Abstract
The epigenome represents a major regulatory interface to the eukaryotic genome. Nucleosome positions, histone variants, histone modifications and chromatin associated proteins all play a role in the epigenetic regulation of DNA function.
Trypanosomes, an ancient branch of the eukaryotic evolutionary lineage, exhibit some highly unusual transcriptional features, including the arrangement of functionally unrelated genes in large, polymerase II transcribed polycistronic transcription units, often exceeding hundreds of kilobases in size. It is generally believed that transcription initiation plays a minor role in regulating the transcript level of genes in trypanosomes, which are mainly regulated post-transcriptionally.
Recent advances have revealed that epigenetic mechanisms play an essential role in the transcriptional regulation of Trypanosoma brucei. This suggested that the modulation of gene activity, particularly that of pol I transcribed genes, is, indeed, an important control mechanism, and that the epigenome is critical in regulating gene expression programs that allow the successful migration of this parasite between hosts, as well as the continuous evasion of the immune system in mammalian hosts.
A wide range of epigenetic signals, readers, writers and erasers have been identified in trypanosomes, some of which have been mapped to essential genetic functions. Some epigenetic mechanisms have also been observed to be unique to trypanosomes. We review recent advances in our understanding of epigenetic control mechanisms in T. brucei, the causative agent of African sleeping sickness, and highlight the utility of epigenetic targets in the possible development of new therapies for human African trypanosomiasis.
Keywords: epigenetics, chromatin, transcription, gene regulation, African sleeping sickness
1. Introduction
Trypanosoma brucei, the causative agent of African sleeping sickness, is an extracellular, flagellated parasite that is transferred into the human host during a blood meal by a Glossina spp. fly. In its initial haemolymphatic phase, bloodstream form (BF) T. brucei invades the bloodstream, interstitial spaces, and lymph system where it divides asexually [1]. With prolonged infection, the parasite crosses the blood brain barrier and enters an encephalitic stage, where the patient exhibits the typical clinical signs of the disease from which the name is derived. Without treatment, African sleeping sickness is lethal.
If the Glossina spp. fly feeds on an infected host, trypanosomes may be taken up, and will transform to a procyclic form (PF) trypomastigote in the insect midgut, where the parasites again multiply by asexual cell division. From the midgut, the parasite moves to the salivary gland of the fly, transforming to a metacyclic form [2], capable of infecting a new mammalian host. The migration of the parasite from a mammalian to an insect host is accompanied by the activation and shutdown of several genes [3]. Many of these genes appear to be regulated by epigenetic mechanisms, implicating chromatin in T. brucei gene regulation.
Chromatin is composed of repetitive arrays of nucleosomes, which are formed by 168 bp of DNA wrapped in two negative supercoils onto a histone octamer and associated with histone H1. Although nucleosomes represent the basic structural unit of chromatin, facilitating the compaction of poly-anionic DNA molecules to a level where it can fit into a cell nucleus, nucleosomes also serve as a molecular message board to genetic processes, and, equally, as a dynamic binding surface for proteins involved in gene regulation and transcriptional control.
Core histones are globular proteins with a characteristic histone fold domain and N-terminal extensions or “tails” emanating from the central fold. An extensive range of post-translational modifications (PTMs) occur on the tails that influence many biological processes, including chromatin condensation and the recruitment of DNA-binding proteins such as chromatin readers, writers and erasers [4]. Extensive studies have shown that histone PTMs can function either singularly or in combination with other PTMs, referred to as histone “cross-talk” [5]. An insight into the organisation of nucleosomes in a genome, as well as the distribution of histone variants and the presence of PTMs, is essential to understand the regulatory role of chromatin in genome function.
The aim of this review is to integrate recent data gathered from the fields of genomics, transcriptomics and proteomics to understand the epigenetic mechanisms that are employed by T. brucei to control its gene expression programs.
2. Genome organisation
The haploid genome of T. brucei is 26 – 35 Mb in size, depending on the strain [3,6], and is composed of 11 megabase chromosomes (MBC) [7], 1–5 intermediate chromosomes (IC) (300 – 900 kb), and approximately 100 minichromosomes (MC) (50 – 150 kb) [8]. MC account for approximately 10% of the nuclear genome, and about half of each MC is composed of 177 bp repeats, as well as silent Variable Surface Glycoprotein (VSG) genes and pseudogenes [9].
The housekeeping portion of the genome, encoded by genes on the MBC, exists as long, non-overlapping, polycistronic transcription units (PTUs). Adjacent PTUs may be located on the same DNA strand, arranged in a head to tail fashion, or on different strands, separated by convergent or divergent strand switching regions (SSRs). The latter refers to the direction of transcription of the adjacent PTUs (Figure 1). The MBCs contain nearly 8800 non-redundant protein-coding genes, including about 500 pseudogenes [10], organised into unidirectional gene clusters that are interrupted by tRNA, snRNA, siRNA and rRNA genes. It is unusual for protein coding genes to be organised in directional PTUs on a genome wide scale [11] as is observed in Trypanosoma. Unlike generic prokaryotic PTUs, which contain genes encoding proteins that act in combination to perform a specific function, genes in trypanosomal polycistrons are usually functionally unrelated. Analysis of the T. brucei transcriptome revealed that RNA polymerase II (pol II) transcription initiates bidirectionally from putative pol II transcription start sites (TSS) at divergent SSRs, as well as a number of positions internal to PTUs [12,13]. Although all genes in a PTU are constitutively expressed, the location of a gene within the PTU was shown to impact on the transcript level of that gene [14].
Figure 1. The epigenetic signals that demarcate transcription units and regulate the expression of genes in T. brucei.
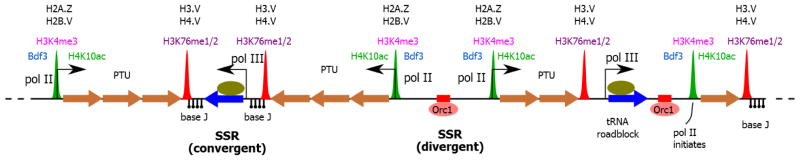
Pol II transcription initiates from weakly defined promoters in divergent SSRs as well as at some internal positions in PTUs. These initiation loci are enriched for TbBDF3, H4K10ac, H3K4me3 and the H2A.Z and H2B.V histone variants. Transcription proceeds through polycistronic units that may span hundreds of kilobases that contain functionally unrelated genes. Transcription terminates in a region enriched for the modified thymidine base J, H3K76me1/2, and the H3.V and H4.V histone variants. TTSs often contain an active pol III transcribed tRNA gene. Replication origins, nucleated by TbORC1, occur at the boundaries of PTUs and have an effect on the expression level of adjacent genes.
3. Transcription in T. brucei
The initiation of transcription represents a key point for controlling the levels of gene products in most eukaryotes. A series of events involving cis- and trans-acting factors binding to specific DNA sequences, collectively functioning to recruit a specific polymerase complex and ultimately initiating mRNA synthesis, is the standard mechanism for regulation of eukaryotic gene expression. However, this paradigm does not seem to apply to leishmanian or trypanosomal protozoa. The lack of classic pol II promoters, activators and co-activators, as well as basal transcription factors, coupled with constitutive polycistronic transcription, suggested that transcription initiation was not a fundamental regulatory event in mRNA synthesis [15]. Although the Leishmania major, T. cruzi and T. brucei genomes encode all five subunits common to the three classes of RNA polymerases [16], T. brucei employs conventional polymerases for alternative functions.
3.1 DNA-dependent RNA polymerases
In T. brucei, pol I, apart from transcribing the rRNA genes, also transcribes two essential, life cycle specific genes that encode cell surface proteins. Procyclin, the major cell surface protein expressed in PF T. brucei, is transcribed from two polycistronic gene loci (GPEET and EP1) [17]. The VSG gene, encoding the BF stage cell surface antigen, is also transcribed by pol I as part of the polycistronic bloodstream expression site, discussed below. Pol I probably allows expression of high levels of a single transcript from a monoallelic transcription unit.
Pol II transcribes the majority of the PTUs, initiating from regions enriched for specific epigenetic features (see Figure 1). Unlike in other eukaryotes, the T. brucei pol II promoter is weakly defined, and lacks a canonical TATA box and initiator sequence [18], although a TBP-like protein, TbTrf4, was identified [19]. Siegel and colleagues reported that oligo[dG]-runs, located between divergent SSRs, may act as an initiator element providing directionality to transcription [20]. The long, resulting, polycistronic RNA is spliced into individual, stable, translatable mRNA molecules by the co-transcriptional trans-splicing of a capped 39 bp spliced leader (SL) RNA, coupled with polyadenylation (reviewed in reference [21]). The SL RNA genes contain the only well-defined promoter of a trypanosomal pol II transcribed gene. The promoter structure consists of a bipartite upstream sequence element and an initiator on which a class II pre-initiation complex nucleates, which contain orthologues of known transcription factors necessary for pol II transcription in other model eukaryotes [22–24]. In L. tarentolae the SL RNA genes and promoters were shown to be depleted of nucleosomes [25].
McAndrew et al. [26] demonstrated α-amanitin sensitive transcription from a T3 polymerase promoter in T. brucei in the presence of T3 polymerase. This was interpreted as the possible opening of the chromatin structure by T3 polymerase, allowing the entry of pol II and subsequent transcription initiation [26]. This, taken together with the lack of identifiable promoter elements and the enrichment of specific histone PTMs and histone variants at SSRs, suggest that epigenetic control mechanisms plays a central role in the modulation of pol II transcription initiation and termination in T. brucei.
Trypanosomal tRNAs, transcribed with other non-coding RNAs by pol III, are interspersed between pol II PTUs. Since the tRNA gene itself may be in the process of transcription by pol III, or associated with regulatory proteins [27], the presence of a tRNA gene presents a kinetic block to a transcribing pol II, and contributes to the termination of transcription, as seen in other eukaryotes [28]. It is therefore possible that the presence of a tRNA gene may contribute to the termination of pol II transcription at the end of a trypanosomal PTU.
3.2 Long non-coding RNA
In one transcriptomic study, 103 transcripts, ranging in size from 154 – 2229 bp, were identified that did not possess recognizable coding potential [13]. Long non-coding RNAs (lncRNAs) have been shown to play a critical role in gene regulation in model eukaryotes [29,30]. LncRNAs can act locally or globally as epigenetic regulators, like the Xist and HOTAIR lncRNAs, respectively [30,31], affecting DNA-protein interactions, chromatin condensation and gene activity. One can speculate that some of the putative T. brucei lncRNAs similarly act at an epigenetic level, adding another layer of control to the regulation of trypanosome gene expression.
3.3 Telomeric silencing and bloodstream expression sites
The BF stage of T. brucei evades clearance by the human host immune system by periodically switching the monoallelically expressed VSG gene, and thus the VSG coat, from a selection of approximately 1500 VSG genes. The active VSG gene is co-transcribed with a set of expression site associated genes (ESAGs) from a single subtelomeric polycistronic unit known as the bloodstream expression site (ES). ESs are transcribed from a single RNA pol I promoter located 30–60 kb upstream of the telomeric repeats. Telomeres are generally associated with transcriptionally repressive heterochromatic structures, where the repressive effect diminishes with distance from the telomeric ends. The ES promoter is preceded by an array of 50 bp repeat sequences stretching for ~10–50 kb [32]. A total of 14 distinct ESs were identified in the Lister 427 T. brucei strain [33], of which the single, active ES is located in a sub-nuclear structure, the expression site body [34]. The canonical structure and associated proteins of the ES and telomeric region are shown in Figure 2. Due to its high relevance to immune clearance in humans and the development of possible therapies, the ES has been the subject of intense research. Unlike the PTUs, which are constitutively transcribed by pol II, the ESs, as well as the procyclin loci, are subject to tight transcriptional regulation. Research has clearly demonstrated the involvement of epigenetic control mechanisms at these genomic loci.
Figure 2. The epigenetic marks that define the transcriptional state of an ES.
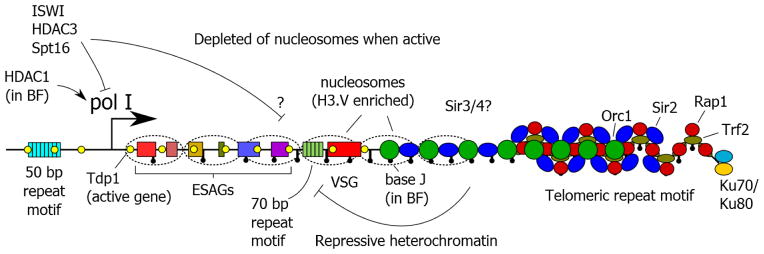
A repressive chromatin structure is formed by TbTRF2 and TbRAP1 (which may recruit Sir2) as well as TbORC1, propagating to sub-telomeric regions. It is not known whether other proteins fulfil the roles of yeast Sir3 and Sir4, for which orthologues are absent in T. brucei. Base J is present at an increasing density towards the telomere termini, and is required for ES silencing. Nucleosomes present on a silent ES are enriched for the transcriptional terminating variant H3.V, and are depleted on an active ES. The HMG box protein TbTDP1 is present on the active ES, and is associated with chromatin decondensation. The histone deacetylase TbHDAC3 and the chromatin remodeller TbISWI is required for efficient ES silencing, and TbHDAC1 is required for activated expression.
The active ES in BF T. brucei was shown to be depleted of nucleosomes compared to silent ESs, a phenomenon probably related to transcriptional activity [35]. TbTDP1, an HMG box protein, was enriched at active ESs [36]. HMG Box proteins are capable of facilitating chromatin decondensation, thus making chromatin more accessible to regulatory factors, and facilitating the recruitment of transcription activators [37,38]. TbTDP1 was also enriched at the 50 bp repeats adjacent to ESs and immediately downstream of the rDNA promoter, binding to the entire rDNA locus. Diminishing TbTDP1 synthesis by RNAi resulted in an increase in histone abundance on pol I transcription units and a concomitant reduction in pol I transcriptional activity, leading to a growth arrest within 24h. TbTDP1 was essential for active pol I transcription, and was enriched at highly transcribed regions which were generally depleted of nucleosomes, including the active ES. This, along with recent results [39,40], strongly suggested the involvement of chromatin remodelling in the regulation of the transcriptional state of an ES. Indeed, the chromatin remodeller TbISWI was shown to play a role in repression of pol I transcribed ESs in both BF and PF stages of T. brucei [41]. TbISWI also contributed to the down regulation of PF-specific procyclin genes, non-transcribed VSG arrays and minichromosomes. The involvement of chromatin remodellers in gene repression was previously shown in the temporal regulation of the pS2 gene [42]. SWI/SNF and NuRD were suggested to be required for resetting the local nucleosomal structures to allow transcriptional shutdown of pS2 in the absence of transcriptional activators [42]. The histone deacetylase, TbHDAC1, antagonised basal telomeric repression in BF cells, and TbHDAC3 was required for VSG ES promoter silencing in both PF and BF cells [43].
In the mammalian telomere complex, TRF2 is bound to duplex telomere DNA, and serves as a recruitment anchor for another telomeric protein, RAP1 [44,45]. T. brucei possesses functional orthologues of both these telomeric proteins, termed TbTRF2 and TbRAP1 [39]. TbRAP1 is found at telomeres, and is essential for growth and critical for ES silencing. Knockdown of TbRAP1 led to a graduated derepression of silent ESs [46]. TbRAP1-mediated silencing increased within the terminal 10 kb of the telomeres, supporting the suggestion that telomere structure were essential for the regulation of VSG expression [39]. It was also shown that telomeric repression decreased with distance from the telomere [39]. In Saccharomyces cerevisiae, the telomere repeat binding protein Rap1, together with the Sir proteins, were shown to be required for telomere proximal silencing as well as for position effect variegation [47]. TbSIR2RP1, a Sir2 related protein in T. brucei, co-localized with telomeric sequences, and appeared to be involved in the establishment of a silencing gradient at the telomeres in the BF parasite [40]. Interestingly, orthologues to the yeast Sir3 and 4 proteins, which are recruited by Rap1 and Sir2 to form propagative, repressive chromatin structures at the telomeres and silent mating type loci in S. cerevisiae, appear absent in T. brucei. This is perhaps not surprizing, since Sir3 only appeared in the S. cerevisiae genome by gene duplication of Orc1 after evolutionary divergence of the trypanosomes [48]. It is not clear what proteins, if any, may function with TbTRF2, TbRAP1 and TbSIR2 to establish a telomere-proximal repressive domain in T. brucei.
In contrast to laboratory strains, T. brucei field strains possess shorter telomeres [49], and switch VSGs more frequently [50,51]. A recent study revealed that telomere length is correlated with VSG switching frequency, and demonstrated that the shorter the telomere structure at an active ES, the more frequently VSG switching occurred [52].
3.4 Base J
One of many unusual epigenetic features found in T. brucei is the modified thymidine residue β-D-glucosyl-hydroxymethyluracil, designated base J, which is found in all kinetoplastids as well as in Dipolonema and Euglena [53]. In T. brucei, J was primarily associated with repetitive DNA elements such as the telomeric, 50, 70, and 177 bp repeats, and was also shown to localize at PTU flanks and at transcription termination sites (TTS) [54]. Base J was particularly enriched at silent VSG expression sites, forming an increasing gradient towards the telomere termini (Figure 1 and 2).
Base J is developmentally regulated, and is only found in the BF stage of the T. brucei life cycle [55]. Two thymidine hydroxylases that are involved in the synthesis of J have been identified: TbJBP1 bound to J DNA and stimulated conversion of adjacent thymine residues to base J, whereas TbJBP2 was capable of de novo J synthesis. Deletion of these enzymes eliminated the first step of J biosynthesis. Although a JBP1 knock-out was lethal in Leishmania [56], T. brucei strains in which both enzymes had been knocked-out exhibited no serious growth defects [57].
In Leishmania the efficient termination of pol II transcription did not occur in the absence of J, unless pol II was terminated by a transcribing pol III [58]. Although the function of J in T. brucei remains unclear, it appears highly likely to interfere with pol II elongation, acting as a transcriptional terminator and epigenetic repressor.
3.5 Replication origin complex and gene silencing
DNA replication, similar to transcription, initiates with the assembly of a pre-replication complex at an origin of replication sequence. This complex is composed of the Origin Recognition Complex (ORC), Cdc6, Cdt1 and MCM [59]. Genome-wide analysis of TbORC1/CDC6 (subsequently referred to as TbORC1) binding sites in T. brucei revealed an overlap between replication origins and the boundaries of PTUs. All functional replication origins occurred in chromosome core regions, associated with transcription initiation and termination [60]. In S. cerevisiae, silent genomic regions such as the silent mating type loci are bordered by A-boxes, sequences recognized and bound by ORC1. ORC1, which contains a nucleosome binding BAH domain, nucleates a complex that is essential in facilitating transcriptional silencing in the adjacent genome [48]. Surprisingly, TbORC1 does not contain an identifiable BAH domain, but was shown to be required for efficient sub-telomeric repression and ES silencing [60–62]. It is not currently known whether all TbORC1-binding sites in T. brucei represent active origins, or whether a subset, specifically those located in sub-telomeric regions or at silent VSG arrays, function exclusively in gene silencing, similar to S. cerevisiae and other yeasts [62]. Indeed, it was shown that the genes that were present adjacent to the boundaries of PTUs, proximal to sites of TbORC1 localization, displayed higher transcript levels following RNAi mediated TbORC1 knock-down [60].
4. Nucleosomal organization
Genome-wide maps of the nucleosome organization in model organisms show a common arrangement of nucleosomes at specific functional locations. A nucleosome free region (NFR), exposing part of the proximal pol II promoter, is seen in yeast [63,64], Caenorhabditis elegans [65], Drosophila [66], and in humans [67]. The NFR is bordered by two well positioned nucleosomes: −1 on the upstream and +1 on the downstream side of the NFR, followed by a nucleosomal array extending over the gene. High levels of histone variants and post-translational modifications are observed for nucleosomes flanking the NFR [68].
A genome-wide map of nucleosome positions in T. brucei has not yet been published. However, looking at the genome-wide map of another protist, Plasmodium falciparum, insight into the possible nucleosomal organisation of T. brucei may be derived. In P. falciparum a nucleosomal organization different from other model eukaryotes was seen [69]. Nucleosomes were found to be associated with coding regions and generally absent from intergenic and promoter regions. The high AT-content of the intergenic regions of Plasmodium may selectively exclude nucleosomes, allowing easy access to polymerases and associated factors [70]. Indeed, it has been suggested that AT-richness in the Plasmodium genome may serve as intrinsic signals for nucleosome positioning [71]. The effect of the many AT-rich repeats, including those associated with centromeres [72], on nucleosome positioning in the T. brucei genome is currently unknown, and may contribute to the epigenetic regulation employed by this kinetoplastid.
5. Histone epigenetic patterns
5.1 H1
Trypanosomal histone H1 differs noticeably from that of other eukaryotes. T. brucei H1 is comprised of a single domain corresponding to the lysine rich C-terminal domain of higher eukaryotic histone H1. This arrangement is similar to Tetrahymena H1 [73], which lacks the central winged helix domain. A recent study demonstrated the involvement of TbH1 in maintaining a condensed state of chromatin at non-transcribed regions, including the silent VSG arrays and inactive VSG ESs. TbH1 is not only required to down-regulate silent VSG ESs, but may also suppress VSG switching [74].
5.2 H2A
Several studies of T. brucei H2A PTMs revealed the absence of modifications that were well conserved in other eukaryotes [75,76]. Additionally, a number of trypanosome-specific PTMs were also identified [75,76].
Analysis of the first 22 amino acid residues of histone H2A revealed 60% monomethylation of A1 and an ~1% acetylation of K4 [76] (Figure 3A). H2A displayed a complex pattern of multiple PTMs of the C-terminus, including 6 acetylated lysines (K115, K119, K120, K122, K125, and K128) of which three (K120, K122, and K128) corresponded to conserved lysine residues with defined epigenetic marks in other species.
Figure 3. Epigenetic modifications of the T. brucei core histone N-terminal tails.
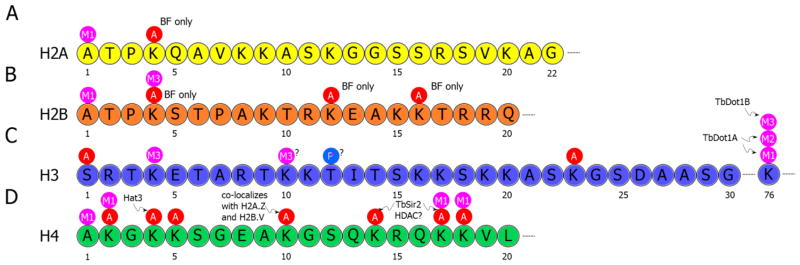
Modifications mapped to specific residues and enzymes involved in the modulation of some modifications are shown. Life stage specific modifications of the parasite are also identified. It is not currently known whether H3K10 or H3K11 is the equivalent of the highly conserved H3K9 present in other eukaryotes, and whether H3T12 is the equivalent of H3S11, a known phosphorylation target. A = Acetylation, Me = Methylation, P = Phosphorylation.
It is possible that T. brucei H2AK122 could be ubiquitinated, since it is the only lysine in the H2A C-terminus adjacent to a potential phosphorylation target, S123. It was suggested that phosphorylation influenced the ubiquitination of neighbouring lysines [77]. In addition, T. brucei H2AK118 and H2AK122 aligns with Drosophila H2AK118 and human H2AK119, sites of ubiquitination associated with transcriptional repression [78,79]. Human H2AK125, corresponding to trypanosomal H2AK128, is a possible target for either acetylation or crotonylation [80,81]. However, neither H2AK118 or K125 appears to be modified in T. brucei [76].
5.3 H2B
H2B is the least conserved of the four core histones [82] in T. brucei, and analysis has revealed only 4 PTMs. The same degree of methylation of A1 and acetylation of K4 was observed as for H2A. Tandem MS analysis showed minor acetylation of K12 and K16 [76] (Figure 3B).
Evidence of life stage-dependent modifications is also seen T. brucei. Acetylated lysine residues are observed at K4 and K122 in H2A and at K4, K12 and K16 of H2B in BF trypanosomes, but not in the procyclic form [76] (Figure 3B).
5.4 H3
Histone H3 as well as its N-terminal tail is highly conserved from human to yeast, where the tail is subjected to an extensive range of PTMs. In T. brucei H3, however, identification of PTMs have been complicated by the acetylated N-terminal serine, which blocks Edman degradation, and only a few PTMs have been mapped to specific residues. MS analyses revealed that S1 and K23 were acetylated, K4 and K32 were tri-methylated, and K76 could be mono-, di- or tri methylated [76] (Figure 3C). Internal sequences of the H3 tail diverge sharply from that of canonical H3, but sequence alignment suggests that T. brucei K19, K23, K32 and K76 could be equivalent to K23, K27, K36, and K79 of other eukaryotes, respectively. Although many of above PTMs have been functionally described in other organisms [4,83], the functional roles of these PTMs in T. brucei, with the exception of K76, is not known.
TbDOT1A is responsible for mono- and di-methylation of H3K76. The RNAi knock-down of TbDOT1A resulted in severe cell cycle defects [84,85]. A clear correlation exists between H3K76 mono- and di-methylation and transcription termination sites (TTSs), suggesting a role in transcription termination [85]. Tri-methylation of H3K76 was mediated by TbDOT1B, which was not essential for viability [85]. Mono-, di-, and tri-methylation of K76 was implicated in several processes, including replication control, antigenic variation, and developmental differentiation [82,85]. K76 di-methylation is only detectable during mitosis [84].
Mandava and co-workers [86] reported that the H2B variant, H2B.V, was present in mononucleosomes enriched for tri-methylated H3K4 and K76, and suggested that H2B.V can replace canonical H2B, permitting H3K4 and K76 methylation. A puzzling feature among kinetoplastids is the absence of the almost universally conserved H3K9, implicated in gene repression in its tri-methylated state [87]. It is not yet clear whether K10 is the equivalent residue, although the sequence context of T. brucei H3K10 is markedly different from that of K9 in other eukaryotes.
5.5 H4
Of all trypanosomal histones, H4 is the most conserved. As in H2A and H2B, H4A1 is also monomethylated to a level of approximately 60% (Figure 3D). K4, K5, K10, and K14 were observed to be acetylated, and K2, K17, and K18 were acetylated or methylated to various extents [76]. Sequence alignment showed the presence of lysine residues at both position 4 and 5 in trypanosomes. In other eukaryotes glycine is the conserved residue at position 4 in H4. H4K4 is the most commonly acetylated histone tail residue in T. brucei [75], suggesting that T. brucei K4 was the functional equivalent of K5 present in other eukaryotes. The histone acetyltransferase TbHAT3 was responsible for H4K4 acetylation in both PF and BF life stages. This non-essential, MYST-type acetyltransferase seemed to acetylate H4 upon import into the nucleus for packaging of newly-replicated DNA [88].
ChIP-seq studies showed twin peaks of acetylated H4K10 at divergent SSRs [20]. A number of single acetylated K10ac peaks were found at non-SSRs, many of which occurred downstream of tRNA genes. Most tRNA genes are located at convergent SSRs, and of those located at non-SSR, all but 3 of 38 were located upstream of a single acetylated K10ac peak. If a tRNA gene represents a roadblock to pol II transcription within a PTU, for example, pol II would need to reinitiate downstream of the tRNA gene, within the region enriched for H4K10ac. This, together with the observation that pol II transcription initiated at divergent SSRs, suggested a link between transcription initiation and acetylated H4K10. Higher levels of this modification were observed upstream of the first and downstream of the last PTU of each chromosome [20]. Distribution profiles of this modification were remarkably similar between parasite life stages, with only two life stage-specific peaks being observed (on chromosome 7 and 11).
The Bromodomain Factor 3 protein (TbBDF3) was shown to bind to acetylated lysines [89]. It co-localises with acetylated H4K10 and is concentrated towards the upstream end of H4K10ac peaks. It was suggested that TbBDF3 is involved in targeting chromatin remodelling complexes to TSSs [89,90]. TbBDF3 was essential for cellular viability, and RNAi mediated knockdown caused an immediate growth defect where most cells died within 48h [20].
6. Histone variants
T. brucei encodes four histone variants: H2A.Z, H2B.V, H3.V and H4.V. Nucleosomes that contain H2A.Z are less stable than nucleosomes containing canonical H2A [20,91], and data also suggested that H2A.Z containing chromatin is less condensed, and thus primed for transcription. H2A.Z was shown to be associated exclusively with H2B.V in T. brucei [92,93], exhibiting virtually identical ChIP profiles and similar genomic distributions during the cell cycle. Both histone variants were shown to be essential for cell viability [92].
ChIP-seq of H2A.Z and H2B.V revealed a genomic distribution almost indistinguishable from that of acetylated H4K10. Distinct matched peaks were observed at divergent SSR as well as single H2B.V peaks at non-SSR, coincident with that of H4K10ac. H2B.V was also shown to be present in nucleosomes that were enriched for trimethylated H3K4 and H3K76, PTMs typically associated with transcriptionally active chromatin [4].
T. brucei encodes two H3 histones, H3 and the variant H3.V, which shares ~60% identity [94,95]. T. brucei appears to lack a centromere-specific CenH3 orthologue [95,96]. H3.V was found to be highly enriched at telomeric repeats and subtelomeric regions (see Figure 2), but not at the 177 bp minichromosome repeat or 5S rDNA loci [94]. Single peaks of H3.V nucleosomes were located at convergent SSR and upstream of all H4K10ac-rich regions not associated with a SSR. Sequence analysis of regions rich in H4K10ac revealed G-rich stretches of 9 to 15 guanine residues at SSRs. Generation of H3.V null strains indicated that H3.V is not essential for viability, mini-chromosome segregation, telomere maintenance or transcriptional silencing of BESs and does not contribute to the open chromatin structure observed at the active BES [94,97].
The distribution of H4.V was found to be similar to that of H3.V throughout the genome [20]. H4.V was, however, less enriched compared to H3.V at sub-telomeric and telomeric sites [20]. Both H3.V and H4.V were found to be significantly enriched immediately downstream of the last coding sequence of a PTU (see Figure 1). This suggested that H3.V-H4.V containing nucleosomes were enriched at presumed pol II TTS, and thus serves as epigenetic markers for the end of transcription units.
Collectively, these findings suggest that putative RNA polymerase transcription start and termination sites are demarcated by specific histone variants and PTMs, likely conferring defined structural states to local chromatin regions, and recruiting functionally important chromatin associated proteins to such regions.
7. Conclusions
The many studies cited in this review have provided ample evidence that in T. brucei, rather than being a constitutive, unregulated process, where transcript levels are only controlled post-transcriptionally, gene expression, particularly of the genes encoding the major cell surface proteins, is closely tied to chromatin. In other areas of the genome, promoters as well as transcription stop sites may be determined, not by DNA sequence, but by chromatin structure, which is established by chromatin remodellers, readers, writers, and erasers. Therefore, although there is little regulatory control at the level of transcription at the PTUs, chromatin plays a key role in delineating T. brucei transcription units, and in controlling the initiation of transcription as well as DNA replication. Furthermore, specific histone variants and histone modification states synergise to provide a rich regulatory interface to control gene expression. This regulation of gene transcription by the epigenome provides the exciting possibility that epigenetic components may represent novel drug targets, and that epigenetic therapies may be developed to treat this lethal disease in future. This is particularly relevant in the light of increasing resistance to widely used drug combinations reported for T. brucei field strains.
8. Where do we go from here?
The sequencing of the T. brucei genome has revealed that many epigenetic modifiers, readers, erasers and chromatin remodelling enzymes are encoded by this kinetoplastid [7]. The localization of specific histone variants, modified histones [21] and glycosylated thymidines [49] at sites of pol II transcription initiation and termination is striking. Also, ORC1, implicated in the nucleation of a repressive heterochromatin at the silent mating type loci in S. cerevisiae, was shown to be recruited to PTU flanks as well as the silent VSG arrays in T. brucei. Thus, there are many bits of enticing data that suggest an important role for chromatin structure and epigenetics in the modulation of gene expression in T. brucei. A number of questions that beg for investigation include the role of histone variants and modified histones in defining regions of pol II initiation. By what mechanism are these proteins and modifications enriched at these loci? Although the pol II promoter remains undefined, is there a sequence recognized by a DNA-binding protein that subsequently recruits additional factors and modifiers?
The silent bloodstream ESs show strong evidence of telomeric repression. The role of TbRAP1 and TbORC1 in this process was clearly shown [34,54]. The repressive effect extends beyond the TTAGGG telomeric repeats. This suggests the propagation of a repressive chromatin structure from the telomeric ends, similar to the process seen in yeast and in Drosophila [59,60,62]. In yeast the Orc1 paralogue, Sir3, together with Sir4 and the histone deacetylase, Sir2, are required for the propagation of the telomeric repression effect. In Drosophila the HP1 protein is involved in heterochromatic structures. What structural proteins contribute to extended telomeric repression in T. brucei?
The pursuit of these questions will deepen our understanding of the epigenome of T. brucei and the mechanisms by which genome function in this kinetoplastid is modulated. It is likely that expansion of our knowledge in this area would include the identification of novel, epigenetic targets that could form part of new therapies directed at the control of African trypanosomiasis.
Highlights.
Pol II genes are arranged as polycistronic units of functionally unrelated genes
Pol II promoters are undefined
H2A.Z and H2B.V and modified H3 and H4 are enriched at transcription start sites
tRNA genes, H3.V, H4.V, base J and H3K76me2/3 co-localizes with termination regions
These links suggest a crucial role for epigenetics in transcriptional control
Acknowledgments
The authors thank Megan Povelones for critical and useful comments on the manuscript. This study was supported by Award Number 1 U01 HG007465-01 (to HGP) administered by the National Human Genome Research Institute as part of the NIH Common Fund H3Africa Initiative.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hide G, Tait A. Molecular epidemiology of African sleeping sickness. Parasitology. 2009;136:1491–1500. doi: 10.1017/S0031182009990333. [DOI] [PubMed] [Google Scholar]
- 2.Barry JD, Graham SV, Fotheringham M, Graham VS, Kobryn K, et al. VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol Biochem Parasitol. 1998;91:93–105. doi: 10.1016/s0166-6851(97)00193-x. [DOI] [PubMed] [Google Scholar]
- 3.Daniels J-P, Gull K, Wickstead B. Cell biology of the trypanosome genome. Microbiol Mol Biol Rev. 2010;74:552–569. doi: 10.1128/MMBR.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/S0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 6.Berriman M, Ghedin E, Hertz-fowler C, Blandin G, Bartholomeu DC, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 7.Melville S, Leech V, Gerrard C. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol Biochem Parasitol. 1998;94:155–173. doi: 10.1016/s0166-6851(98)00054-1. [DOI] [PubMed] [Google Scholar]
- 8.Ersfeld K. Nuclear architecture, genome and chromatin organisation in Trypanosoma brucei. Res Microbiol. 2011;162:626–636. doi: 10.1016/j.resmic.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Wickstead B, Ersfeld K, Gull K. The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 2004;14:1014–1024. doi: 10.1101/gr.2227704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–62. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- 12.Martínez-Calvillo S, Yan S, Nguyen D, Fox M. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 2003;11:1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 13.Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, et al. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly S, Kramer S, Schwede A, Maini PK, Gull K, et al. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biol. 2012;2:120033. doi: 10.1098/rsob.120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palenchar JB, Bellofatto V. Gene transcription in trypanosomes. Mol Biochem Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Kelly S, Wickstead B, Gull K. An in silico analysis of trypanosomatid RNA polymerases: insights into their unusual transcription. Biochem Soc Trans. 2005;33:1435–1437. doi: 10.1042/BST20051435. [DOI] [PubMed] [Google Scholar]
- 17.Günzl A, Bruderer T, Laufer G, Tu L, Chung H, et al. RNA polymerase i transcribes procyclin genes and Variant Surface Glycoprotein gene expression sites in Trypanosoma brucei Eukaryot. Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo H, Gilinger G, Mukherjee D, Bellofatto V. Transcription initiation at the TATA-less Spliced Leader RNA gene promoter requires at least two DNA-binding proteins and a tripartite architecture that includes an initiator element. J Biol Chem. 1999;274:31947–31954. doi: 10.1074/jbc.274.45.31947. [DOI] [PubMed] [Google Scholar]
- 19.Ruan J, Arhin GK, Ullu E, Tschudi C. Functional characterization of a Trypanosoma brucei TATA-binding protein-related factor points to a universal regulator of transcription in trypanosomes. Mol Cell Biol. 2004;24:9610–9618. doi: 10.1128/MCB.24.21.9610-9618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, et al. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang X, Haritan A, Uliel S, Michaeli S. Trans and cis splicing in Trypanosomatids: Mechanism, Factors, and Regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Jung HS, Günzl A. Transcriptionally active TFIIH of the early-diverged eukaryote Trypanosoma brucei harbors two novel core subunits but not a cyclin-activating kinase complex. Nucleic Acids Res. 2009;37:3811–3820. doi: 10.1093/nar/gkp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JH, Nguyen TN, Schimanski B, Günzl A. Spliced leader RNA gene transcription in Trypanosoma brucei requires transcription factor TFIIH. Eukaryot. Cell. 2007;6:641–649. doi: 10.1128/EC.00411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schimanski B, Brandenburg J, Nguyen TN, Caimano MJ, Günzl A. A TFIIB-like protein is indispensable for spliced leader RNA gene transcription in Trypanosoma brucei. Nucleic Acids Res. 2006;34:1676–1684. doi: 10.1093/nar/gkl090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hitchcock R, Thomas S, Campbell D, Sturm NR. The promoter and transcribed regions of the Leishmania tarentolae spliced leader RNA gene array are devoid of nucleosomes. BMC Microbiol. 2007;7:44. doi: 10.1186/1471-2180-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAndrew M, Graham S, Hartmann C, Clayton C. Testing promoter activity in the trypanosome genome: isolation of a metacyclic-type VSG promoter, and unexpected insights into RNA polymerase II transcription. Exp Parasitol. 1998;76:65–76. doi: 10.1006/expr.1998.4317. [DOI] [PubMed] [Google Scholar]
- 27.Nagarajavel V, Iben JR, Howard BH, Maraia RJ, Clark DJ. Global “bootprinting” reveals the elastic architecture of the yeast TFIIIB-TFIIIC transcription complex in vivo. Nucleic Acids Res. 2013;41:8135–8143. doi: 10.1093/nar/gkt611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hull MW, Erickson J, Johnston M, Engelke DR. tRNA Genes as Transcriptional Repressor Elements. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/MCB.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 32.Sheader K, Berberof M, Isobe T, Borst P, Rudenko G. Delineation of the regulated Variant Surface Glycoprotein gene expression site domain of Trypanosoma brucei. Mol Biochem Parasitol. 2003;128:147–156. doi: 10.1016/S0166-6851(03)00056-2. [DOI] [PubMed] [Google Scholar]
- 33.Hertz-Fowler C, Figueiredo LM, Quail Ma, Becker M, Jackson A, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PloS One. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 35.Stanne TM, Rudenko G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot. Cell. 2010;9:136–147. doi: 10.1128/EC.00281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayanan MS, Rudenko G. TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res. 2013;41:2981–2992. doi: 10.1093/nar/gks1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Reeves R. Nuclear functions of the HMG proteins. Biochim Biophys Acta. 2010;1799:3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Figueiredo LML, Espinal A, Okubo E, Li B. RAP1 Is essential for silencing telomeric Variant Surface Glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudenko G. Epigenetics and transcriptional control in African trypanosomes. Essays Biochem. 2010;48:201–219. doi: 10.1042/bse0480201. [DOI] [PubMed] [Google Scholar]
- 41.Stanne TM, Kushwaha M, Wand M, Taylor JE, Rudenko G. TbISWI regulates multiple polymerase I (Pol I)-transcribed loci and is present at Pol II transcription boundaries in Trypanosoma brucei. Eukaryot. Cell. 2011;10:964–976. doi: 10.1128/EC.05048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Métivier R, Penot G, Hübner MR, Reid G, Brand H, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 43.Wang Q, Kawahara T, Horn D. Histone deacetylases play distinct roles in telomeric VSG expression site silencing in African trypanosomes. Mol Microbiol. 2010;77:1237–1245. doi: 10.1111/j.1365-2958.2010.07284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 45.Li B, Oestreich S, de Lange T. Identification of Human Rap1. Cell. 2000;101:471–483. doi: 10.1016/S0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 46.Pandya UM, Sandhu R, Li B. Silencing subtelomeric VSGs by Trypanosoma brucei RAP1 at the insect stage involves chromatin structure changes. Nucleic Acids Res. 2013;41:7673–7682. doi: 10.1093/nar/gkt562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, et al. The clustering of telomeres and colocalization with Rapl, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–136. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hickman M, Rusche LN. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc Natl Acad Sci U S A. 2010;107:19384–19389. doi: 10.1073/pnas.1006436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dreesen O, Cross GAM. Telomere length in Trypanosoma brucei. Exp Parasitol. 2008;118:103–110. doi: 10.1016/j.exppara.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamont GS, Tucker RS, Cross GAM. Analysis of antigen switching rates in Trypanosoma brucei. Parasitology. 1986;92:355–367. doi: 10.1017/s003118200006412x. http://dx.doi.org/10.1017/S003118200006412X. [DOI] [PubMed] [Google Scholar]
- 51.Robinson NP, Burman N, Melville SE, Barry JD. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol Cell Biol. 1999;19:5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hovel-Miner Ga, Boothroyd CE, Mugnier M, Dreesen O, Cross GAM, et al. Telomere length affects the frequency and mechanism of antigenic variation in Trypanosoma brucei. PLoS Pathog. 2012;8:e1002900. doi: 10.1371/journal.ppat.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Leeuwen F, Kieft R, Cross M, Borst P. Tandemly repeated DNA is a target for the partial replacement of thymine by β-d-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol Biochem Parasitol. 2000;109:133–145. doi: 10.1016/s0166-6851(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 54.Cliffe LJ, Siegel TN, Marshall M, Cross GAM, Sabatini R. Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei. Nucleic Acids Res. 2010;38:3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Leeuwen F, Wijsman ER, Kieft R, van der Marel GA, van Boom JH, et al. Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997;11:3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Genest P-A, ter Riet B, Dumas C, Papadopoulou B, van Luenen HGAM, et al. Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania. Nucleic Acids Res. 2005;33:1699–1709. doi: 10.1093/nar/gki304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cliffe LJ, Kieft R, Southern T, Birkeland SR, Marshall M, et al. JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res. 2009;37:1452–1462. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Luenen HGAM, Farris C, Jan S, Genest P, Tripathi P, et al. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012;150:909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 60.Tiengwe C, Marcello L, Farr H, Dickens N, Kelly S, et al. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep. 2012;2:185–197. doi: 10.1016/j.celrep.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benmerzouga I, Concepción-Acevedo J, Kim H-S, Vandoros AV, Cross GM, et al. Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol Microbiol. 2013;87:196–210. doi: 10.1111/mmi.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tiengwe C, Marques Ca, McCulloch R. Nuclear DNA replication initiation in kinetoplastid parasites: new insights into an ancient process. Trends Parasit. 2014;30:27–36. doi: 10.1016/j.pt.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 63.Lee W, Tillo D, Bray N, Morse RH, Davis RW, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 64.Brogaard K, Xi L, Wang J-P, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westenberger SJ, Cui L, Dharia N, Winzeler E, Cui L. Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes. BMC Genomics. 2009;10:610. doi: 10.1186/1471-2164-10-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ponts N, Harris EY, Prudhomme J, Wick I, Eckhardt-Ludka C, et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 2010;20:228–238. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Echeverry MC, Bot C, Obado SO, Taylor MC, Kelly JM. Centromere-associated repeat arrays on Trypanosoma brucei chromosomes are much more extensive than predicted. BMC Genomics. 2012;13:29. doi: 10.1186/1471-2164-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen X, Yu L, Weir JW, Gorovsky MA. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 74.Povelones ML, Gluenz E, Dembek M, Gull K, Rudenko G. Histone H1 plays a role in heterochromatin formation and VSG expression site silencing in Trypanosoma brucei. PLoS Pathog. 2012;8:e1003010. doi: 10.1371/journal.ppat.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Janzen CJ, Fernandez JP, Deng H, Diaz R, Hake SB, et al. Unusual histone modifications in Trypanosoma brucei. FEBS Lett. 2006;17:2306–2310. doi: 10.1016/j.febslet.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 76.Mandava V, Fernandez JP, Deng H, Janzen CJ, Hake SB, et al. Histone modifications in Trypanosoma brucei. Mol Biochem Parasitol. 2007;156:41–50. doi: 10.1016/j.molbiopara.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 78.Wang H, Zhai L, Xu J, Joo H-Y, Jackson S, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 79.Gutiérrez L, Oktaba K, Scheuermann JC, Gambetta MC, Ly-Hartig N, et al. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development. 2012;139:117–127. doi: 10.1242/dev.074450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan M, Luo H, Lee S, Jin F, Yang JS, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Basu A, Rose KL, Zhang J, Beavis RC, Ueberheide B, et al. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci U S A. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Figueiredo LM, Cross GAM, Janzen CJ. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat Rev Microbiol. 2009;7:504–513. doi: 10.1038/nrmicro2149. [DOI] [PubMed] [Google Scholar]
- 83.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 84.Janzen CJ, Hake SB, Lowell JE, Cross GAM. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 85.Gassen A, Brechtefeld D, Schandry N, Arteaga-Salas JM, Israel L, et al. DOT1A-dependent H3K76 methylation is required for replication regulation in Trypanosoma brucei. Nucleic Acids Res. 2012:1–10. doi: 10.1093/nar/gks801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mandava V, Janzen CJ, Cross GAM. Trypanosome H2Bv replaces H2B in nucleosomes enriched for H3 K4 and K76 trimethylation. Biochem Biophys Res Commun. 2008;368:846–851. doi: 10.1016/j.bbrc.2008.01.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 88.Siegel TN, Kawahara T, Degrasse JA, Janzen CJ, Horn D, et al. Acetylation of histone H4K4 is cell cycle regulated and mediated by HAT3 in Trypanosoma brucei. Mol Microbiol. 2008;67:762–771. doi: 10.1111/j.1365-2958.2007.06079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winston F, Allis CD. The bromodomain: a chromatin-targeting module ? Nat. Struct Mol Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 90.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 91.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lowell JE, Kaiser F, Janzen CJ, Cross GAM. Histone H2AZ dimerizes with a novel variant H2B and is enriched at repetitive DNA in Trypanosoma brucei. J Cell Sci. 2005;118:5721–5730. doi: 10.1242/jcs.02688. [DOI] [PubMed] [Google Scholar]
- 93.Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Mol Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- 94.Lowell JE, Cross GAM. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J Cell Sci. 2004;117:5937–5947. doi: 10.1242/jcs.01515. [DOI] [PubMed] [Google Scholar]
- 95.Talbert PB, Henikoff S. Chromatin-based transcriptional punctuation. Genes Dev. 2009;23:1037–1041. doi: 10.1101/gad.1806409. [DOI] [PubMed] [Google Scholar]
- 96.Akiyoshi B, Gull K. Discovery of unconventional kinetochores in kinetoplastids. Cell. 2014;156:1247–1258. doi: 10.1016/j.cell.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Figueiredo LM, Cross GM. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot Cell. 2010;9:148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


