Abstract
Extremely low abundance microorganisms (members of the “rare biosphere”) are believed to include dormant taxa, which can sporadically become abundant following environmental triggers. Yet, microbial transitions from rare to abundant have seldom been captured in situ, and it is uncertain how widespread these transitions are. A bloom of a single ribotype (≥99% similarity in the 16S ribosomal RNA gene) of a widespread betaproteobacterium (Janthinobacterium sp.) occurred over 2 weeks in Arctic marine waters. The Janthinobacterium population was not detected microscopically in situ in January and early February, but suddenly appeared in the water column thereafter, eventually accounting for up to 20% of bacterial cells in mid February. During the bloom, this bacterium was detected at open water sites up to 50 km apart, being abundant down to more than 300 m. This event is one of the largest monospecific bacterial blooms reported in polar oceans. It is also remarkable because Betaproteobacteria are typically found only in low abundance in marine environments. In particular, Janthinobacterium were known from non-marine habitats and had previously been detected only in the rare biosphere of seawater samples, including the polar oceans. The Arctic Janthinobacterium formed mucilagenous monolayer aggregates after short (ca. 8 h) incubations, suggesting that biofilm formation may play a role in maintaining rare bacteria in pelagic marine environments. The spontaneous mass occurrence of this opportunistic rare taxon in polar waters during the energy-limited season extends current knowledge of how and when microbial transitions between rare and abundant occur in the ocean.
Keywords: Arctic, betaproteobacteria, biofilm, bloom, Janthinobacterium, rare biosphere
Introduction
Widespread reports of the large number of rare microorganisms in the environment challenge our understanding of microbial ecology and biogeography. The search for the mechanisms that maintain this “rare biosphere” has become a focus of intense research (Pedrós-Alió, 2012). Rare microbes have been proposed to have important ecological roles such as stabilizing ecosystem processes after disturbance or maintaining critical biogeochemical functions. However, a large fraction of the rare biosphere detected in high throughput sequencing studies may be composed of dead microbes, or dormant/inactive microbial species. These non-dividing microbes are prone to extinction over time, but they may act as a reservoir of taxa able to proliferate under specific conditions, as suggested by the “seed bank” hypothesis, with key ecological implications (Lennon and Jones, 2011). While dormancy is increasingly assumed to play a key role in fostering and maintaining microbial diversity, testing hypotheses related to rare and dormant microbes in natural ecosystems is still challenging.
By comparing ratios of 16S ribosomal RNA to 16S ribosomal RNA genes (from RNA and DNA, respectively) of rare and abundant bacterial taxa, some rare microbes appear to be highly active in aquatic environments (Jones and Lennon, 2010; Campbell et al., 2011). A likely explanation for this observation is that microbial rank abundances are dynamic and characterized by transitions between dormant and active states (Lennon and Jones, 2011). Examples of microbial shifts from rare to abundant are typically found in experimental manipulations such as bottle incubations (Fuchs et al., 2000), after the addition of specific organic compounds (Teira et al., 2007) or under changes in experimental conditions (Bouvier and del Giorgio, 2007; Sjöstedt et al., 2012). Yet, there are extremely few examples of such microbial transitions taking place in the environment (Piccini et al., 2006).
In the ocean, transient blooms of rare Flavobacteria populations have been found associated with phytoplankton blooms in highly productive waters (Teeling et al., 2012). Episodic increases in the abundance of fast-growing Gammaproteobacteria such as Glaciecola (Alonso-Sáez et al., 2007) or Alteromonas (Bano and Hollibaugh, 2002) have also been reported, with no clear link with any obvious environmental trigger. However, reports of temporal shifts of bacterial taxa from rare to abundant are still scarce in the surface ocean. A pyrosequencing study in Arctic waters did not find evidence for such transitions (Kirchman et al., 2010), and a 6-year time-series study in the English Channel only reported a single massive bloom of a rare species (Vibrio sp.) at a time point coincident with maximum concentrations of nitrogen, carbon and chlorophyll (Gilbert et al., 2012).
Such transitions are by definition ephemeral. Therefore, the monthly resolution of most temporal studies may largely miss bacterial bloom events if they take place over timescales spanning days to weeks. Thus, more highly resolved temporal sampling programmes could be critical for detecting blooms of rare taxa. Finding out what types of microorganisms are prone to increase rapidly, how often these blooms occur in situ, and identifying environmental thresholds for these transitions would greatly enhance our understanding of the ecology of rare microorganisms. In the present study, we illustrate this case by reporting a transient bloom of a rare betaproteobacterium in Arctic seawater, detected by weekly sampling during winter under the ice cover.
Materials and methods
Seawater samples down the depth profile were collected weekly in different stations from the Amundsen Gulf, Western Arctic, from January to March 2008 on board the CCGS Amundsen (Figure 1). Samples for Catalyzed Reporter Deposition Fluorescence in situ Hybridization (CARD-FISH) and DNA analyses were collected directly from 12-L Niskin-type bottles mounted on a Carousel Rosette. Profiles of temperature and salinity were obtained using a SeaBird 911 + CTD mounted on the rosette, with additional probes for oxygen and chlorophyll fluorescence. Concentrations of nitrate were determined using standard colorimetric methods (Grasshoff et al., 1999) adapted for the AutoAnalyzer 3. Surface samples analyzed in this study were collected at a depth of 12 m to avoid any potential ship-associated contamination. We also analyzed an additional seawater sample collected at 5 m depth on February 5th through a hole in the ice ca. 450 m upstream from the ship.
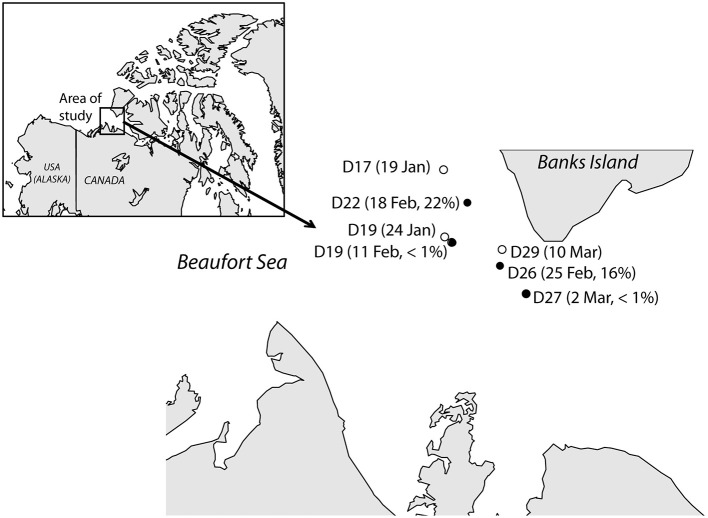
Figure 1.
Map showing the location of the stations in this study and the dates when they were sampled in 2008. Two different samples were collected at station D19 while the research vessel was passively drifting with the ice. Filled dots represent stations where Janthinobacterium cells were detected in situ by CARD-FISH, and their contribution (in percentage of total cells) are shown.
Microbial biomass for DNA analysis was collected by sequentially filtering 7 L of sample through a 3-μm pore size polycarbonate filter (Poretics) and a 0.2-μm pore size Sterivex filter unit using a peristaltic pump. Contents on both filters were preserved with 1.8 mL of lysis buffer (50 mM Tris-HCl pH 8.3, 40 mM EDTA pH 8.0, 0.75 M sucrose) and kept at −80°C. A seawater sample collected in February 18th at station D22 (71°18.649 N, −124°29.798 W) was selected for DNA cloning and sequencing. Two clone libraries were generated with samples collected in the size-fraction above and below 3 μm (large and small size-fraction, respectively). Nucleic acids were extracted using a standard salt protocol and 16 S rRNA genes were amplified by PCR with the universal primers 8F and 1492R. DNA extraction, cloning and sequencing were performed as described in Harding et al. (2011). Sixty-nine clones were analyzed in each clone library. Sequences were checked using the Chimera check software at Ribosomal Data Project (RDP) and chimeras were removed from further analysis. The remaining sequences were manually trimmed and taxonomically classified using the RDP Classifier (Wang et al., 2007). Sequences affiliated with Janthinobacterium were aligned using MUSCLE, and the alignment was manually edited using Jalview v. 2.8.0b1. Maximum-likelihood (ML) analysis was carried out with RAxML v. 7.5.6 (Stamatakis et al., 2005). The ML tree was identified following 300 alternative runs on distinct starting trees under the GTR + gamma + I model. A bootstrap analysis was also conducted in RAxML using 1000 replicates. Nucleotide sequences have been deposited in GenBank (Accession numbers KJ365318–KJ365399).
Samples for CARD-FISH (Pernthaler et al., 2004) were fixed with formaldehyde (3.7% final conc., overnight at 4°C), then filtered onto 0.22-μm pore size polycarbonate filters and frozen (−20°C) until analysis. Cells were permeabilized with lysozyme (1 h) and achromopeptidase (30 min) at 37°C. A specific oligonucleotide probe was designed (Jan64 5′-CAAGCTCCGTGCTGCCGTT-3′) that targets a large number (>2000) of sequences affiliated with cultured and uncultured Janthinobacterium sp., including the genotypes from the Arctic clone libraries. An online ProbeCheck analysis (Loy et al., 2008) against the current SILVA reference database (Pruesse et al., 2007) revealed that only 16 non-target sequences matched this probe even if 3 base pair mismatches were accounted for. The probe Jan64 was applied for sample analysis using 55% formamide in the hybridization buffer. Additionally, specific probes for Alphaproteobacteria (Alf968), Gammaproteobacteria (Gam42a), Bacteroidetes (CF319a), and the general probe set for Bacteria (Eub338-II-III) were used as previously described (Alonso-Sáez et al., 2007). The filter sections were counter-stained with DAPI (1 μg mL−1). At least 300 DAPI cells were counted manually in a minimum of 10 fields.
For MAR-CARD-FISH (Microautoradiography combined with CARD-FISH) analysis, surface samples (20 mL) were incubated with [3H]-leucine (0.5 nM, Perkin Elmer, NET460A005, 140 Ci/mmol) in the dark, in ice-cold seawater for ca. 8 h. After incubation, samples were fixed with formaldehyde (1.8%, overnight at 4°C) and filtered onto 0.2-μm polycarbonate filters (Millipore). Filters were hybridized following the CARD-FISH protocol, and subsequently processed for microautoradiography as previously described (Alonso-Sáez et al., 2008). Microautoradiography preparations were imaged by fully automated microscopy (Zeder and Pernthaler, 2009; Zeder et al., 2011) using the automated image analysis software ACMEtool Tool (www.technobiology.ch). Since aggregates were present as monolayers and the nucleic acid stain signals of individual cells were well separated, the quantification of their abundance in the samples (i.e., cells hybridized with probe Jan64 from total DAPI-stained cells) was possible by this automated procedure (Supplementary Figure 1).
Results
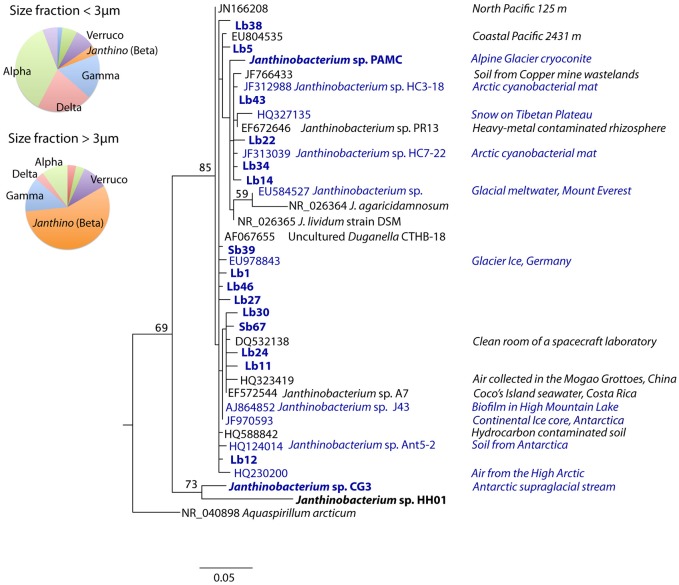
Samples for CARD-FISH and DNA analyses were collected weekly under the ice in Arctic coastal waters from January to March 2008 (Figure 1). During this period the microorganisms experienced extremely low light availability and the surface water temperature (−1.7°C) was close to freezing point (Figure 2). Clone libraries from a sample collected on February 18th recovered an unusually high abundance of sequences affiliated with a single betaproteobacterial species (Janthinobacterium sp.). These sequences were mostly recovered in the size-fraction >3 μm accounting for 57% of the clones, and they formed a tight cluster (>99% similarity) closely related with the isolate Janthinobacterium lividum and the genome-sequenced psychrotolerant Janthinobacterium sp. PAMC 25,724 (Figure 3). Based on the sequencing information, a specific oligonucleotide probe targeting a large diversity of Janthinobacterium, including the sequences from the Arctic clone libraries, was designed and used to follow the winter temporal dynamics of this population in surface Arctic waters.
Figure 2.
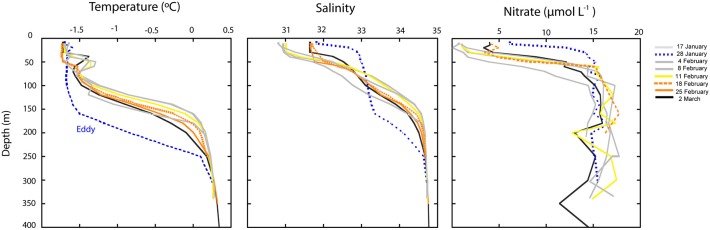
Depth profiles of temperature, salinity and concentration of nitrate during the study. The dotted blue line corresponds to the temperature profile on January 28th, when an oceanographic eddy crossed the Arctic area of study. The yellow line correspond to the station where Janthinobacterium was first detected in situ and orange lines correspond to the stations where maximal abundances of Janthinobacterium were found by CARD-FISH.
Figure 3.
Maximum-likelihood phylogenetic tree of sequences affiliated with Janthinobacterium including the environmental clones retrieved from Arctic seawater (highlighted in bold) from the clone libraries constructed with DNA from different size-fractions. Janthinobacterium representatives with their genomes sequenced have also been highlighted in bold, and those isolated from cold environments (including ice, snow, glacier melt waters and high-mountain lakes) appear in blue. Isolation sources are shown to the right. The tree was rooted with a clade containing sequences of Burkholderia spp. (full tree is shown in Supplementary Figure 4), and bootstrap values based on 1000 replicates (>50%) are indicated on the branches. The pie charts show the proportion of clones affiliated with different bacterial phylogenetic groups as identified by the RDP classifier in the small and large size-fraction clone libraries (Verruco: Verrucomicrobia, Alpha: Alphaproteobacteria, Beta: Betaproteobacteria, Gamma: Gammaproteobacteria, Delta: Deltaproteobacteria, Janthino: Janthinobacterium).
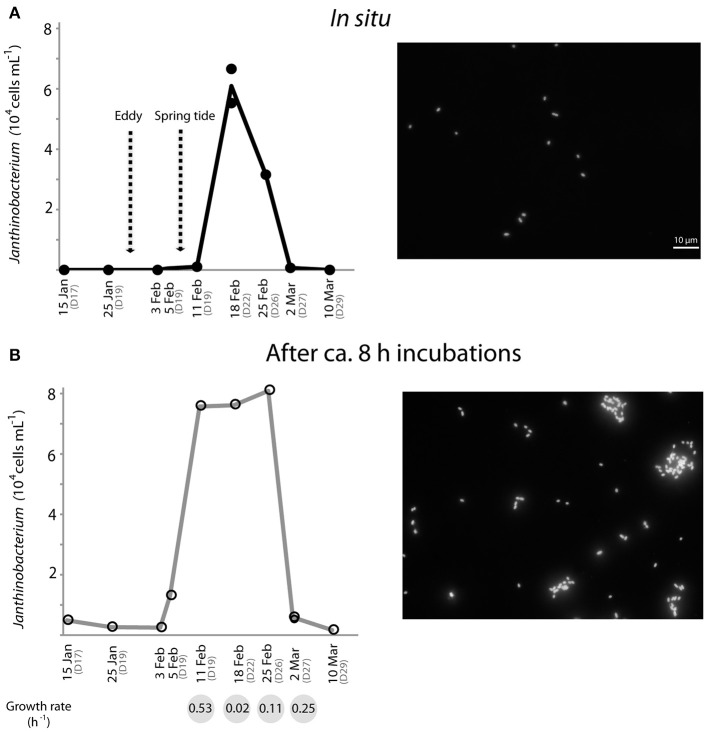
Janthinobacterium were below CARD-FISH detection levels in samples collected in January and early February (Figure 4A). These microorganisms were first detected on February 11th, when they contributed <1% of bacterial cells, and subsequently increased over the two following weeks with maximum concentrations on February 18th (22% of surface bacteria). The targeted cells appeared mostly as single individuals or within small aggregates (Figure 4A) and compared to other ambient marine bacteria they were unusually large (Figure 5). The length of Janthinobacterium cells was 1.48 ± 0.76 μm (n = 268) and 2.13 ± 1.12 μm (n = 89) in surface samples collected on February 18th and 25th, respectively (Figure 5). For comparison, the average length of other ambient bacteria in the samples was 0.86 ± 0.58 μm. High abundances of this population were detected in samples collected ca. 50 km apart (stations D22 and D26, Figure 1), indicating a non-local spatial distribution of this bloom. Moreover, high abundances of Janthinobacterium were also found in deeper samples at the time of the bloom (Figure 6), reaching 28% of cells below 100 m on February 18th. In early March (March 2nd and March 10th), when the ship returned to a location close to where the bloom had been first discovered (station D29, Figure 1), Janthinobacterium were again rare (i.e., below 1% of cells).
Figure 4.
Temporal dynamics of the in situ abundance of Janthinobacterium (probe Jan64) in surface Arctic seawater as analyzed by CARD-FISH (A), and abundances reached by the same population after 8 h incubations (for MAR-CARD-FISH analysis) of the corresponding environmental samples (B). Fluorescence microscope images of Arctic Janthinobacterium cells in situ (where they appeared mostly as single-cells) and in the MAR-CARD-FISH incubations (where they formed mucilaginous aggregates) are shown on the right side. The arrows in (A) indicate the day that an eddy crossed the Arctic area of study and the presence of strong spring tides due to the full moon on February 7th. Specific growth rates (h−1) of Janthinobacterium estimated in the incubations from February 11th to March 2nd are shown in the gray circles below the graph in (B). All samples were collected onboard at 12 m depth, except for the MAR-CARD-FISH sample from February 5th, which was collected at 5 m depth through a hole in the ice. Replicate counts are shown when available.
Figure 5.
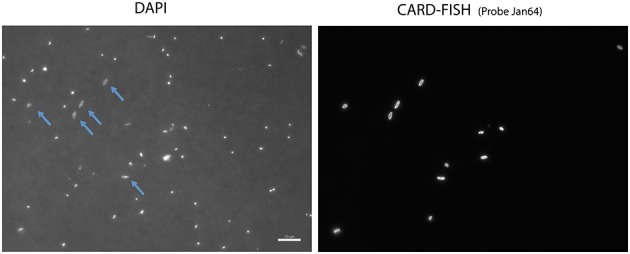
Microscope images of the Arctic Janthinobacterium population (right) and all cells stained with DAPI (left) in a sample collected on February 25th. Some of the hybridized cells are highlighted with arrows in the DAPI images.
Figure 6.
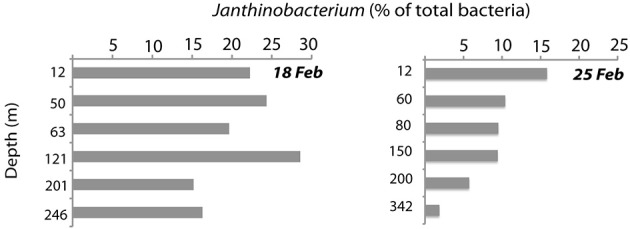
Abundance of Janthinobacterium down the depth profile on the two dates when maximal abundances were found (February 18th and 25th), as analyzed by CARD-FISH.
The single-cell in situ activity of Janthinobacterium in the uptake of an amino acid (leucine) from January to March was analyzed by MAR-CARD-FISH. We found that the Arctic Janthinobacterium were not actively incorporating leucine (<1% active cells) at any of the sampling points. Yet, increases in the total cell abundance of Janthinobacterium were found during the relatively short MAR-CARD-FISH incubations, by comparing their abundance in situ (in samples immediately fixed after collection) and after ca. 8 h of incubation. These results confirm their capacity to grow in Arctic winter seawater. Janthinobacterium were detected at low abundance (<1% of DAPI cells) in the incubations performed in January and late March (March 26th, Figure 4B). However, a steady increase in their abundance was observed in the incubations from the 5th to the 25th of February, while the contribution of other major bacterial groups did not show drastic changes (Supplementary Figure 2). Estimated specific growth rates of Janthinobacterium in the incubations were higher at times of low in situ abundance as compared to the time of the bloom, suggesting that the population had reached its carrying capacity (Figure 4). Interestingly, while growing in the vials, the Arctic Janthinobacterium population developed mucilaginous biofilm-like aggregates, indicating their ability to switch between free-living and attached lifestyles (Figure 4B and Supplementary Figure 3).
Over the sampling period, temperature and salinity profiles varied in the region, particularly in surface waters (Figure 2). The most conspicuous change in physico-chemical parameters occurred at the end of January and was due to water column mixing associated with the passage of a cold-core eddy (Figure 2). Nutrient concentrations significantly increased in surface waters during this event (Figure 2), and after ca. a week (4th of February), the water column was again stratified. During this post-eddy period Janthinobacterium were first detected in MAR-CARD-FISH incubations and in situ (5–11th February). The following weeks temperature and salinity profiles did not show substantial changes, suggesting that the water masses were relatively stable. In the upper 50 m, salinity and nitrate concentration were elevated at the stations where maximum abundances of Janthinobacterium were found (D22 and D26, sampled on the 18th and 25th February, respectively), and also at station D27 (sampled on 2nd March), when Janthinobacterium were again rare.
Discussion
Janthinobacterium have been found in numerous habitats and have attracted substantial interest for biotechnological applications (Du et al., 2007; Shi et al., 2008; Johnsen et al., 2010). Interestingly, isolates of J. lividum and the strain PAMC 25,724, closely related to the Arctic population studied here, are able to produce violacein (Pantanella et al., 2007; Kim et al., 2012), a pigment with anti-grazing properties that may provide a survival advantage in the environment (Matz et al., 2004). We present the first report of a natural bloom of Janthinobacterium and, to our knowledge, also the first report of a betaproteobacterium bloom in seawater. While abundant and active in freshwater systems (Glöckner et al., 1999; Salcher et al., 2013), the concentration of Betaproteobacteria sharply decreases when exposed to marine salinities (Cottrell and Kirchman, 2003; Garneau et al., 2006). A few marine betaproteobacterial taxa have shown high activity, such as the OM43 clade, but their abundance is usually low, rarely exceeding 2% of cells (Morris et al., 2006; Sowell et al., 2011). The large proportion (up to 22% of total cells) and spatial extent of the Janthinobacterium bloom found in this study indicates the capacity of this rare microorganism to proliferate under particular conditions. Ribosomal gene sequences with similarity ≥94% to J. lividum were found in 15 stations of the Global Ocean Sampling (GOS, search using the ncRNA database, http://portal.camera.calit2.net), but only seven hits with similarity >97% were found. Some sequences affiliated with Janthinobacterium were also found in Arctic and Antarctic waters (VAMPS database, http://vamps.mbl.edu/), but being always rare (<0.01%).
Despite their low abundances, the wide range of environments where Janthinobacterium has been found is striking (Figure 3). In an early study, a phylotype closely related to J. lividum (99% similarity for the 16S rRNA gene) was found in a negative control of a DNA extraction (AF067655, “uncultured Duganella clone CTHB-18”) leading to the suggestion that it might be a PCR contaminant (Tanner et al., 1998). This possibility can be ruled out in our study since Janthinobacterium were detected at high concentrations both in situ and in seawater incubations by the PCR-independent CARD-FISH approach. At the global scale, Janthinobacterium have been retrieved in soils, lakes, seawater, and the cold biosphere (Gillis and De Ley, 2006), including mountain glaciers, polar ice, and snow, where high abundances have been occasionally found (Segawa et al., 2005). Their cosmopolitan distribution suggests a very effective dispersal capacity and implies physiological flexibility to colonize new environments. Interestingly, Janthinobacterium have also been found in dust and air, including some samples from the High Arctic (HQ230200, Harding et al., 2011), indicating that aerial transport may be a major dissemination pathway for this cosmopolitan microbe.
The fact that Janthinobacterium were abundant down the depth profile during the bloom despite stratification, raises the question about the mechanisms that would have facilitated their distribution throughout the water column. Actually, oceanographic eddies actively traveled under the ice in this region during the winter of 2008. On January 28th, a cold-core eddy crossed the area of study, followed by strong tidal currents due to a new moon on Feburary 7th. Both phenomena generated turbulence and enhanced vertical mixing (Gratton et al., 2012). Remarkably, an increase in the reproductive activity of small copepods was observed at the same time, consistent with an unusual winter increase in the biological production of some components of the Arctic marine food web (Darnis et al., 2012). The latter authors suggested that the passage of eddies may have brought particulate organic matter susceptible to exploitation by attached bacterial communities, which may have subsequently been exploited for food by small detritivorous copepods.
Evidence of the use of pathways for exploiting high-molecular weight compounds such as humic acids or carbon polymers has been found in the J. lividum isolate OW6/RT-3 (Freese et al., 2010), and in the genome of the psychrotrophic Janthinobacterium sp. HH01 (Hornung et al., 2013), respectively. While we did not test the activity of Arctic Janthinobacterium on polymers, they did not actively take up the amino acid leucine, which is a typical small monomer. In addition, seawater incubations carried out during the Janthinobacterium bloom showed that this microbe formed aggregates (Figure 4B), which favors the use of exoenzymes needed to break down more complex organic matter (Huston and Deming, 2002). Overall these lines of evidence are consistent with the possibility of Janthinobacterium profiting from an influx of complex substrates (Freese et al., 2010).
The growth of Janthinobacterium in the relatively short (8 h) incubations in ice-cold waters provided additional clues about their activity and lifestyle. First, large increases in total cell numbers occurred in the incubations carried out between early February and March, with an unusually short doubling time of 1.3 h on February 11th. Comparably fast growth rates have been reported for some Janthinobacterium isolates with a doubling time of 1.1 h (Hornung et al., 2013). In a cold non-marine environment (mountain snow) rapid temporal increases of Janthinobacterium abundance (ca. 105-fold by quantitative PCR) have been reported during the melting season, preceding the growth of snow algae (Segawa et al., 2005). These results support the view that this psychrophilic group, able to produce bursts of growth, does not rely on labile photosynthetic products as substrates. The different cell abundance reached by Janthinobacterium in the incubations from January to March seemed consistent with a temporal change in their activity. The lower activity outside of the blooming period (Figure 4) may be due to a lack of availability of suitable substrates. An alternative explanation may have been a release from top-down pressures. In a previous study with samples off the coast of Oregon, rare members of betaproteobacteria appeared to be tightly controlled by viruses (Bouvier and del Giorgio, 2007). Changes in physico-chemical conditions associated with the passage of eddies may have disrupted interactions between Janthinobacterium and their grazers, allowing this group to bloom. Unfortunately, species-specific viral data is not available, and there is currently no evidence supporting such viral control.
The production of biofilms associated with autoinducing signals (i.e., quorum sensing) has been described for the isolate J. lividum, and appears to be regulated by changes in the carbon source (Pantanella et al., 2007). The latter authors postulated that alternations between free-living and attached life-styles could represent a response to environmental stress, and a key factor in the survival of J. lividum and their ability to colonize new environments. The Arctic Janthinobacterium population developed mucilaginous biofilm-like aggregates possibly triggered by confinement in vials during MAR-CARD-FISH incubations (Figure 4B). While the presence of mechanisms for quorum sensing in psychrophiles has been largely unexplored (Montgomery et al., 2013), our results suggest that biofilm formation is a strategy for some rare Arctic marine bacteria, and support the hypothesis that signaling compounds may have a role in dormant microbes (Epstein, 2009).
Whether the Janthinobacterium population was part of the local “seed bank” of Arctic waters or whether they were inocula from another ecosystem such as ice, snow or air before the beginning of sampling, cannot be confirmed based on available data. However, the possibility of advection of a large population of Janthinobacterium cells from another region rather than an in situ bloom is unlikely due to several reasons. First, the bloom was not local (Figure 1) and occurred throughout the water column. Despite some variability in surface samples, the physico-chemical properties of the water masses were relatively constant shortly before, during and after the bloom (from the beginning of February to March, Figure 2). Second, the fast growth rates detected in the incubations demonstrate that Janthinobacterium could actively grow in Arctic winter waters outcompeting other Arctic marine bacterial groups at low temperatures. As mentioned earlier, a plausible explanation to their mass occurrence in mid February involves the passage of an oceanographic eddy facilitating their distribution with depth and carrying a supply of particulate carbon that may have promoted their growth over the following weeks.
Finally, in a recent comparison of the marine bacterial composition between poles, Betaproteobacteria were always rare, but their contribution was higher in the Arctic as compared to the Antarctic Ocean (Ghiglione et al., 2012). While it has been suggested that such a difference is due to the higher freshwater inputs to the Arctic Ocean, we hypothesize that the ability of some Arctic Betaproteobacteria to produce episodic bursts of growth could also contribute to their higher abundance in Arctic marine “seed banks.”
In summary, our study provides one of the first examples of how a widespread member of the rare biosphere (Janthinobacterium sp.) produces a transient bloom in oceanic waters. The fact that the environmental setting of the bloom was the Arctic during winter, a cold and energy-limited environment, broadens current knowledge of how and when transitions of bacterial taxa between rare and abundant occur in the ocean. It is also the first report of a transient bloom of a single betaproteobacterial species to become highly abundant in seawater (over 20% of total counts). Janthinobacterium have a widespread distribution and the capability to survive and colonize new environments, features that presumably characterize some members of the rare biosphere that appear to be persistent in oceanic seed-banks (Gibbons et al., 2013). Their ability to produce mucilaginous (biofilm-like) aggregates and to switch between free-living and attached lifestyles, may contribute to the survival of this and other rare microbes, and to their capacity to produce ephemeral blooms. Finally, our results highlight the fact that low-frequency sampling can overlook relevant microbial events such as the blooms of rare microbes, with potentially important impacts on our understanding of oceanic biogeochemistry.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the crew, officers and fellow scientists of the CCCG Amundsen for their support during the CFL cruise. We also thank I. Forn and N. Arandia for help with sample analysis, J. Gagnon and J. E. Tremblay for nutrient data, P. Guillot and Y. Gratton for conductivity–temperature–depth (CTD) data, and Laura Eme for providing in-house scripts useful for the phylogenetic analysis. This work is a contribution to the International Polar Year—Circumpolar Flaw Lead system study (IPY-CFL 2007/2008) supported through grants from the Canadian IPY Federal Program Office, the Natural Sciences and Engineering Research Council (NSERC), and grants BOREAL (CLG2007-28872-E/ANT) and GEMMA (CTM2007-63753-C02-01/MAR) from the Spanish Ministry of Science and Innovation to Carlos Pedrós-Alió. The Fond Québécois de Recherches Nature et Technologies to Québec Océan and the NSERC support for ArcticNet are acknowledged for administrative and logistics support. Laura Alonso-Sáez was supported by a Marie Curie Intraeuropean Fellowship (CHEMOARC PIEF-GA-2008-221121) and the Swedish Research Council (grant to Stefan Bertilsson). Tommy Harding was supported by a NSERC student fellowship.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00425/abstract
References
- Alonso-Sáez L., Balagué V., Sa E. L., Sánchez O., González J. M., Pinhassi J., et al. (2007). Seasonality in bacterial diversity in North-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH. FEMS Microbiol. Ecol. 60, 98–112 10.1111/j.1574-6941.2006.00276.x [DOI] [PubMed] [Google Scholar]
- Alonso-Sáez L., Sánchez O., Gasol J. M., Balagué V., Pedrós-Alió C. M. (2008). Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ. Microbiol. 10, 2444–2454 10.1111/j.1462-2920.2008.01674.x [DOI] [PubMed] [Google Scholar]
- Bano N., Hollibaugh J. T. (2002). Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68, 505–518 10.1128/AEM.68.2.505-518.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier T., del Giorgio P. A. (2007). Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ. Microbiol. 9, 287–297 10.1111/j.1462-2920.2006.01137.x [DOI] [PubMed] [Google Scholar]
- Campbell B. J., Yua L., Heidelberg J. F., Kirchman D. L. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. U.S. A. 108, 12776–12781 10.1073/pnas.1101405108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell M. T., Kirchman D. L. (2003). Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48, 168–178 10.4319/lo.2003.48.1.0168 [DOI] [Google Scholar]
- Darnis G., Robert D., Pomerleau C., Link H., Archambault P., Nelson R. J., et al. (2012). Current state and trends in Canadian Arctic marine ecosystems: II. Heterotrophic food web, pelagic-benthic coupling, and biodiversity. Clim. Change 115, 179–205 10.1007/s10584-012-0483-8 [DOI] [Google Scholar]
- Du G., Liu L., Song Z., Hua Z., Zhu Y., Chen J. (2007). Production of polyvinyl alcohol-degrading enzyme with Janthinobacterium sp. and its application in cotton fabric desizing. Biotechnol. J. 2, 752–758 10.1002/biot.200600121 [DOI] [PubMed] [Google Scholar]
- Epstein S. (2009). Microbial awakenings. Nature 457:1083 10.1038/4571083a [DOI] [PubMed] [Google Scholar]
- Freese H. M., Eggert A., Garland J. L., Schumann R. (2010). Substrate utilization profiles of bacterial Strains in plankton from the river Warnow, a humic and eutrophic river in North Germany. Microb. Ecol. 59, 59–75 10.1007/s00248-009-9608-7 [DOI] [PubMed] [Google Scholar]
- Fuchs B. M., Zubkov M. V., Sahm K., Burkill P. H., Amann R. (2000). Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2, 191–201 10.1046/j.1462-2920.2000.00092.x [DOI] [PubMed] [Google Scholar]
- Garneau M.-È., Vincent W. F., Alonso-Sáez L., Gratton Y., Lovejoy C. (2006). Prokaryotic community structure and heterotrophic production in a river-influenced coastal arctic ecosystem. Aquat. Microb. Ecol. 42, 27–40 10.3354/ame042027 [DOI] [Google Scholar]
- Ghiglione J.-F., Galand P. E., Pommier T., Pedrós-Alió C., Maas E. W., Bakker K., et al. (2012). Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl. Acad. Sci. U.S. A. 109, 17633–17638 10.1073/pnas.1208160109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons S. M., Caporaso J. G., Pirrung M., Field D., Knight R., Gilbert J. A. (2013). Evidence for a persistent microbial seed bank throughout the global ocean. Proc. Natl. Acad. Sci. U.S. A. 110, 4651–4655 10.1073/pnas.1217767110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Steele J. A., Caporaso J. G., Steinbrück L., Reeder J., Temperton B., et al. (2012). Defining seasonal marine microbial community dynamics. ISME J. 6, 298–308 10.1038/ismej.2011.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis M., De Ley J. (2006). The Genera Chromobacterium and Janthinobacterium. Prokaryotes 5, 737–746 10.1007/0-387-30745-1_32 [DOI] [Google Scholar]
- Glöckner F. O., Fuchs B. M., Amann R. (1999). Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65, 3721–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasshoff K., Kremling K., Ehrhardt M. G. (1999). Methods of Seawater Analysis, 3rd Edn. Weinheim: VCH Publishers [Google Scholar]
- Gratton Y., Bourgault D., Galbraith P., Prieur L., Tsarev V., Barette J., et al. (2012). Physical oceanography, in On The Edge: From knowledge to Action During the Fourth International Polar Year Circumpolar Flaw Lead System Study (2007–2008), eds Barber D. G., Tjaden T., Leitch D., Barber L., Chan W. (Winnipeg, MB: University of Manitoba; ), 34–50 [Google Scholar]
- Harding T., Jungblut A. D., Lovejoy C., Vincent W. F. (2011). Microbes in high Arctic snow and implications for the cold biosphere. Appl. Environ. Microbiol. 77, 3234–3243 10.1128/AEM.02611-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung C., Poehlein A., Haack F. S., Schmidt M., Dierking K., Pohlen A., et al. (2013). The Janthinobacterium sp. HH01 genome encodes a homologue of the V. cholerae CqsA and L. pneumophila LqsA autoinducer synthases. PLoS ONE 8:e55045 10.1371/journal.pone.0055045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston A. L., Deming J. W. (2002). Relationships between microbial extracellular enzymatic activity and suspended and sinking particulate organic matter: seasonal transformations in the North Water. Deep-Sea Res. II Top. Stud. Oceanogr. 49, 5211–5225 10.1016/S0967-0645(02)00186-8 [DOI] [Google Scholar]
- Johnsen M. G., Hansen O. C., Stougaart P. (2010). Isolation, characterization and heterologous expression of a novel chitosanase from Janthinobacterium sp. Strain 4239. Microb. Cell Fact. 9:5 10.1186/1475-2859-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. E., Lennon J. T. (2010). Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U.S. A. 107, 5881–5886 10.1073/pnas.0912765107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Shin S. C., Hong S. G., Lee Y. M., Lee J., Choi I. G., et al. (2012). Genome sequence of Janthinobacterium sp. strain PAMC 25724, isolated from Alpine glacier cryoconite. J. Bacteriol. 194, 2096–2096 10.1128/JB.00096-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman D. L., Cottrell M., Lovejoy C. (2010). The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16SrRNA genes. Environ. Microbiol. 12, 1132–1143 10.1111/j.1462-2920.2010.02154.x [DOI] [PubMed] [Google Scholar]
- Lennon J. T., Jones S. E. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130 10.1038/nrmicro2504 [DOI] [PubMed] [Google Scholar]
- Loy A., Arnold R., Tischler P., Rattei T., Wagner M., Horn M. (2008). probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity Environ. Microbiol. 10, 2894–2898 10.1111/j.1462-2920.2008.01706.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C., Deines P., Boenigk J., Arndt H., Eberl L., Kjelleberg S., et al. (2004). Impact of violacein-producing bacteria on survival and feeding of bacteriovorans nanoflagellates. Appl. Environ. Microbiol. 70, 1593–1599 10.1128/AEM.70.3.1593-1599.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery K., Charlesworth J. C., LeBard R., Visscher P. T., Burns B. P. (2013). Quorum sensing in extreme environments. Life 3, 131–148 10.3390/life3010131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R. M., Longnecker K., Giovannoni S. J. (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ. Micriobiol. 8, 1361–1370 10.1111/j.1462-2920.2006.01029.x [DOI] [PubMed] [Google Scholar]
- Pantanella F., Berlutti F., Passariello C., Sarli S., Morea C., Schippa S. (2007). Violacein and biofilm production in Janthinobacterium lividum. J. Appl. Microbiol. 102, 992–999 10.1111/j.1365-2672.2006.03155.x [DOI] [PubMed] [Google Scholar]
- Pedrós-Alió C. (2012). The rare bacterial biosphere. Annu. Rev. Mar. Sci. 4, 449–466 10.1146/annurev-marine-120710-100948 [DOI] [PubMed] [Google Scholar]
- Pernthaler A., Pernthaler J., Amann R. (2004). Sensitive multi-color fluorescence hybridization for the identification of environmental microorganisms, in Molecular Microbial Ecology Manual, 2nd Edn., eds Kowalchuk G. A., De Bruijin F. J., Head I. M., Akkermans D. L., van Elsas J. D. (Dordrecht: Kluwer Academic Publishers; ), 711–726 [Google Scholar]
- Piccini C., Conde D., Alonso C., Sommaruga R., Pernthaler J. (2006). Blooms of single bacterial species in a coastal lagoon of the southwestern Atlantic ocean. Appl. Environ. Microbiol. 72, 6560–6568 10.1128/AEM.01089-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W., Peplies J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcher M. M., Posch T., Pernthaler J. (2013). In situ substrate preferences of abundant bacterioplankton populations in a prealpine freshwater lake. ISME J. 7, 896–907 10.1038/ismej.2012.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa T., Miyamoto K., Ushida K., Agata K., Okada N., Kohshima S. (2005). Seasonal change in bacterial flora and biomass in mountain snow from the Tateyama Mountains, Japan, analyzed by 16S rRNA gene sequencing and Real-Time PCR. Appl. Environ. Microbiol. 71, 123–130 10.1128/AEM.71.1.123-130.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.-L., Lu X.-Z., Yu W.-G. (2008). A new β-agarase from marine bacterium Janthinobacterium sp. SY12. World J. Microbiol. Biotechnol. 24, 2659–2664 10.1007/s11274-008-9792-5 [DOI] [Google Scholar]
- Sjöstedt J., Koch-Schmidt P., Pontarp M., Canbäck B., Tunlid A., Lundberg P., et al. (2012). Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance. Appl. Environ. Microbiol. 78, 1361–1369 10.1128/AEM.05542-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell S. M., Abraham P. E., Shah M., Verberkmoes N. C., Smith D. P., Barofsky D. F., et al. (2011). Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J. 5, 856–865 10.1038/ismej.2010.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Ludwig T., Meier H. (2005). RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21, 456–463 10.1093/bioinformatics/bti191 [DOI] [PubMed] [Google Scholar]
- Tanner M. A., Goebel B. M., Dojka M. A., Pace N. R. (1998). Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64, 3110–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- Teira E., Lekunberri I., Gasol J. M., Nieto-Cid M., Álvarez-Salgado X. A., Figueiras P. G. (2007). Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ. Microbiol. 9, 2551–2562 10.1111/j.1462-2920.2007.01373.x [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder M., Ellrott A., Amann R. (2011). Automated sample area definition for high-throughput microscopy. Cytometry A 79A, 306–310 10.1002/cyto.a.21034 [DOI] [PubMed] [Google Scholar]
- Zeder M., Pernthaler J. (2009). Multispot live-image autofocusing for high-throughput microscopy of fluorescently stained bacteria. Cytometry A 75A, 781–788 10.1002/cyto.a.20770 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.