that make them potentially amenable to locoregional antitumor therapy. Current clinical data show that they are potentially curable with surgical resection or/and radiotherapy. Yet, mechanisms of progression from primary tumor to oligometastasis, rather than to polymetastases, is lacking in detail. In the current review we focus on the role of micro-RNAs in the regulation of metastases development and the role they may play in the differentiation of oligometastatic from polymetastatic progression. We also discuss the analyses of metastatic samples from oligo-and polymetastatic patients, which suggest that oligometastasis is a distinct biologic entity regulated in part by micro-RNAs. In addition, a review of the known functions of oligometastatic-specific micro-RNAs suggest that they regulate multiple steps in the metastatic cascade, including epithelial–mesenchymal transition, tumor invasion, intravasation, distant vascular extravasation and proliferation in a distant organ. Understanding the role of micro-RNAs and their target genes in oligometastatic disease may allow for the development of targeted therapies to effectively conrol the spread of metastases.
Keywords: Oligometastases, Metastases, Micro-RNA, Breast cancer, Lung cancer, Colorectal cancer
Introduction
Current approaches to the staging of cancer patients are based on the separation of primary or locoregional disease and distant metastases. Such separation depends on the ability to apply curative surgical or radiotherapy approaches to the primary tumors or locoregional disease, while distant metastases are commonly treated by palliative systemic therapy [1, 2]. The heterogeneity of metastatic spread is generally underestimated and many clinical regimens are not based on the extent of metastatic dissemination. The term “oligometastases” was introduced in 1995 and describes an intermediate state of metastatic spread between localized disease and diffuse metastases [3]. The initial definition of oligometastases was introduced mostly based on the limited number of secondary lesions, but more recently, the rate of metastases development was taken in consideration with introduction of the concept of “oligorecurrence” [4–6]. The concept of oligometastatic disease has significant clinical value because a limited number of lesions can be treated with the same potentially ablative approaches as spatially confined primary tumors. Previously, the feasibility of this approach was demonstrated mostly with surgical intervention in limited number of cancers [7, 8]. More recently, rapid advances in imaging techniques and stereotactic body radiotherapy (SBRT) allow earlier detection of oligometastases and non-invasive or minimally invasive intervention in cancers with different histologies and different secondary sites [9–11]. Results from large-scale randomized controlled studies are not available, but recent comprehensive reviews of the literature suggest that approximately 20% of patients may represent an oligometastatic phenotype where local treatment of metastases by resection, ablation or radiotherapy yields outcomes that are substantially improved when compared to non-treated patients [12–15]. Yet, little is known so far about the molecular basis of oligometastases and the pathways that can be responsible for differentiating between polymetastatic or oligometastatic spread.
In the current report we discuss the potential role of micro-RNAs in oligometastatic disease based on known examples of regulation of the metastatic cascade by micro-RNAs, along with our initial investigations of micro-RNA expression in the secondary lesions of oligometastatic and polymetastatic patients.
Oligometastases versus polymetastases: are there genome-driven mechanisms?
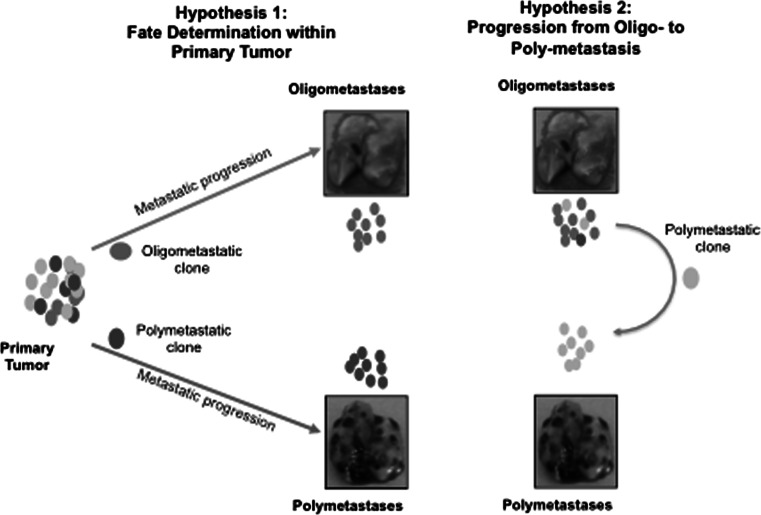
Despite advances in the understanding of the clinical significance of oligometastasis, comparative molecular and genomic investigations of oligometastatic and polymetastatic lesions are limited. Two extreme possibilities implicate either a clonal origin with inherited molecular and phenotypic differences of oligo-or polymetastases, or sequential development of the metastatic process with oligometastases as an early but transient phase of polymetastatic disease (Fig. 1). Current data based on genomics approaches are consistent with both suggestions. The clonal nature of metastases and their origin from topographically and genetically distinct regions of tumors has been demonstrated in the next-generation sequencing of the primary and secondary tumor samples taken from the same patients [16–18]. Distinct geographic regions of primary pancreatic tumors represented different clonal structure and gave origin to the distinct metastatic clones [16, 17]. In clear-cell renal carcinoma it was demonstrated that metastatic clones could share common mutational events with ancestor clones in the primary tumor [19]. In breast cancer, application of single-cell sequencing approaches revealed a clonal sub-structure of primary tumors and the initiation of liver metastases as the result of expansion of individual primary clones [18, 20]. These studies demonstrated that secondary tumors are derived from individual clones of heterogeneous primary tumors and follow the rules of Darwinian evolution [21, 22]. Therefore it is reasonable to suggest that secondary lesions with an oligometastatic phenotype may originate from the tumor clones with genetic properties different from clones potentially “programmed” towards polymetastatic dissemination. Detection of these differences can be pivotal in identifying prognostic/predictive markers of metastatic disease and detection of new therapy targets. In addition, constant evolution and “re-seeding” have been demonstrated [17, 23]. This raises the possibility that oligometastases may be a transient stage of general metastatic progression (Fig. 1). The time scale of spontaneous metastases development, estimated from genomic differences between matched primary and secondary tumors, varies between 1 year to 5 decades [24], providing a variety of opportunities for the evolution from restricted oligometastatic clones to disseminated disease either through tertiary metastases or progressive evolution of the primary tumor [17]. Genomic and transcriptomic profiling of oligo-and polymetastatic lesions may reveal recurrent mutations/patterns common for both types of secondary tumors or identify distinguishing characteristics that separate biologic processes responsible for distinct phenotypes.
Fig. 1.
Pathways of oligo-and polymetastases development. Two hypotheses of Oligometastastic Disease: Hypothesis 1 Oligometastases and Polymetastases may be distinct metastasis phenotypes determined by dissemination of clonal populations with differing metastatic potential. Hypothesis 2 Metastasis may be a continuum of phenotypes identified early (oligometastases) or late (polymetastases) in the progression of disease
Multi-step nature of metastatic cascades as the basis for differences in oligo-and polymetastatic progression
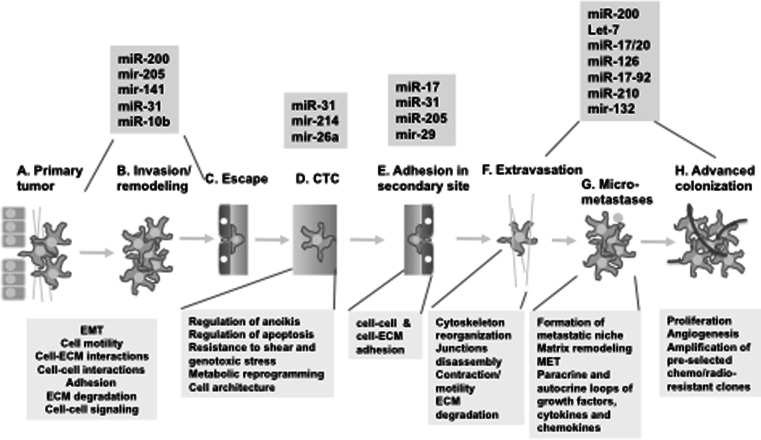
Based on advances of high-throughput genomic, transcriptomic and proteomic approaches, the last decade has brought important breakthroughs in our understanding of the metastasis process. It is commonly accepted now that metastases formation is a multi-step process with defined steps (Fig. 2). Understanding of the specific biological processes and pathways underlying each step is rapidly evolving (reviewed in [25–28]). This multi-step nature of metastatic cascades suggests that divergence between oligometastases and polymetastases may be initiated on different steps of metastases development. Each step can provide a bifurcation between the evolutionary pathways of these metastatic types, though it is possible to speculate that bifurcations in the earlier steps (such as invasion/remodeling, Fig 2b) can lead to more pronounced differences between oligo-and polymetastatic phenotypes as compared with bifurcations at the later steps (Fig. 2g, h). This multi-step nature of metastatic cascades can induce heterogeneity even within these two metastatic phenotypes. Indeed, recently Tree et al. [12] noticed that at least three different cohorts of patients were reported as oligometastatic: those who present at time of diagnosis with oligometastatic disease (“synchronous” oligometastases), those who had truly oligometastatic relapse after curative locoregional therapy of primary disease (metachronous oligometastases) and those with “induced” oligometastases after cytoreductive therapy. These groups probably have different prognoses and benefit from different therapeutic approaches. Detailed investigations of these steps in the context of oligometastases and polymetastases development are necessary for detection of prognostic markers and potential therapeutic targets with potential clinical applications.
Fig. 2.
Multiple steps of metastases development and role of micro-RNA in their regulation. Sequential steps of metastases development are presented based on the current reviews [25, 26, 28, 51]. Note that final selection of metastatic clones can lead to the formation of radio/chemoresistant secondary tumors, as was described elsewhere [33], presumably due to selection against cytotoxic factors of host microenvironment (see g–h). Boxes below diagram represent major functions involved in each step, while boxes above illustrate some micro-RNAs involved in the regulation of each step (see text for explanations and references)
Examination of biological processes implicated in the different stages of metastases development leads to an important conclusion: if the most prominent hallmark of cancer growth—sustained proliferation [29]—is critical for the development of primary tumors and advanced colonization (Fig. 2a, h), all intermediate steps of metastases development are based on the processes not directly related to the regulation of tumor growth. Rather, they involve multiple interactions of metastatic cells with microenvironment either through direct heterotypic cell–cell and cell–ECM contacts (as in steps 2b–2f) or formation of multiple paracrine and autocrine loops between tumor and stromal cells via secreted ligands and their cognate receptors (Fig. 2g) [30, 31, 32]. Of note, formation of these auto-paracrine loops is often associated with development of tumor clones resistant to genotoxic stress [33–36]. These processes involve constant alterations of the cytoskeleton and cell architecture associated with motility, invasion and circulation in blood. This suggests that metastases development might be regulated through mechanisms different from traditional oncogenic signaling, and instead adopted to the tumor plasticity typical for metastatic cascades. Accumulating evidences indicate that one of the most important mechanisms of regulation of the broad set of pathways involved in metastases regulation is based on micro-RNAs.
Micro-RNAs as systemic regulators of metastases development
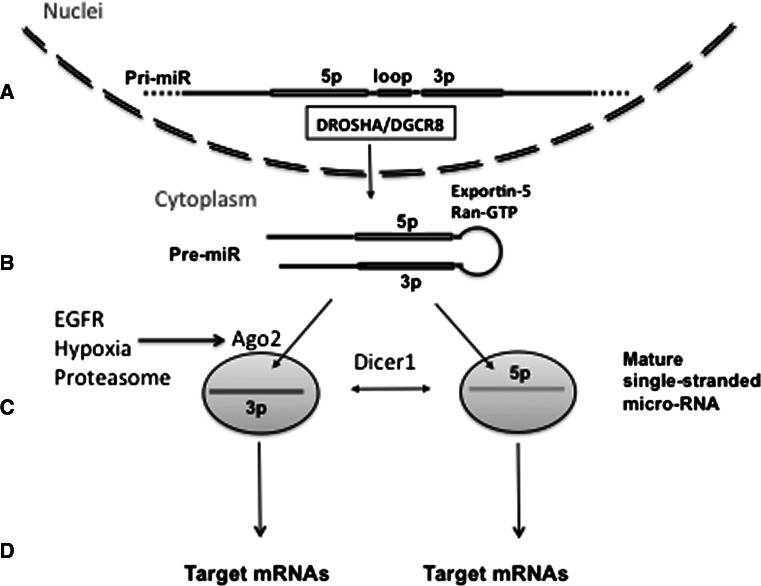
MicroRNAs (miRs) are small (19–22 nucleotide) RNAs involved in the regulation of gene expression. A full description of the biogenesis and complex functions of miRs is beyond the scope of this review and is available elsewhere [37–39]. Briefly, precursors of miRs are encoded by intronic regions of protein encoding genes or by RNA encoding regions of the genome known as primary microRNAs (pri-miRs). They are processed and translocated into the cytoplasm in the form of double-stranded stem-loop structures known as precursor microRNAs (pre-miRs) with the involvement of multiple nucleases including the Drosha, Dicer and Argonaut families (Fig. 2). Importantly, recent investigations detected systemic changes in micro-RNA processing, with imbalance between pre-miRs and mature miRs, and between 3p and 5p mature miRs, due to Ago2 protein modifications by EGFR and other regulators [40, 41] in addition to tumor-related somatic mutations of Dicer1 [42]. Suppression of Ago2 can lead to the decrease of the ratio of mature miRs to stem-loop precursors, while mutations in Dicer1 are associated with the decrease of 5p to 3p ratio of mature single-stranded miRs (Fig. 3). These observations indicate that functional or mutational changes of single proteins involved in the processing of miRs can affect hundreds of down-stream genes. They also indicate that in the analysis of micro-RNA expression, it is important to account for the balance between pre-miRs and mature miRs and the balance between 5p and 3p mature miRs. Each mature single-stranded miR binds to target mRNAs through complimentary seed sequences 6 to 8 nucleotides in length. Longer mRNA complimentary sites and the presence of adenosine immediately 5’ of the site may increase mRNA suppression (reviewed in [43] ). Upon binding, primarily to the 3’ untranslated region (UTR) of the mRNA, miRs induce either suppression of mRNA translation or degradation of the mRNA itself. Each miR is able to bind to hundreds of target mRNAs, thus they represent a potent group of regulators of gene expression.
Fig. 3.
Processing and maturation of micro-RNAs. Genomic sequence of primary micro-RNA (pri-miR) encodes ~80 nt sequence of the stem-loop pre-micro-RNA (Fig. 3a). This stem-loop structure (pre-miR) is recognized and excised from transcribed RNA by multiple proteins including Drosha and DGCR8. The stem-loop pre-miR is exported in cytoplasm by Exportin-5/Ran-GTP complexes (Fig. 3b) and is further processed by Ago2/Dicer1 nucleases into the mature single-stranded 5p and 3p miRs (Fig. 3c). The 5p and 3p mature single-stranded miRs will bind to different down-stream target mRNAs and can lead to different functional outcomes (Fig. 3d). Therefore the balance of micro-RNA precursors and mature forms can determine different phenotypes of normal and tumor cells. Current data indicate that this balance is susceptible to post-translational modifications and/or mutations of enzymes, responsible for miRs processing, including Ago2 and Dicer1 (Fig. 3c). These mutations and post-translational modifications of Ago2, associated with EGFR signaling, hypoxia and proteasome functions are described elsewhere [18–20]
Micro-RNAs’ extensive regulatory ability has engendered intensive investigation in the context of metastases development. Micro RNAs can mediate the formation of the flexible networks involved in the regulation of the multistep metastatic cascades. Micro-RNAs have also been shown to be regulated by p53 and myc, leading to either inhibition or activation of pro-survival mechanisms necessary for initial proliferation of the primary tumor [44]. This step can also be modulated by regulation of DNA repair and epigenetic modifications to DNA by micro-RNAs, and the converse regulation of micro-RNAs by these mechanisms, as has been highlighted in cancers with micro-satellite instability [45].
Proceeding along the metastatic cascade, many micro-RNAs have been implicated in regulation of epithelial to mesenchymal transition (EMT). One of the most studied families of micro-RNAs is the miR-200 family, which has been implicated with the maintenance of an epithelial phenotype (preventing EMT) via its gene targets Zeb1 and Zeb2, the transcriptional suppressors of E-cadherin [46, 47, 48]. This suppression prevents cancer cells in primary tumors from initiating the process of metastasis. Mir-205 is also implicated in EMT suppression through a feedback loop between Zeb1 and TGFB1 [49]. A similar function can be performed by mir-203 through formation of negative feedback loop with Snail [50]. It is clear that epithelial cell plasticity plays an important role in metastatic cascades, and the role of other micro-RNAs in this process is under intensive investigations as detailed below.
Concurrent with EMT is invasion of tumor cells through the basement membrane, a process extensively regulated by many micro-RNAs through regulation of Rho-mediated actin cytoskeleton regulation, integrin-mediated cell-cell adhesion and alterations in the extra-cellular matrix itself (reviewed in [51]). Another well-studied example of regulation at this step is miR-31, which suppresses invasion/migration of tumor cells, thereby suppressing the initial stages of metastases [52]. Proceeding along the metastatic cascade, the step of adhesion in the second site is critical for determination of site-specificity of a given tumor. Interestingly, due to the targeting of RhoA and ITGA5, miR-31 is critical in adhesion of circulating tumor cells, therefore providing one of the few known “master oncomirs” [52]. Similarly, miR-214 through suppression of AP2-transcription factor TFAP2C and Integrin alpha-3, can also promote both invasion and survival in anoikis [53]. This step of survival in circulation can also be promoted by miR-26a through Rb1-E2F1 pathway [54]. Some examples of metastases-related miRs are presented in Fig. 2. The final stages of metastases formation include initial colonization and subsequent proliferation in distant sites. These steps involve multiple miRs regulating the mesenchymal-to-epithelial transition (MET), formation of a pre-metastatic niche, adaptation of surviving tumor cells to the new microenvironment, secondary tumor growth and angiogenesis (Fig. 2g, h). Multiple miRs can regulate the phenotype of tumor cells in these stages. The oncogenic miR-17-92 cluster can be up-regulated by c-Myc, E2F and Hedgehog [55, 56] and suppresses thrombospondin 1, thereby promoting angiogenesis [57]. Let-7 and miR-126 can inhibit proliferation through HMGA2 and PIK3 signaling [58, 59], while the hypoxia-inducible miR-210 is involved in mitochondria stabilization and regulation of cell survival/proliferation (reviewed by [60]). An understanding of both the pro- and anti-metastatic effects of micro-RNA regulated networks is leading to an appreciation of the role of both tumor cells and the micro-environment in metastasis (further reviewed in [61]). These data suggest that analysis of miRs may lead to the identification of the biological basis of oligometastases and perhaps differentiate between patients who are truly oligometastatic or those who are at risk for widespread distant disease.
Detection of micro-RNAs associated with oligometastases development (oligomiRs)
As described above, microRNAs are potent regulators of metastatic cascades, providing flexible regulatory networks for metastases development. Yet, due to the limited number of studies comparing oligometastatic to polymetastatic clinical samples, little is known about their role in oligometastases development. To detect microRNAs associated with oligometastases and discriminate them from polymetastases, we investigated archival samples from two clinical databases of oligometastatic and polymetastatic patients [6, 62]. These investigations focused on the direct comparison of oligo-and polymetastatic secondary lesions rather than on comparison of primary and secondary tumors. This led us to the detection of microRNA patterns specific to oligometastatic lesions and suggested the differential regulation of specific molecular pathways that guide oligometastatic vs. polymetastatic development of secondary tumors. We will further name these oligometastases-specific microRNAs as oligomiRs.
Micro-RNA signatures of surgically resected lung metastasis with oligo-and polymetastatic progression
Sixty-three patients with available metastatic samples were considered oligometastatic at the time of pulmonary resection, with 5 or less metastases. Based on the analysis of recurrence patterns in the lung following resection, 32 patients were designated as slow progressors, corresponding to the oligometastatic phenotype and 16 were classified as rapid progressors, corresponding to the polymetastatic phenotype. These patients had dramatic differences in survival, as demonstrated by Kaplan–Meier analysis, in favor of the slow progressors [6].
Statistical analysis revealed that 39 micro-RNAs were differentially expressed between oligometastatic (slow progressors) and polymetastatic (rapid progressors) samples. Only three micro-RNAs were down regulated in oligometastases as compared to polymetastases; the remainder were up regulated (Fig. 4b). As described below, several miRNAs up regulated in oligometastases have been characterized as tumor suppressors, implying that their up-regulation may repress oncogenic genes/pathways and limit metastatic spread (Table 1). Therefore these experiments revealed that oligometastases of different primary origin that develop in the same secondary site (lungs) express patterns of tumor-suppressive micro-RNAs, which differentiates them from polymetastases.
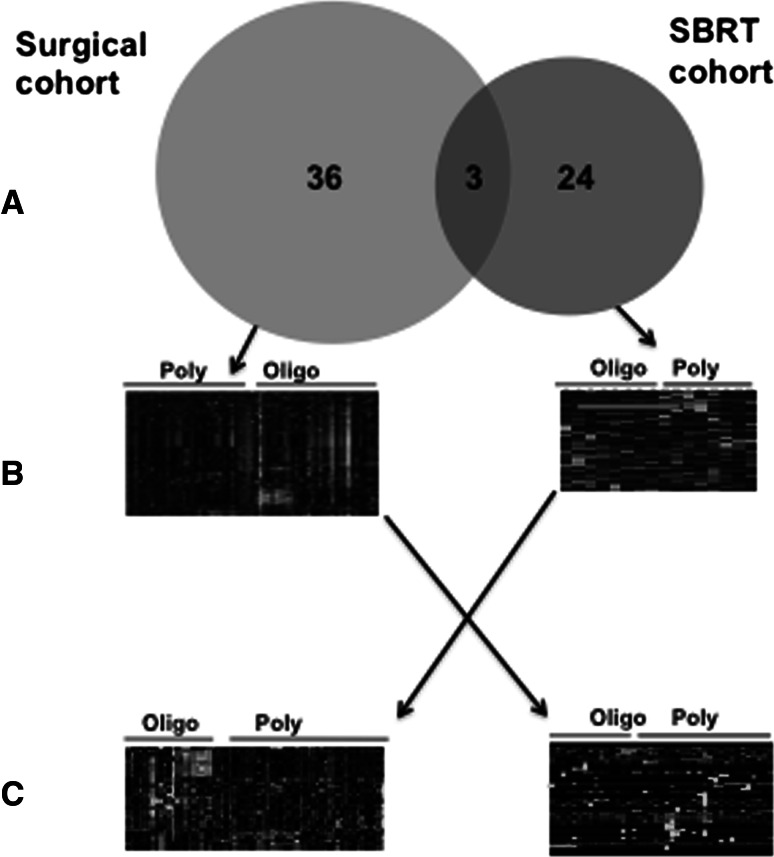
Fig. 4.
Cross-talk between micro-RNA patterns obtained in surgical and stereotactic body radiotherapy (SBRT) cohorts. a Only 3 overlapping micro-RNAs were identified in the surgical and SBRT cohorts (miR-328, miR-502-5p and miR-199b-5p (Table 1). b Unsupervised clustering of patients in the surgical and SBRT cohorts based on differentially expressed micro-RNAs successfully segregated patients with oligo-and polymetastatic disease independently on clinical parameters of disease progression (heat maps represent normalized CT values; green are up- and red are down-regulated micro-RNAs). c Application of the SBRT micro-RNA signature to surgical patients successfully separated them into oligo-and polymetastatic clusters (left panel); the same was true for application of the surgery micro-RNA signature to SBRT cohort
Table 1.
Known functions and down-stream targets of oligomiRs detected in clinical databases of oligometastatic patients
| miRNA | Dataset | Oncomir or suppressor | Controlled biological processes | Down-stream targets | References |
|---|---|---|---|---|---|
| Mir-655 | Surgery | Suppressor | EMT | TGFBR2,ZEB1 | [88] |
| mir-154 | Surgery | Suppressor | Proliferation, motility, cell cycle, EMT | TLR2, HMGA2, CCND2 | [89–91] |
| mir-329 | Surgery | Suppressor | Proliferation | E2F1 | [84] |
| mir-330 | Surgery | Mixed | Proliferation, invasion, cell cycle, adhesion, apoptosis | SP1, CDC42, SH3GL2 | [92, 93] |
| mir-485 | Surgery | Suppressor | Proliferation, migration | MAT1A | [76] |
| mir-380 | Surgery | Oncomir | Proliferation, cell cycle | p53 | [94] |
| mir-298 | Surgery | Suppressor | Drug sensitivity | P-gp | [95] |
| mir-153 | Surgery | Mixed | Proliferation, apoptosis, drug resistance | PTEN, FOXO3a, FOXO1 | [96–98] |
| mir-296 | Surgery | Mixed | Proliferation, cell cycle, apoptosis, drug resistance, angiogenesis | ICAM1, CDX1, EAG1, Scrib, HMGA1, HGS | [99–102] |
| mir-448 | Surgery | Suppressor | EMT | SATB1 | [103] |
| mir-133a | Surgery | Suppressor | Invasion, proliferation | FSCN1, PNP, TALGN2 | [104–106] |
| mir-328 | Both | Suppressor | Invasion, drug resistance, cytokinesis, migration, cell cycle, proliferation, ECM degradation | ABCG2, MMP16, KIF23, PTPRJ | [78, 107–110] |
| mir-502 | Both | Suppressor | Proliferation, cell cycle | Rab1B | [85] |
| miR-199b-5p | Both | Suppressor | Proliferation, drug resistance, invasion | HER2, HES1 | [111–113] |
| miR-125a-3p | SBRT | Suppressor | Invasion, proliferation, apoptosis, cell cycle arrest | FYN, RHOA, BCL2, ALDH1A3, MCLl1, VEGF, MMP11, TNFAIP3 | [74, 79, 114–118] |
| miR-140-5p | SBRT | Suppressor | EMT, proliferation | TGFBR1, FGF9 | [119] |
| miR-29c-3p | SBRT | Suppressor | Proliferation, invasion, adhesion, angiogenesis, drug resistance, apoptosis, cell cycle arrest | MMP2, ITGB1, SIRT1, TIAM1, TNFAIP3, CCNE1, p85, CDC42 | [80, 120–124] |
| miR-489 | SBRT | Suppressor | Proliferation | PTPN11 | [125] |
| miR-331-5p | SBRT | Suppressor | Cell cycle | E2F1 | [83] |
| miR-193a-3p | SBRT | Suppressor | Apoptosis, proliferation, EMT, drug resistance, cell cycle | MCL1, EGFR, PLAU, KRAS, p73, E2F6 | [82, 126–129] |
| miR-363-3p | SBRT | Mixed | EMT, invasion, proliferation | MBP1, PDPN | [75, 130, 131] |
Functions that are up-regulated by a micro-RNA are italicized
OligomiRs differentially expressed in patients treated with Stereotactic Body Radiotherapy (SBRT)
Stereotactic body radiotherapy is a form of radiotherapy that exploits advances in radiotherapy technology, computing and imaging to deliver high doses of radiation to local tumors. Patients enrolled in a clinical trial, reported by Lussier et al. [14, 63], had diverse primary and metastatic sites, received adjuvant therapy or/and surgery prior to enrollment and had controlled primary tumors with five or less metastases. Patients were classified into two groups based on radiographic response after completion of SBRT: polymetastatic patients had (1) progression with development of 5 new metastatic tumors in less than 4 months from time of first metastatic progression, or (2) progression within a body cavity suggesting the presence of diffuse metastatic disease. In contrast, oligometastatic patients had either no evidence of progression or experienced progression that did not satisfy the above criteria for designation of polymetastases. The expression patterns of micro-RNAs from metastatic tumors correctly classified seven of nine oligometastatic patients and five of five poly-metastatic patients (Fig. 4b). Twenty-nine differentially expressed oligomiRs were identified in this dataset—16 were down-regulated and 13 up-regulated in oligometastases relative to polymetastases (Table 1). Importantly, in five patients with available matched primary and metastatic tumors, samples of primary and secondary tumors were co-clustered. This suggests that secondary tumors inherit patterns of micro-RNAs expressed in primary tumors (Fig. 1a). On the other hand, analysis of the primary tumors was unable to statistically significantly separate the oligo-and polymetastatic patients based on primary micro-RNA patterns. This lack of discrimination may be due to the heterogeneity of primary tumors, wherein the micro-RNA expression patterns of those clones that can metastasize are masked by numerous non-metastasizing clones (Fig. 1). In genetically more homogenous metastatic tumors, these patterns can be revealed and potentially used for prognostic/predictive purposes. Development of micro-RNA signatures associated with types of metastases progression and/or response to local therapy can lead to improved prognostic tools for oligometastatic patients.
Cross-talk between oligomiRs patterns in two independent databases
Patterns of oligomiRs detected in each cohort of patients were able to stratify patients in oligo-and polymetastatic phenotype independently from clinical parameters (7, 24). In the surgery cohort, microRNA expression successfully classified 13 of 16 (81.3%) polymetastatic patients and 20 of 32 (62.5%) oligometastatic [odds ratio = 7.22; p = 0.006, Fisher’s exact test (FET), 24]. Similarly, in SBRT cohort patterns of oligomiRs successfully classified 8 of 10 (80%) oligometastatic patients and 6 of 6 (100%) polymetastatic (p = 0.007, FET, [7]). These separations are schematically depicted in Fig. 4b. At the same time, both patterns did not overlap significantly; only three microRNA were common between oligomiRs from surgery and SBRT cohorts (miR-328, miR-502-5p and miR-199b-5p; see Fig. 4a). Yet, when we used 39 oligomiRs detected in the surgery cohort to classify patients in the SBRT cohort, we obtained significant separation of oligo-and polymetastatic patients (OR = 7.2, p = 0.047, FET; Fig. 4c). Similarly, when we applied 27 oligomiRs from the SBRT cohort to classify patients in the surgery cohort we observed divergence of oligo-and polymetastatic phenotypes (OR = 3.9, p = 0.032, FET). These findings suggest that both sets of detected oligomiRs are actually co-expressed in oligometastatic lesions from two independent clinical cohorts, but cannot be simultaneously detected due to the limited statistical power of these smaller sample sets. Further studies incorporating larger numbers of samples and more sensitive profiling platforms will lead to extended list of oligomiRs commonly shared between different clinical groups of patients.
Biological validation of clinical observations-controversial functions of mir-200c in oligometastases development
Detection of oligomiRs differentially expressed in oligometastases as compared with polymetastases posed questions about their functional significance. We found in the SBRT cohort three up-regulated micro-RNAs in polymetastases that were members of the mir-200 family (mir-200b, mir-200c and mir-141). We suggested that if these differentially expressed micro-RNAs are involved in the regulation of metastatic phenotypes, their ectopic expression in oligometastatic tumor may switch its phenotype to polymetastatic. To test this concept two models were used: a syngeneic mouse melanoma B16 model with oligo and poly-metastatic clones derived as described elsewhere [33], and a human MB-435 breast carcinoma model with oligo- and poly-metastatic clones selected in vivo. Over-expression of mir-200c was sufficient to switch the oligometastatic phenotypes to polymetastatic phenotypes in both models. These findings indicated that oligomiRs detected in our clinical datasets may be causal in the regulation of oligo- vs. poly-metastatic development (7). At the same time these were paradoxical observations, since extensive literature indicate that mir-200c is a tumor suppressor (Fig. 2) [26, 27]. However, our data suggested that cells that had escaped the primary tumor were more metastatic when expressing microRNA 200c. Currently these results are supported by those of Dykxhoorn et al. [64] and Korpal et al. [65], both of which demonstrated that expression of 200c increases the metastatic phenotype in tumor cells that escape the primary tumor by enforcing the epithelial phenotype and/or suppressing tumor suppressive proteins induced by Sec23a, a target of mir-200c. These results demonstrate that micro-RNAs’ effects on metastasis progression may be different in primary tumor and distant metastases.
Multiple steps of the metastatic cascade are targeted by oligometastatic miRs
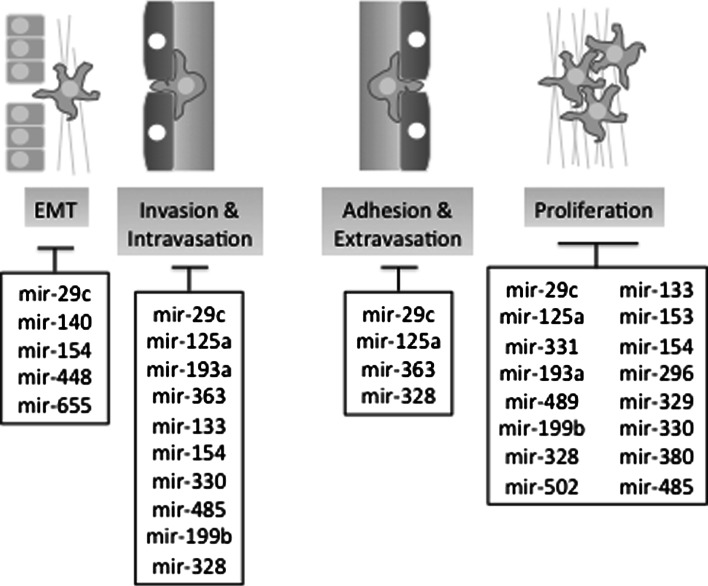
Almost all of the micro-RNAs over-expressed in the surgery oligometastatic samples, and many of those up-regulated in the SBRT oligometastatic samples, have been shown to be tumor-suppressors (Table 1). This suggests that oligometastases development may be associated with miR-based negative regulatory loops which are suppressed in polymetastases evolution. Examination of the molecular pathways predicted to be regulated by oligomiRs from both datasets suggests that they may regulate proliferation, EMT, invasion and motility through multiple down-stream targets (Table 1). Accordingly they may be implicated into the regulation of the distinct metastatic steps (Fig. 5).
Fig. 5.
Potential functions of oligomiRs. Multiple micro-RNAs up-regulated in oligometastases datasets (black SBRT, red Surgery and blue both datasets) are each capable of inhibiting multiple points along the metastatic cascade. These include epithelial–mesenchymal transition (EMT); invasion through parenchyma and intravasation into vasculature; adhesion to distant vasculature and extravasation into the distant organ parenchyma; and proliferation in the distal organ. Table 1 details biological processes/functions and down-stream targets of oligomiRs presented in this figure. (Color figure online)
Initial formation of pro-metastatic cells predisposed to successful accomplishment of metastatic cascade is commonly associated with epithelial–mesenchymal transition (EMT)— a morphological transformation necessary for escape from the primary tumor and subsequent invasion and metastatic growth. As discussed above, the mir-200c family is a well-known regulator of this mechanism through targeting of the TGF-beta pathway [66–69]. Upstream modulators of ZEB1 and mir-200c expression such as WASF3 and OSM have also been identified to play a role in EMT progression and tumor metastases [70, 71]. Another prominent regulator of EMT is mir-29c, which is up-regulated in oligometastatic patient samples and found to also suppress EMT [72]. Finally, the most highly differentially expressed micro-RNA in the pulmonary oligometastatic samples, mir-655, has recently shown to target ZEB1 and TGFBR2, resulting in similar inhibition of EMT [73]. These data suggest that limited metastatic dissemination seen in oligometastatic patients may partially depend on the control of EMT in the primary tumors (Fig. 5).
Several miRNAs over-expressed in oligometastatic samples have been shown to inhibit tumor cell adhesion and migration. Though these miRNAs have varied targets, their effects converge on mechanisms that are essential to regulation of the cell cytoskeleton and interaction with the extra-cellular matrix. Inhibition at this step can suppress metastasis progression by preventing invasion through the basement membrane in the primary tumor, or adhesion and migration of circulating tumor cells that could otherwise form metastases. Mir-125a is capable of inhibiting Fyn and RhoA, kinases involved in promoting cell adhesion and invasion through cytoskeletal re-arrangements and integrin signaling [74]. Mir-363 has similar effects on neuroblastoma cells and squamous cell carcinomas, inhibiting invasion in vitro and metastasis in vivo partly through suppression of podoplanin, a regulator of adhesion and cytoskeletal structure [75]. Similar findings have been observed in micro-RNAs up regulated in the pulmonary oligometastatic samples. Mir-485 can inhibit breast cancer migration, in addition to inhibition of colony formation in vitro [76]. Mir-296 inhibits Scrib, a regulator of cell motility, thus inhibiting growth in vivo, and repression of this micro-RNA increases tumor invasiveness in vitro [77]. Invasion and adhesion to extracellular matrix is an essential mechanism for metastasis development, and inhibition of this step may result in the low total metastatic burden observed in oligometastases by preventing colonization of distant organs by circulating tumor cells (Fig. 5).
Finally, regulation of proliferation may be critical on the late stages of metastatic cascades, leading to successful colonization (Figs. 2, 5, Table 1). Several micro-RNAs found to be up-regulated in both the SBRT trial and pulmonary resection oligometastases groups are implicated in such regulation. These include, mir-125a & b, mir-29c, mir-133a, mir-193a, mir-331, mir-329 and mir-502 (Table 1) [78–85].The ability of these oligomiRs to inhibit tumor growth is mediated by direct targeting of transcription factors, including the E2F family and cell-checkpoint proteins such as Cyclins D1, D2 and E1 in a variety of carcinoma types. Cyclin pathways, especially Cyclin D1 and its downstream effectors CDK4 and CDK6, represent important targets of tumor-suppressing micro-RNAs. Though the majority of known cell-cycle suppressing micro-RNAs appear to inhibit progression through G1, a few “oncomir” families have been identified that suppress key inhibitors of the cyclin/CDK complexes, p21 and p27 [86, 87]. Tumor cell growth inhibition through cell-cycle arrest may be a significant mechanism of the formation of locally confined slow-growing oligometastases that are amenable to treatment with localized therapy or resection.
Conclusion
Oligometastasis is a unique phenotype observed in some cases of metastatic cancer. This type of tumor spread is amenable to localized therapy including radiotherapy and resection, resulting in improved outcomes compared to patients with poly-metastatic disease. Analysis of patients’ tumor samples in two different datasets revealed differential micro-RNA expression profiles correlating with metastasis progression, regardless of primary tumor histology. Many micro-RNAs identified in these experiments were up-regulated in oligometastatic lesions and were associated with tumor-suppressive functions. Such “oligomiRs” may be involved in regulation of oligometastatic phenotype during multistep process of metastases development. Biological effects of oligomiRs identified in both datasets substantially overlapped and converged on cell proliferation, migration, adhesion and EMT. These functions are consistent with continuous interactions of the tumor cells with the host microenvironment during dissemination of metastatic cells from the primary tumor to distant sites. The ability to derive microRNA signatures from oligo/polymetastases, combined with an understanding of the pathways regulated by these oligoMirs, may help identify patients with oligometastatic progression with more accuracy than current clinical predictions. An understanding of the biologic basis of oligometastases has the potential to improve patient care through diagnostics and therapeutics. In addition, exploration of the functions of microRNAs linked to oligometastases may give more insight into the underlying mechanisms of metastatic progression and reveal new previously unidentified targets for future therapy.
Acknowledgments
This work was supported in part by The Virginia and D.K. Ludwig Fund for Cancer Research, Lung Cancer Research Foundation (LCRF) and Prostate Cancer Foundation (PCF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that they have no conflict of interest.
Contributor Information
Abhineet Uppal, Email: abhineet.uppal@uchospitals.edu.
Nikolai N. Khodarev, Email: n-khodarev@uchicago.edu
Ralph R. Weichselbaum, Email: rrw@radonc.uchicago.edu
References
- 1.Damin DC. Evolving treatment strategies for colo. World J Gastroenterol. 2014;20:877–887. doi: 10.3748/wjg.v20.i4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giordano SH. Update on locally advanced breast cancer. Oncologist. 2003;8(6):521–530. doi: 10.1634/theoncologist.8-6-521. [DOI] [PubMed] [Google Scholar]
- 3.Hellman S, Weichselbaum RR. Oligometastases. J Clin Oncol. 1995;13:8–10. doi: 10.1200/JCO.1995.13.1.8. [DOI] [PubMed] [Google Scholar]
- 4.Niibe Y, Chang JY, Onishi H, Salama J, Hiraki T, Yamashita H (2013) Oligometastases/oligo-recurrence of lung cancer. Pulm Med 438236–3. doi: 10.1155/2013/438236 [DOI] [PMC free article] [PubMed]
- 5.Withers HR, Lee SP. Modeling growth kinetics and statistical distribution of oligometastases. Semin Radiat Oncol. 2006;16:111–119. doi: 10.1016/j.semradonc.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Lussier YA, Khodarev NN, Regan K, Corbin K, Li H, Ganai S, Khan SA, Gnerlich J, Darga TE, Fan H, Karpenko O, Paty PB, Posner MC, Chmura SJ, Hellman S, Ferguson MK, Weichselbaum RR. Oligo-and polymetastatic progression in lung metastasis(es) patients is associated with specific MicroRNAs. PLoS One. 2012;7:e50141. doi: 10.1371/journal.pone.0050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–318. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adam R, De Gramont A, Figueras J, Guthrie A, Kokudo N, Kunstlinger F, Loyer E, Poston G, Rougier P, Rubbia-Brandt L, Sobrero A, Tabernero J, Teh C, Van Cutsem E, Jean-Nicolas Vauthey of the EGOSLIM (Expert Group on OncoSurgery management of LIver Metastases) group (2012) The oncosurgery approach to managing liver metastases from colorectal cancer: a multidisciplinary international consensus. In: Oncologist. pp 1225–1239 [DOI] [PMC free article] [PubMed]
- 9.Salah S, Watanabe K, Park JS, Addasi A, Park JW, Zabaleta J, Ardissone F, Kim J, Riquet M, Nojiri K, Gisabella M, Kim SY, Tanaka K. Repeated resection of colorectal cancer pulmonary oligometastases: pooled analysis and prognostic assessment. Ann Surg Oncol. 2013;20:1955–1961. doi: 10.1245/s10434-012-2860-y. [DOI] [PubMed] [Google Scholar]
- 10.Milano MT, Katz AW, Muhs AG, Philip A, Buchholz DJ, Schell MC, Okunieff P. A prospective pilot study of curative-intent stereotactic body radiation therapy in patients with 5 or fewer oligometastatic lesions. Cancer Res. 2007;112:650–658. doi: 10.1002/cncr.23209. [DOI] [PubMed] [Google Scholar]
- 11.Fumagalli I, Bibault J-E, Dewas S, Kramar A, Mirabel X, Prevost B, Lacornerie T, Jerraya H, Lartigau E (2012) A single-institution study of stereotactic body radiotherapy for patients with unresectable visceral pulmonary or hepatic oligometastases. Radiat Oncol (London, England) 7:164. doi: 10.1186/1748-717X-7-164 [DOI] [PMC free article] [PubMed]
- 12.Tree AC, Khoo VS, Eeles RA, Ahmed M, Dearnaley DP, Hawkins MA, Huddart RA, Nutting CM, Ostler PJ, van As NJ. Stereotactic body radiotherapy for oligometastases. Lancet Oncol. 2013;14:e28–e37. doi: 10.1016/S1470-2045(12)70510-7. [DOI] [PubMed] [Google Scholar]
- 13.Weichselbaum RR, Hellman S. Oligometastases revisited. Nat Rev Clin Oncol. 2011;8:378–382. doi: 10.1038/nrclinonc.2011.44. [DOI] [PubMed] [Google Scholar]
- 14.Corbin KS, Hellman S, Weichselbaum RR. Extracranial oligometastases: a subset of metas. J Clin Oncol. 2013;31:1384–1390. doi: 10.1200/JCO.2012.45.9651. [DOI] [PubMed] [Google Scholar]
- 15.Ashworth A, Rodrigues G, Boldt G, Palma D. Is there an oligometastatic state in non-small cell lung cancer? A systematic review of the literature. Lung Cancer. 2013;82:197–203. doi: 10.1016/j.lungcan.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin M-L, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein CA. Selection and adaptation during metastatic cancer progression. Nature. 2013;501:365–372. doi: 10.1038/nature12628. [DOI] [PubMed] [Google Scholar]
- 21.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 22.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 23.Kim MY, Oskarsson T, Acharyya S, Nguyen DX. Tumor self-seeding by circulating cancer cells. Cell. 2009;139(7):1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Chiang AC, Massagué J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reymond N, d’Água BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Khodarev NN, Yu J, Labay E, Darga T, Brown CK, Mauceri HJ, Yassari R, Gupta N, Weichselbaum RR. Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J Cell Sci. 2003;116:1013–1022. doi: 10.1242/jcs.00281. [DOI] [PubMed] [Google Scholar]
- 31.Geng Y, Chandrasekaran S, Hsu J-W, Gidwani M, Hughes AD, King MR. Phenotypic switch in blood: effects of pro-inflammatory cytokines on breast cancer cell aggregation and adhesion. PLoS One. 2013;8:e54959. doi: 10.1371/journal.pone.0054959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rückert F, Grützmann R, Pilarsky C. Feedback within the inter-cellular communication and tumorigenesis in carcinomas. PLoS One. 2012;7:e36719. doi: 10.1371/journal.pone.0036719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khodarev NN, Roach P, Pitroda SP, Golden DW, Bhayani M, Shao MY, Darga TE, Beveridge MG, Sood RF, Sutton HG, Beckett MA, Mauceri HJ, Posner MC, Weichselbaum RR. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4:e5821. doi: 10.1371/journal.pone.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Efimova EV, Liang H, Pitroda SP, Labay E, Darga TE, Levina V, Lokshin A, Roizman B, Weichselbaum RR, Khodarev NN. Radioresistance of Stat1 over-expressing tumour cells is associated with suppressed apoptotic response to cytotoxic agents and increased IL6-IL8 signalling. Int J Radiat Biol. 2009;85:421–431. doi: 10.1080/09553000902838566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitroda SP, Zhou T, Sweis RF, Filippo M, Labay E, Beckett MA, Mauceri HJ, Liang H, Darga TE, Perakis S, Khan SA, Sutton HG, Zhang W, Khodarev NN, Garcia JGN, Weichselbaum RR. Tumor endothelial inflammation predicts clinical outcome in diverse human cancers. PLoS One. 2012;7:e46104. doi: 10.1371/journal.pone.0046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khodarev NN, Roizman B, Weichselbaum RR. Molecular pathways: interferon/stat1 pathway: role in the tumor resistance to genotoxic stress and aggressive growth. Clin: Can. Res; 2012. [DOI] [PubMed] [Google Scholar]
- 37.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 38.Melo SA, Esteller M. Disruption of microRNA nuclear transport in human cancer. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim S-O, Du Y, Wang Y, Chang W-C, Chen C-H, Hsu JL, Wu Y, Lam YC, James BP, Liu X, Liu C-G, Patel DJ, Hung M-C. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J-S, Smibert P, Westholm JO, Jee D, Maurin T, Lai EC. Intertwined pathways for Argonaute-mediated microRNA biogenesis in Drosophila. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heravi-Moussavi A, Anglesio MS, Cheng S-WG, Senz J, Yang W, Prentice L, Fejes AP, Chow C, Tone A, Kalloger SE, Hamel N, Roth A, Ha G, Wan ANC, Maines-Bandiera S, Salamanca C, Pasini B, Clarke BA, Lee AF, Lee C-H, Zhao C, Young RH, Aparicio SA, Sorensen PHB, Woo MMM, Boyd N, Jones SJM, Hirst M, Marra MA, Gilks B, Shah SP, Foulkes WD, Morin GB, Huntsman DG. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med. 2012;366:234–242. doi: 10.1056/NEJMoa1102903. [DOI] [PubMed] [Google Scholar]
- 43.Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin Oncol. 2011;38:724–733. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O’Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, Urban N, Knudsen BS, Tewari M. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 2010;116:117–125. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’Amato NC, Howe EN, Richer JK. MicroRNA regulation of epithelial plasticity in cancer. Cancer Lett. 2013;341:46–55. doi: 10.1016/j.canlet.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 49.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 50.Ding X, Park SI, McCauley LK, Wang C-Y. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial–mesenchymal transition and tumor metastasis. J Biol Chem. 2013;288:10241–10253. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011;124:999–1006. doi: 10.1242/jcs.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pittà C, Pinatel E, Stadler MB, Provero P, Bernengo MG, Osman I, Taverna D. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011;30:1990–2007. doi: 10.1038/emboj.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y-F, Zhang A-R, Zhang B-C, Rao Z-G, Gao J-F, Lv M-H, Wu Y-Y, Wang S-M, Wang R-Q, Fang D-C. MiR-26a regulates cell cycle and anoikis of human esophageal adenocarcinoma cells through Rb1-E2F1 signaling pathway. Mol Biol Rep. 2013;40:1711–1720. doi: 10.1007/s11033-012-2222-7. [DOI] [PubMed] [Google Scholar]
- 55.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci USA. 2008;105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Northcott PA, Fernandez-L A, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fish JE, Srivastava D (2009) MicroRNAs: opening a new vein in angiogenesis research. Sci Signal 2:pe1–pe1. doi: 10.1126/scisignal.252pe1 [DOI] [PMC free article] [PubMed]
- 58.Guo C, Sah JF, Beard L, Willson JKV, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosom Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Q, Lv G-D, Qin X, Gen Y-H, Zheng S-T, Liu T, Lu X-M. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol Biol Rep. 2012;39:1239–1246. doi: 10.1007/s11033-011-0854-7. [DOI] [PubMed] [Google Scholar]
- 60.Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lussier YA, Xing HR, Salama JK, Khodarev NN, Huang Y, Zhang Q, Khan SA, Yang X, Hasselle MD, Darga TE, Malik R, Fan H, Perakis S, Filippo M, Corbin K, Lee Y, Posner MC, Chmura SJ, Hellman S, Weichselbaum RR. MicroRNA expression characterizes oligometastasis(es) PLoS One. 2011;6:e28650. doi: 10.1371/journal.pone.0028650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salama JK, Hasselle MD, Chmura SJ, Malik R, Mehta N, Yenice KM, Villaflor VM, Stadler WM, Hoffman PC, Cohen EEW, Connell PP, Haraf DJ, Vokes EE, Hellman S, Weichselbaum RR. Stereotactic body radiotherapy for multisite extracranial oligometastases. Cancer Res. 2011;118:2962–2970. doi: 10.1002/cncr.26611. [DOI] [PubMed] [Google Scholar]
- 64.Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celià-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y. Direct targeting of Sec23a by miR-200 s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshino H, Enokida H, Itesako T, Tatarano S, Kinoshita T, Fuse M, Kojima S, Nakagawa M, Seki N. Epithelial–mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J Hum Genet. 2013;58:508–516. doi: 10.1038/jhg.2013.31. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Chen X, Wang R, Xiao P, Xu Z, Chen L, Hang W, Ruan A, Yang H, Zhang X (2013) microRNA-200c modulates the epithelial-to-mesenchymal transition in human renal cell carcinoma metastasis. Oncol Rep 30:643–650. doi:10.3892/or.2013.2530 [DOI] [PubMed]
- 68.Kurashige J, Kamohara H, Watanabe M, Hiyoshi Y, Iwatsuki M, Tanaka Y, Kinoshita K, Saito S, Baba Y, Baba H. MicroRNA-200b regulates cell proliferation, invasion, and migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg Oncol. 2012;19(Suppl 3):S656–S664. doi: 10.1245/s10434-012-2217-6. [DOI] [PubMed] [Google Scholar]
- 69.Schliekelman MJ, Gibbons DL, Faca VM, Creighton CJ, Rizvi ZH, Zhang Q, Wong C-H, Wang H, Ungewiss C, Ahn Y-H, Shin D-H, Kurie JM, Hanash SM. Targets of the tumor suppressor miR-200 in regulation of the epithelial–mesenchymal transition in cancer. Cancer Res. 2011;71:7670–7682. doi: 10.1158/0008-5472.CAN-11-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teng Y, Mei Y, Hawthorn L, Cowell JK. WASF3 regulates miR-200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells. Oncogene. 2014;33:203–211. doi: 10.1038/onc.2012.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo L, Chen C, Shi M, Wang F, Chen X, Diao D, Hu M, Yu M, Qian L, Guo N. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial–mesenchymal transition. Oncogene. 2013;32:5272–5282. doi: 10.1038/onc.2012.573. [DOI] [PubMed] [Google Scholar]
- 72.Castilla MÁ, Moreno-Bueno G, Romero-Pérez L, De Vijver KV, Biscuola M, López-García MÁ, Prat J, Matías-Guiu X, Cano A, Oliva E, Palacios J. Micro-RNA signature of the epithelial–mesenchymal transition in endometrial carcinosarcoma. J Pathol. 2010;223:72–80. doi: 10.1002/path.2802. [DOI] [PubMed] [Google Scholar]
- 73.Harazono Y, Muramatsu T, Endo H, Uzawa N, Kawano T, Harada K, Inazawa J, Kozaki K-I. miR-655 Is an EMT-suppressive MicroRNA targeting ZEB1 and TGFBR2. PLoS One. 2013;8:e62757. doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiu X. MiRNA-125a-3p is a negative regulator of the RhoA-actomyosin pathway in A549 cells. Int J Oncol. 2013;42:1734–1742. doi: 10.3892/ijo.2013.1861. [DOI] [PubMed] [Google Scholar]
- 75.Qiao J, Lee S, Paul P, Theiss L, Tiao J, Qiao L, Kong A, Chung DH. miR-335 and miR-363 regulation of neuroblastoma tumorigenesis and metastasis. Surgery. 2013;154:226–233. doi: 10.1016/j.surg.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anaya-Ruiz M, Bandala C, Perez-Santos JLM. miR-485 acts as a tumor suppressor by inhibiting cell growth and migration in breast carcinoma T47D cells. Asian Pac J Cancer Prev. 2013;14:3757–3760. doi: 10.7314/apjcp.2013.14.6.3757. [DOI] [PubMed] [Google Scholar]
- 77.Vaira V, Faversani A, Dohi T, Montorsi M, Augello C, Gatti S, Coggi G, Altieri DC, Bosari S. miR-296 regulation of a cell polarity-cell plasticity module controls tumor progression. Oncogene. 2012;31:27–38. doi: 10.1038/onc.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Z, Sun L, Wang H, Yao J, Jiang C, Xu W, Yang Z. MiR-328 expression is decreased in high-grade gliomas and is associated with worse survival in primary glioblastoma. PLoS One. 2012;7:e47270. doi: 10.1371/journal.pone.0047270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS, Lee JY, Borlak J, Nam SW. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 80.Ding DP, Chen ZL, Zhao XH, Wang JW, Sun J, Wang Z, Tan FW, Tan XG, Li BZ, Zhou F, Shao K, Li N, Qiu B, He J. miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis. 2011;32:1025–1032. doi: 10.1093/carcin/bgr078. [DOI] [PubMed] [Google Scholar]
- 81.Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 82.Nakano H, Yamada Y, Miyazawa T, Yoshida T. Gain-of-function microRNA screens identify miR-193a regulating proliferation and apoptosis in epithelial ovarian cancer cells. Int J Oncol. 2013;42:1875–1882. doi: 10.3892/ijo.2013.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo X, Guo L, Ji J, Zhang J, Zhang J, Chen X, Cai Q, Li J, Gu Q, Liu B, Zhu Z, Yu Y. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem Biophys Res Commun. 2010;398:1–6. doi: 10.1016/j.bbrc.2010.05.082. [DOI] [PubMed] [Google Scholar]
- 84.Xiao B, Tan L, He B, Liu Z, Xu R. MiRNA-329 targeting E2F1 inhibits cell proliferation in glioma cells. J Transl Med. 2013;11:172. doi: 10.1186/1479-5876-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhai H, Song B, Xu X, Zhu W, Ju J. Inhibition of autophagy and tumor growth in colon cancer by miR-502. Oncogene. 2012;32:1570–1579. doi: 10.1038/onc.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim Y-K, Yu J, Han TS, Park S-Y, Namkoong B, Kim DH, Hur K, Yoo M-W, Lee H-J, Yang H-K, Kim VN. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–145. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harazono Y, Muramatsu T, Endo H, Uzawa N, Kawano T, Harada K, Inazawa J, Kozaki K-I. miR-655 Is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One. 2013;8:e62757. doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xin C, Zhang H, Liu Z. miR-154 suppresses colorectal cancer cell growth and motility by targeting TLR2. Mol Cell Biochem. 2014;387:271–277. doi: 10.1007/s11010-013-1892-3. [DOI] [PubMed] [Google Scholar]
- 90.Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai H, Li P, Cao Q, Ju X, Meng X, Wang M, Zhang Z, Qin C, Hua L, Yin C, Shao P. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem. 2013;379:69–75. doi: 10.1007/s11010-013-1628-4. [DOI] [PubMed] [Google Scholar]
- 91.Zhu C, Shao P, Bao M, Li P, Zhou H, Cai H, Cao Q, Tao L, Meng X, Ju X, Qin C, Li J, Yin C. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol Oncol. 2014;32(31):e9–e16. doi: 10.1016/j.urolonc.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 92.Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z, Liu Y. MicroRNA-330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS One. 2012;7:e46010. doi: 10.1371/journal.pone.0046010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Jeyapalan Z, Deng Z, Shatseva T, Fang L, He C, Yang BB. Expression of CD44 3’-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–3041. doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swarbrick A, Woods SL, Shaw A, Balakrishnan A, Phua Y, Nguyen A, Chanthery Y, Lim L, Ashton LJ, Judson RL, Huskey N, Blelloch R, Haber M, Norris MD, Lengyel P, Hackett CS, Preiss T, Chetcuti A, Sullivan CS, Marcusson EG, Weiss W, L’Etoile N, Goga A. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med. 2010;16:1134–1140. doi: 10.1038/nm.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bao L, Hazari S, Mehra S, Kaushal D, Moroz K, Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am J Pathol. 2012;180:2490–2503. doi: 10.1016/j.ajpath.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Anaya-Ruiz M, Cebada J, Delgado-López G, Sánchez-Vázquez ML, Perez-Santos JLM. miR-153 silencing induces apoptosis in the MDA-MB-231 breast cancer cell line. Asian Pac J Cancer Prev. 2013;14:2983–2986. [PubMed] [Google Scholar]
- 97.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho K-K, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW-F. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao S, Deng Y, Liu Y, Chen X, Yang G, Mu Y, Zhang D, Kang J, Wu Z. MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Mol Biol Rep. 2013;40:2789–2798. doi: 10.1007/s11033-012-2278-4. [DOI] [PubMed] [Google Scholar]
- 99.Yoon A-R, Gao R, Kaul Z, Choi I-K, Ryu J, Noble JR, Kato Y, Saito S, Hirano T, Ishii T, Reddel RR, Yun C-O, Kaul SC, Wadhwa R. MicroRNA-296 is enriched in cancer cells and downregulates p21WAF1 mRNA expression via interaction with its 3’ untranslated region. Nucleic Acids Res. 2011;39:8078–8091. doi: 10.1093/nar/gkr492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Savi F, Forno I, Faversani A, Luciani A, Caldiera S, Gatti S, Foa P, Ricca D, Bulfamante G, Vaira V, Bosari S. MiR-296/Scribble axe is deregulated in human breast cancer and miR-296 restoration reduces tumor growth in vivo. Clin Sci. 2014;127:233–242. doi: 10.1042/CS20130580. [DOI] [PubMed] [Google Scholar]
- 101.Bai Y, Liao H, Liu T, Zeng X, Xiao F, Luo L, Guo H, Guo L. MiR-296-3p regulates cell growth and multi-drug resistance of human glioblastoma by targeting ether-à-go-go (EAG1) Eur J Cancer. 2013;49:710–724. doi: 10.1016/j.ejca.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 102.Wei JJ, Wu X, Peng Y, Shi G, Olca B, Yang X, Daniels G, Osman I, Ouyang J, Hernando E, Pellicer A, Rhim JS, Melamed J, Lee P. Regulation of HMGA1 expression by MicroRNA-296 affects prostate cancer growth and invasion. Clin Cancer Res. 2011;17:1297–1305. doi: 10.1158/1078-0432.CCR-10-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li Q-Q, Chen Z-Q, Cao X-X, Xu J-D, Xu J-W, Chen Y-Y, Wang W-J, Chen Q, Tang F, Liu X-P, Xu Z-D. Involvement of NF-κB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial–mesenchymal transition of breast cancer cells. Cell Death Differ. 2011;18:16–25. doi: 10.1038/cdd.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu Z-S, Wang C-Q, Xiang R, Liu X, Ye S, Yang X-Q, Zhang G-H, Xu X-C, Zhu T, Wu Q. Loss of miR-133a expression associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion. BMC Cancer. 2012;12:51. doi: 10.1186/1471-2407-12-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, Seki N. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106:405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pan Y-Z, Morris ME, Yu A-M. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75:1374–1379. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu XT, Xu Q, Tong JL, Zhu MM, Nie F, Chen X, Xiao SD, Ran ZH. MicroRNA expression profiling identifies miR-328 regulates cancer stem cell-like SP cells in colorectal cancer. Br J Cancer. 2012;106:1320–1330. doi: 10.1038/bjc.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paduano F, Dattilo V, Narciso D, Bilotta A, Gaudio E, Menniti M, Agosti V, Palmieri C, Perrotti N, Fusco A, Trapasso F, Iuliano R. Protein tyrosine phosphatase PTPRJ is negatively regulated by microRNA-328. FEBS J. 2013;280:401–412. doi: 10.1111/j.1742-4658.2012.08624.x. [DOI] [PubMed] [Google Scholar]
- 110.Li W-Q, Li Y-M, Tao B-B, Lu Y-C, Hu G-H, Liu H-M, He J, Xu Y, Yu H-Y. Downregulation of ABCG2 expression in glioblastoma cancer stem cells with miRNA-328 may decrease their chemoresistance. Med Sci Monit. 2010;16:HY27–HY30. [PubMed] [Google Scholar]
- 111.Fang C, Zhao Y, Guo B. MiR-199b-5p targets HER2 in breast cancer cells. J Cell Biochem. 2013;114:1457–1463. doi: 10.1002/jcb.24487. [DOI] [PubMed] [Google Scholar]
- 112.Garzia L, Andolfo I, Cusanelli E, Marino N, Petrosino G, De Martino D, Esposito V, Galeone A, Navas L, Esposito S, Gargiulo S, Fattet S, Donofrio V, Cinalli G, Brunetti A, Vecchio LD, Northcott PA, Delattre O, Taylor MD, Iolascon A, Zollo M. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 2009;4:e4998. doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andolfo I, Liguori L, De Antonellis P, Cusanelli E, Marinaro F, Pistollato F, Garzia L, De Vita G, Petrosino G, Accordi B, Migliorati R, Basso G, Iolascon A, Cinalli G, Zollo M. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro-oncology. 2012;14:596–612. doi: 10.1093/neuonc/nos002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ninio-Many L, Grossman H, Shomron N, Chuderland D, Shalgi R. microRNA-125a-3p reduces cell proliferation and migration by targeting Fyn. J Cell Sci. 2013;126:2867–2876. doi: 10.1242/jcs.123414. [DOI] [PubMed] [Google Scholar]
- 115.Chen J, Chen Y, Chen Z. MiR-125a/b regulates the activation of cancer stem cells in paclitaxel-resistant colon cancer. Cancer Invest. 2013;31:17–23. doi: 10.3109/07357907.2012.743557. [DOI] [PubMed] [Google Scholar]
- 116.Sun Y-M, Lin K-Y, Chen Y-Q. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bi Q, Tang S, Xia L, Du R, Fan R, Gao L, Jin J, Liang S, Chen Z, Xu G, Nie Y, Wu K, Liu J, Shi Y, Ding J, Fan D. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One. 2012;7:e40169. doi: 10.1371/journal.pone.0040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim S-W, Ramasamy K, Bouamar H, Lin A-P, Jiang D, Aguiar RCT. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) Proc Natl Acad Sci USA. 2012;109:7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor β receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–217. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- 120.Fan Y-C, Mei P-J, Chen C, Miao F-A, Zhang H, Li Z-L. MiR-29c inhibits glioma cell proliferation, migration, invasion and angiogenesis. J Neurooncol. 2013;115:179–188. doi: 10.1007/s11060-013-1223-2. [DOI] [PubMed] [Google Scholar]
- 121.Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang L, Zhang H, Chen X, Yang Y, Liu G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin β1 and matrix metalloproteinase2 (MMP2) PLoS One. 2013;8:e70192. doi: 10.1371/journal.pone.0070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bae HJ, Noh JH, Kim JK, Eun JW, Jung KH, Kim MG, Chang YG, Shen Q, Kim S-J, Park WS, Lee JY, Nam SW (2013) MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene. doi: 10.1038/onc.2013.216 [DOI] [PubMed]
- 123.Wang C-M, Wang Y, Fan C-G, Xu F-F, Sun W-S, Liu Y-G, Jia J-H. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun. 2011;411:586–592. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 124.Liu N, Tang L-L, Sun Y, Cui R-X, Wang H-Y, Huang B-J, He Q-M, Jiang W, Ma J. MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett. 2013;329:181–188. doi: 10.1016/j.canlet.2012.10.032. [DOI] [PubMed] [Google Scholar]
- 125.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103:877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uhlmann S, Mannsperger H, Zhang JD, Horvat E-Á, Schmidt C, Küblbeck M, Henjes F, Ward A, Tschulena U, Zweig K, Korf U, Wiemann S, Sahin O. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 2012;8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iliopoulos D, Rotem A, Struhl K. Inhibition of miR-193a expression by Max and RXRα activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 2011;71:5144–5153. doi: 10.1158/0008-5472.CAN-11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ory B, Ramsey MR, Wilson C, Vadysirisack DD, Forster N, Rocco JW, Rothenberg SM, Ellisen LW. A microRNA-dependent program controls p53-independent survival and chemosensitivity in human and murine squamous cell carcinoma. J Clin Invest. 2011;121:809–820. doi: 10.1172/JCI43897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kozaki KI, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive MicroRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 130.Hsu K-W, Wang A-M, Ping Y-H, Huang K-H, Huang T-T, Lee H-C, Lo S-S, Chi C-W, Yeh T-S. Downregulation of tumor suppressor MBP-1 by microRNA-363 in gastric carcinogenesis. Carcinogenesis. 2014;35:208–217. doi: 10.1093/carcin/bgt285. [DOI] [PubMed] [Google Scholar]
- 131.Sun Q, Zhang J, Cao W, Wang X, Xu Q, Yan M, Wu X, Chen W. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. Int J Biochem Cell Biol. 2013;45:513–520. doi: 10.1016/j.biocel.2012.12.004. [DOI] [PubMed] [Google Scholar]