Abstract
Pancreatic ductal adenocarcinoma remains one of the most deadly types of tumor. Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) is a safe, cost-effective, and accurate technique for evaluating and staging pancreatic tumors. However, EUS-FNA may be inconclusive or doubtful in up to 20% of cases. This review underlines the clinical interest of the molecular analysis of samples obtained by EUS-FNA in assessing diagnosis or prognosis of pancreatic cancer, especially in locally advanced tumors. On EUS-FNA materials DNA, mRNA and miRNA can be extracted, amplified, quantified and subjected to methylation assay. Kras mutation assay, improves diagnosis of pancreatic cancer. When facing to clinical and radiological presentations of pseudo-tumorous chronic pancreatitis, wild-type Kras is evocative of benignity. Conversely, in front of a pancreatic mass suspected of malignancy, a mutated Kras is highly evocative of pancreatic adenocarcinoma. This strategy can reduce false-negative diagnoses, avoids the delay of making decisions and reduces loss of surgical resectability. Similar approaches are conducted using analysis of miRNA expression as well as Mucin or markers of invasion (S100P, S100A6, PLAT or PLAU). Beyond the diagnosis approach, the prediction of response to treatment can be also investigated form biomarkers expression within EUS-FNA materials.
Keywords: Pancreatic ductal adenocarcinoma, Endoscopic ultrasound-guided fine-needle aspiration, Solid pancreatic mass, KRAS-mutation assay, qPCR analysis, Micro-RNA, Chronic pancreatitis
Core tip: This review depicts the widespread potential for the molecular analysis of samples obtained by ultrasound-guided fine needle aspiration in assessing diagnosis or prognosis of pancreatic adenocarcinoma, as well as translational studies on new markers and epigenetic alterations. Among these markers, Kras oncogene assay appears now the most robust for improvement of positive and differential diagnosis of pancreatic cancer. Clinical implication of miRNA, Mucins and markers of invasion is still debated. Future molecular developments may open windows towards personalized treatments after molecular characterization of a single patient.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most deadly types of tumor. The 5-year survival rate after diagnosis is < 3.5%[1]. The only curative treatment is surgical resection but this surgery is possible in only 10% to 15% of cases. The remaining cases with locally advanced and/or metastatic pancreatic cancer are treated in a palliative way with chemotherapy (Gemcitabine or FOLFIRINOX) or best supportive cares[1]. This dismal prognostic is partly due to the lack of robust markers for the early diagnosis of PDAC that may jeopardize treatment efficacy in a subset of patients. Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) is a rapid, safe, cost-effective, and accurate technique for evaluating and staging pancreatic tumors[2-6]. In addition, EUS-FNA is the main clinical appliance for cytological and histological material collection from locally advanced PDAC that represents 85% of pancreatic cancer patients. However, its accuracy for the diagnosis of malignancy varies widely with a sensitivity ranging from 65% to 95%, and with a mean accuracy of 85% (negative predictive value ranging from 50% to 70%). Despite the miniaturization of histological samples provided by the FNA using 22 Gauge needle, immunohistochemistry can be achieved when micro biopsies are collected, fixed and embedded in paraffin. In our experience, micro-biopsies can be thus obtained in near 80% of cases. These Immunodiagnostic can be useful to differentiate for instance PDAC from endocrine tumors. It is harder to differentiate malignant from inflammatory lesions of exocrine pancreas. Despite modern imaging techniques, difficulties persist to early diagnose PDAC and to differentiate PDAC from benign diseases such as chronic pancreatitis especially in its pseudotumoral form[2-5]. It is indeed critical to avoid unnecessary resection of benign lesions (such as focal lesions of chronic pancreatitis or autoimmune pancreatitis) or to delay the treatment of PDAC in a subset of patients. Finally EUS-FNA may be inconclusive or doubtful in up to 20% of cases[2-7]. An explanation for an inconclusive cytopathology is multiple: (1) in PDAC the presence of desmoplastic reaction often associated with poor cellularity; (2) distinguishing well-differentiated PDAC and reactive atypia is difficult to appreciate in small samples; (3) small tumors are often not easy to biopsy and performances of cytopathology are lower[7]; and (4) well vascularized tumors that have a high risk of coagulating within the FNA materials. In cases where there is an inconclusive biopsy, a doubt persists regarding the presence or not of malignancy. Some technical improvements have been developed such as contrast ultrasound, elastography, new generations of needle (pro-core biopsy needle), or transport media for samples[8-11]. However, a subset of samples remained inconclusive and accuracy of EUS-FNA is still around 80%-85%. In parallel, the improvement of molecular biology techniques including DNA and RNA amplification permits the analysis and the quantification of molecular markers in cytological samples, especially from EUS-guided FNA of pancreatic lesions[12-17]. In addition, EUS-FNA that allows sampling of biological material and molecular biology is mandatory not only for pathologists but also for scientists to discover new molecular biomarkers for this disease. This review depicts the widespread potential for the molecular analysis of samples obtained by EUS-FNA in assessing diagnosis or prognosis of PDAC, as well as translational studies on new markers and epigenetic alterations.
POTENTIAL OF MOLECULAR ANALYSIS ON EUS-FNA MATERIALS
DNA extraction
Despite using fine needles, sufficient materials can be obtained for cytology and histology. A portion of this material, collected following air or saline flushing of the needle once the core biopsies have been reclaimed for histopathology, can be used for further molecular analysis. A mean of 550 nanograms of DNA (range 100 nanograms to 1.5 mg) is obtained and DNA amplification is possible in 98 to 100 of cases[18]. For comparison, previous studies and protocols conducted on pure pancreatic juice attested for a lack of extraction/amplification in almost 13% of samples[19-21]. Thereafter, purified DNA authorizes PCR followed by Restriction Fragment Length Polymorphism or sequencing. Recently we developed an allelic discrimination assay on material sampled on EUS-FNA as well as specific Methylation-Specific PCR assay[22]. All these procedures are successful in almost 100% of the cases, in the absence of DNA pre-amplification. This is of prime importance because DNA amplification generates mutations especially when using a low amount of starting material that can eventually bias subsequent analysis. In addition, new development of large-scale sequencing allows analysis of 400 genes simultaneously with a minimal quantity of DNA of 50 ng DNA. High volume for sequencing is also offered with a mean value of 1.5 μg. That opens a window to large-scale molecular analysis from a single EUS-FNA materials and from a single patient.
RNA extraction
While material collected from pancreatic tumor or inflammatory tissue is less exposed to RNAse digestion as compared to normal pancreatic tissue, the risk of degradation is very high if one wants to analyze high-quality RNA. From a practical point of view, cytological samples should be immediately stored in transport medium (such as RNable) and frozen at -25 °C until use. After centrifugation, total RNA can be extracted using Micro kits (for example RNeasy from Qiagen) followed by DNAse treatment. At this crucial stage, RNA quality and quantity should be determined with specific bioanalyzer (for example Biorad Experian analyser and Agilent Technologies). RNA samples that are highly degraded (RNA 18S/28S ratio less than 1) or with a quantity lower than 5 ng should be discarded. Indeed, degraded RNA are not suitable for reverse transcription or amplification. In our experience, near 50% of FNA materials appears non available for a reliable mRNA analysis. For assay of qPCR for 3 to 5 molecular markers an amplification is theoretically not required but if analysis on a larger panel of molecular targets is mandatory, amplification should be performed. Using 5 ng of total RNA (not degraded) is sufficient to perform RNA amplification kits (for instance Full Spectrum Kit) that permit up to 500-fold amplification with satisfactory reproducibility and reliability. In other terms, the RNA amplification from EUS-FNA material preserves the pattern of gene expression and is not influenced by the origin of the sample[23]. We had thus apply the technology of Taqmann Low Density Array to assess simultaneously the quantitative expression of 20 to 50 genes from EUS-FNA cellular materials (see below).
Micro RNA extraction
Interestingly, microRNAs are small molecules (19-25 nucleotides) of non coding RNA with high stability (less prompted to be degraded by RNase) in tissues and fluids. Moreover, they can be quantified in very low amounts of material and in highly degraded samples. Prior to microRNA analysis, tissues can be stored either frozen, or formalin-fixed and paraffin-embedded or in specific medium such as RNARetain[24]. It is important to mention that microRNA analysis of pancreatic FNA samples is possible but still in its infancy and may prove essential to help clinicians for the diagnosis of pancreatic lesions.
GENETIC MARKERS
Kras oncogene
The molecular mechanisms underlying pancreatic oncogenesis remain partially understood. However, several genetic alterations are well characterized in PDAC such as codon-12 Kras mutation (75% to 95%) and to a less extend p16 (CDKN2A, INK4), DPC4 and p53 gene mutations[25,26] associated to a loss of heterozygosity of respectively 9p21, 18q and 17p. These somatic genetic alterations are also detected in pre-cancerous lesion of PDAC as intraepithelial neoplasias (PanIN) and intraductal papillary mucinous neoplasm (IPMN)[26]. Previous studies conducted by we and others on pure pancreatic juice obtained by ERCP concluded that Kras mutation was found in 60% to 65 % of PDAC[19,20]. Moreover, additional p16 and DPC4 mutations analysis in pure pancreatic juice did not improve the sensitivity and specificity of Kras mutation analysis alone for diagnosis of PADC and to differentiate PDAC from CP[21]. Several research groups, including ours, have demonstrated that KRAS mutations can be detected in cellular materials obtained by EUS-FNA[27-37]. Kras-mutation analysis after EUS-FNA appears to be highly accurate at differentiating benign vs malignant pancreatic solid lesions[27-35].
We have conducted a multicenter prospective study to assess whether combining EUS-FNA with KRAS-mutation analysis could facilitate a differential diagnosis between PDAC and CP in a subgroup of patients with pseudo-tumorous forms[28]. We concluded that, when facing to clinical and radiological presentations of pseudo-tumorous CP, both pathological analyses (inflammation, fibrosis) and wild-type Kras are evocative of benignity. Based on the combination of cytopathological (including a second biopsy in case of negative results at the first biopsy) and Kras mutation analysis a medical or surgical conservative treatment can be applied. Otherwise, unnecessary pancreatic resection could be avoided. Conversely, when facing a clinical and radiological presentation of CP the presence of mutated Kras at EUS-FNA may justify a second biopsy and a follow up to rule out a PADC.
Whether the combination of EUS-FNA plus the Kras-mutation assay can improve diagnosis of malignant pancreatic tumors is still debatable. However, several studies have suggested that combining cytopathology and Kras-mutation analysis, improves the diagnosis of PDAC and malignancy (Table 1). This appears crucial in case of inconclusive or doubtful diagnosis at cytopathology. Inconclusive specimens were defined as the presence of coagulum with normal cells or acellular samples. Doubtful samples can be defined by the presence of atypia and/or low-grade dysplasia. Even if molecular biology cannot replace histology, the presence of a Kras mutation in EUS-FNA material indicates several possibilities: either immediate re-reading of the cytopathology (especially if doubtful) or a rapid indication from a second FNA, or surgery. In addition, from a clinical point of view, reducing false-negative diagnoses avoids the delay of making decisions, improves patients’ treatment, and reduces loss of surgical resectability. Conversely, in cases where there is an inconclusive EUS-FNA specimen, the presence of wild-type Kras may be evocative of benignity. Figure 1, integrates these conclusions in a proposed algorithm that include Kras mutation assay in the diagnosis approach of pancreatic solid masses using EUS-guided FNA. Because Kras analysis is now widely available, due to its use as a predictive marker for anti-EGFR therapy in colon cancer, this diagnostic tool could also be applied to help clinicians manage solid pancreatic masses. Kras assay has been improved by means of Taqmann Allelic discrimination that is cheaper, faster and more selective than other previous methods[38]. We have conducted recently a prospective study that included 186 patients with a pancreatic mass (including 104 PDAC, 22 other malignant pancreatic tumors and 60 benign lesions). Cytopathology and Kras mutations, using TaqMan® allelic discrimination, were performed on EUS-guided FNA material. We concluded that EUS-guided FNA plus Kras-mutation analysis, using allelic discrimination, is accurate and improves the diagnosis of pancreatic adenocarcinoma when pathology is inconclusive or doubtful (Table 1)[39]. In addition, we also confirmed that, when facing a clinical, radiological presentation of pseudo-tumorous chronic pancreatitis (including an evocative cytopathology), identification of wild-type Kras can rule out malignant transformation[39]. A retrospective study that included PDAC patients but also patients with an autoimmune pancreatitis reported also that a Kras mutation in EUS-guided FNA material from a pancreatic mass is associated with malignancy and may help discriminate from benign conditions such as autoimmune pancreatitis. In the study from Khalid et al[36] all of autoimmune pancreatitis cases had a wild type Kras.
Table 1.
Main studies investigating Kras mutation assay on specimens obtained by endoscopic ultrasound-guided fine-needle aspiration for the differential diagnosis between pancreatic carcinoma and pseudo-tumorous chronic pancreatitis
| Ref. | Patient | Sensitivity (%) | Specificity (%) | Overall accuracy (%) |
| PC/CP | Cytopathology alone/Kras + cytoP | Cytopathology alone/Kras + cytoP | Cytopathology alone/Kras + cytoP | |
| Tada et al[31] | 28/8 | 62/81 | 100/100 | 71/85 |
| Pellisé et al[35] | 33/24 | 97/97 | 100/100 | 84/98 |
| Takahashi et al[34] | 62/15 | 84/94 | 100/100 | CytoP alone: 58 |
| Maluf-Filho et al[33] | 57/11 | 82/90 | 97/47 | 59/89 |
| Bournet et al[28] | 129/27 | 83/88 | 100/100 | 72/90 |
| Reicher et al[30]1 | 34/16 | 88 | 94 | 90 |
| Ogura et al[29] | 307/47 | 87/93 | 100/100 | 89/94 |
| Ginestà et al[32] | 43/18 | 76/86 | 100/100 | 82/90 |
| Bournet et al[39]2 | 104/72 | 71/90 | 100/99 | 84/94 |
1Combination of Kras + cytoP + FISH/Fluorescence in situ hybridization; 2PC vs others malignant and benign pancreatic lesions. PC: Pancreatic carcinoma; CP: Chronic pancreatitis; CytoP: Cytopathology.
Figure 1.
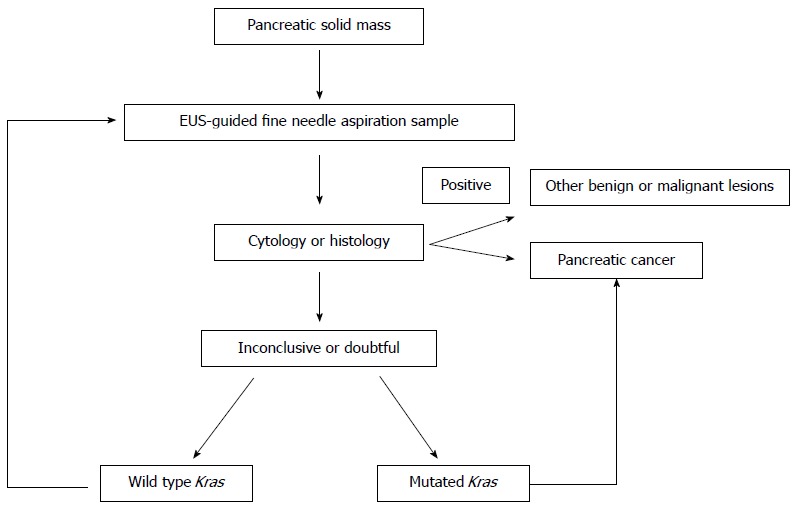
Integration of Kras mutation assay coupled to cytopathology in the algorithm of diagnosis of pancreatic cancer using endoscopic ultrasound-guided fine needle aspiration. EUS: Endoscopic ultrasound.
Several groups, including ours, investigated whether presence or not of Kras mutation can influence prognosis of PDAC, especially in advanced tumors that are only investigated through EUS-FNA. Four recent studies have reported Kras mutations in advanced PDAC, though no correlation was found between Kras mutations and patient survival[40-43]. Conversely, three other published studies (one that included patients with resected PDAC, and two that included mixed populations with resected and locally advanced/metastatic PDAC) suggest that the presence of Kras mutations in tumor tissues have a significant adverse impact on median survival time[44-46]. Therefore, it is still difficult to conclude that the presence of Kras mutations influences or does not influence the prognosis of advanced PDAC. To gain further insights and to obtain a definitive conclusion, investigations on a larger cohort of similar patients (to allow strong multivariate analysis) are needed.
Others molecular alterations
Itoi et al[47] conducted a p53 immunohistochemical analysis in FNA biopsy specimens obtained from CP and pancreatic cancers. They reported that p53 protein overexpression was observed in 67% of the samples with pancreatic cancer, but not in samples with chronic pancreatitis, and they found that by using the combination of p53 protein overexpression and conventional histological examination, the diagnosis of pancreatic cancers improved as follows: 90% sensitivity, 91% specificity, and 92% accuracy, whereas the conventional histological EUS-FNA diagnostic test statistics for the pancreatic masses were as follows: 76% sensitivity, 91% specificity, and 79% accuracy. Jahng et al[48] reported that the combination of p53 and cytology to detect malignancy increased the sensitivity to 51% with 100% specificity, whereas cytology alone had 41% sensitivity and 100% specificity. Salek et al[49] reported also that EUS-guided FNA cytology combines with screening of Kras mutations and allelic losses of tumor suppressor p16 and DPC4 represents a very sensitive approach particularly in cases where FNA has been inconclusive. Another group recently investigated with the same issue the quantitative analysis of MMR genes[50].
MICRORNA
MicroRNA: from basics to clinics?
MicroRNAs are small non coding RNA that functions as translation inhibitors of messenger RNA mainly following binding to 3’-untranslated region[51-53]. This mechanism is conserved from plants to humans. These molecules are tightly involved in the regulation of many physiological processes. Indeed they regulate more than 30% of mammalian gene products. In addition, microRNAs play important roles in many diseases, including cancer, cardiovascular disease, and immune disorders. Besides their high stability in tissues and fluids, microRNAs can repress the expression of dozens or hundreds of genes, making them an attractive therapeutic target.
MicroRNA expression is finely regulated by epigenetic modification (DNA methylation of promoters encoding for microRNA), change in DNA copy numbers, and genetic mutations[54]. For example miR-21 production is increased by Kras (G12D) or EGFR and decreased by TGF-b[55]. For epigenetic regulation Choi et al[56] described in 2012 the DNA methylation of promoter encoding for many microRNAs as a physiological process for mesenchymal stem cell differentiation. As described previously, microRNAs are very stable in tissues and fluids (urine, plasma, saliva). This is a key advantage as compare to protein or mRNA. That is why microRNAs are an emerging class of biomarkers in physiological and pathological processes, including pancreatic diseases.
MicroRNA and pancreatic cancer
microRNA expression is profoundly altered in cancer or is strongly modulated during carcinogenesis. MicroRNAs can be organized in two classes; the oncomicroRNAs which are upregulated in cancer (miR-155, miR-21)[57] and the tumor suppressor microRNAs (let-7 family) which are down regulated in cancer cells[58].
Concerning pancreatic cancer, many articles described that there is an early aberrant microRNA production in pancreatic carcinogenesis and more precisely in the development of precancerous lesions called PanIN. Indeed, the production of miR-21, miR-221, miR-222, and let-7a increased with human PanIN grade, with peak production occurring in hyperplastic PanIN-2/3 lesions[55]. Epigenetic regulation of microRNA is also described to modulate microRNA expression during pancreatic carcinogenesis. For example, miR-148 is down regulated due to an hypermethylation of its DNA[59]. These early disturbances in the expression of microRNA persist then partly in advanced pancreatic cancer stages. In addition, many recent reports describe the alteration of microRNA expression in IPMNs, well-described non-invasive precursor lesions of pancreatic cancer[60]. Such approach may aid in diagnosis and surgical treatment decisions for patients with pancreatic cystic lesions. Taken together, microRNAs could be the ultimate biomarkers at the clinical level for the early diagnosis of pancreatic cancer and would thus allow tumor resection that is usually associated with the best prognosis.
Wang et al[61] were the first to report the detection of microRNA in the blood of pancreatic cancer patients. Interestingly, microRNA profiling in plasma can differentiate pancreatic cancer patients from healthy controls. They demonstrate that the plasma levels of panel of four microRNAs (miR-21, miR-210, miR-196a and miR-155) reveal a sensitivity of 64% and a specificity of 89% for pancreatic cancer. In addition, expression profiles of microRNAs may also be very informative not only to discriminate pancreatic cancer from the normal pancreas, but also for the differential diagnosis of chronic pancreatitis. This shows the interest of microRNAs as diagnostic tool in biological fluids in a non-invasive manner.
MicroRNA have been described as key players in pancreatic cancer development but above in pancreatic cancer cell chemoresistance. The mechanisms involved in drug resistance of cancer cell include alteration of drug target, altered regulation of the cell cycle and apoptosis, increased DNA damage repair and ejection of the drug from the cell by drug efflux pumps. Interestingly, microRNAs can influence the drug response by regulating all of these cellular processes[62]. MiR-21, miR-146, miR205, miR10b, miR-7 and many others microRNAs are implicated in these phenomenon. In this context, microRNAs can serve not only as a valuable therapeutic target but also as a predictive marker for a large number of diseases including pancreatic cancer. The study of microRNA expression in tumors may also lead to the identification of different molecular subtypes of pancreatic cancer that may provide insight into selection of patients likely to benefit from therapies. Nevertheless, whether this will translate into clinical applications is still highly debated.
Some microRNAs are not only predictive and diagnostic markers but also prognostic markers. Indeed, Bloomston et al[63] originally reported that 6 microRNAs (miR-452, miR-105, miR-127, miR-518a-2, miR-187, miR-30a-3p) are over-expressed in the patients with a longer survival (greater than 2 years). In addition, Yu et al[64] reported that patients with high levels of miR-200c expression present with significantly better survival rates than those with low levels of miR-200c expression.
To conclude microRNA expression signature may be informative for the diagnosis, prognosis and predictive of pancreatic cancer[65]. In other words microRNAs appear to be reliable biomarkers to assist clinicians in all stages of care for patients with pancreatic cancer. Thus, microRNAs are expected in the future to prove specific and/or sensitive as a long-awaited screening tool for pancreatic cancer.
MicroRNA detection EUS-FNA
Nowadays, the vast majority of pancreatic cancer patients have metastatic and/or locally advanced diseases at the time of diagnosis; in other words, these patients are not eligible for curative resection which explains the limited access to pancreatic tissue specimens. As explain before, endoscopic ultrasound-guided fine needle aspiration-biopsy is the most widely used approach for cytological and histological material sampling in this situation. Szafranska et al[24] revealed that microRNA and more precisely, miR-196a and miR-217 expression analyses from FNA material can discriminate pancreatic cancer from benign lesion with a high sensitivity (90%) and specificity (100%). These results paved the way to the first development of a molecular test using microRNA for the differential diagnosis of pancreatic cancer[66].
Preis’s[67] group has demonstrated that miR-10b and miR-21, two well-characterized onko microRNAs, are over expressed in the FNA material from pancreatic cancer patients. MiR-10b is now classified as a novel and powerful diagnostic biomarker for pancreatic cancer. In addition, reduced expression of miR-10b is associated with improved response to multimodality neoadjuvant therapy, surgical resection, time to metastasis, and increased survival. Thus, miR-10b may serve as a novel diagnostic and prognostic biomarker in PDAC and as a tool for predicting response to therapy.
Then, we recently demonstrated that let-7a tumor suppressor microRNA expression is repressed[58] in FNA material of pancreatic cancer and that the measurement of hypermethylation of microRNA miT-148a encoding DNA region is potentially useful to differentiate pancreatic cancer and pseudo-tumor forms of chronic pancreatitis[22].
To conclude, microRNAs are very promising emerging molecular markers in pancreatic cancer that can be quantified in EUS-FNA specimens. Table 2 resumes clinical applications of microRNAs in FNA material. However, forthcoming prospective studies are needed to ask whether microRNAs may translate into reliable biomarkers for pancreatic cancer management.
Table 2.
Main studies with expression of miRNA in endoscopic ultrasound-guided fine needle aspiration specimens
| Ref. | miRNA | FNA | Possible clinical implication |
| Torrisani et al[58] | Let-7a | X ↓ | Diagnosis |
| Hanoun et al[22] | miR-148b | X ↓ | Diagnosis |
| Szafranska et al[24] | |||
| Szafranska et al[24] | miR-196a | X ↑ | Diagnosis |
| Szafranska-Schwarzbach et al[66] | |||
| Szafranska et al[24] | miR-217 | X ↓ | Diagnosis |
| Szafranska-Schwarzbach et al[66] | |||
| Du Rieu et al[55] | miR-21 | X ↑ | Diagnosis, prognosis, response to treatment |
| Bloomston et al[63] | |||
| Preis et al[67] | miR-10b | X ↑ | Prognosis, response to treatment |
X ↑: Up regulated; X ↓: Down regulated. FNA: Fine needle aspiration.
MUCINS AND MARKERS OF INVASION
Expression of mucins
Mucine (MUC) are the main components of mucus. They are synthesized and secreted by specialized cells of the epithelium and in some case, by non mucin-secreting cells. MUC are membrane-tethered high molecular weight glycoprotein, and frequently overexpressed in PDAC. Mucins have been implicated in tumorigenicity, invasiveness, metastasis and drug resistance through their characteristic O-linked and N-linked oligosaccharides, extended structures and unique domains. MUC are classified into three categories, membrane associated mucins with MUC1, MUC3 or MUC4, gel-forming mucins with MUC 2 or MUC5AC and soluble mucin with MUC7. MUC are expressed in normal pancreatic tissue, PDAC or precusors lesions as IPMN or PanIN[68]. The MUC expression profile has a hight value for the diagnosis of PDAC (especially MUC1) and several studies implicated these markers in the prognosis and outcome of patient. From samples obtained under EUS FNA, MUC can be detected using immunohistochemistry[69,70]. By this way, Nagata et al[68] investigated the expression of MUC in various pancreatic tissues. MUC1 and MUC6 are expressed in normal pancreatic tissues while MUC 2 and MUC 5AC are never expressed. The expression profile of MUC in IPMN is different between the different subtypes of IPMN. IPMN of intestinal type display a high expression of MUC2 and MUC5 AC while IPMN of pancreaticobiliary type has a low expression of MUC2 and a high expression of MUC 5AC. In PDAC tissues, MUC1 has a high expression but no expression of MUC2. Wang et al[71], after immunohistochemistry on EUS-FNA samples demonstrated the expression of MUC1, MUC2 and MUC5AC in PDAC and in benign pancreatic disease samples. They investigated the diagnosis value of cytology analysis alone vs combination of cytology together with the presence of MUC1 or MUC 5AC. They concluded that the combination of cytology and MUC1+ or MUC5AC+ provide higher sensibility and accuracy for the diagnosis of PDAC (Table 3).
Table 3.
Main studies investigated expression of Mucin and molecular markers for the diagnosis of pancreatic cancer using endoscopic ultrasound-guided fine needle aspiration materials
| Molecular markers | Methods for analysis | Sensitivity (%) | Specificity (%) | Accuracy (%) | Ref. |
| MUC1+/MUC2-/MUC5AC+ | IHC | 70 | 100 | 75 | 70 |
| CytoP + MUC5AC | IHC | 90 | 93 | 91 | 71 |
| CytoP + MUC1 | 85 | 100 | 89 | ||
| MSLN, UPAR | qRT-PCR | 100 | 94 | - | 77 |
| S100P | IHC | 90 | 90 | 87 | 78 |
| MSLN | 62 | 74 | 66 | ||
| S100P + KRT7 | qRT-PCR | 80 | 77 | - | 23 |
| cytoP alone | qRT-PCR | 68/88 | 100/90 | 75/89 | 73 |
| cytoP alone/cytoP + S100A6 |
CytoP: Cytopathology; IHC: Immunohistochemistry; qRT PCR: Quantitative reverse transcription polymerase chain reaction; MUC: Mucin; MSLN: Mesothelin; UPAR: Urokinase plasminogen activator receptor.
Expression of factors implicated in tumor invasion
The identification of others biomarkers has been proposed from samples of pancreatic tissue obtained by EUS-FNA. Indeed the quality and the amount of cellular pancreatic samples obtained by EUS-FNA allow immunohistochemistry thanks to cellblocks but also the extraction of RNA to perform Real Time Reverse Transcription Polymerase Chain Reaction.
We previously performed an expression study using cDNA macro array of pancreatic cancer cells and PDAC tissues. Following this DNA chip study, RT-QPCR validated the increased expression of LCN2 (lipocalin 2) and for the first time PLAT (tissue-type plasminogen activator or tPA) in PDAC as compared with normal pancreas. Following, PLAT and LCN2 transcripts obtained through EUS-guided FNA from patients with PDAC showed a significant increased expression levels in comparison with those found in normal tissues, indicating that a sufficient amount of high quality RNA can be obtained with this technique[72].
Subsequently we conducted a prospective multicenter study using dedicated TaqMan Low Density Array technology on EUS-FNA materials[23]. We quantified candidate gene expression in biopsies sampled from 44 locally advanced and/or metastatic pancreatic carcinoma and from 17 pseudotumoural chronic pancreatitis. We found that eight genes (S100P, PLAT/Plasminogen Activator Tissue, PLAU/PLasmine Activator Urokinase, MSLN/Mesothelin, Matrix MetalloProteins 7 and 11, KRT7 and 17/Keratin) were significantly over expressed in pancreatic cancer samples when compared to pseudotumoural chronic pancreatitis. The area under receiver operating curve establishes the clinical validity of the potential diagnostic markers identified in this study (values ranging from 0.69 to 0.76). In addition, combination of S100P (calcium binding protein P) and KRT7 gave better diagnosis performances (Table 3). We demonstrate that molecular studies on EUS-guided FNA material are feasible for the identification and quantification of markers in PDAC patients diagnosed with non-resectable tumours. Using low-density array, we isolated a molecular signature of advanced pancreatic carcinoma including mostly cancer invasion-related genes[23]. Zihao et al[73] demonstrated the interest of S100A6 for the diagnosis of pancreatic cancer. The material used RNA extracted for quantification of gene expression by RT PCR and S100A6 immunohistochemistry to validate the expression. Deng et al[74] demonstrated also a nuclear or cytoplasmic staining for S100P and it was specific for pancreatic cancer with 100% diagnostic sensibility. Kosarac et al[75] obtained similar results. These different biomarkers as calcium binding proteins such as S100P (that are associated to metastase and poor prognosis) may contributed in positive diagnosis of pancreatic cancer but also in differential diagnostic with benign pancreatic disease[76-78].
MARKERS FOR THE TREATMENT EFFICACY
Gemcitabine is transported into cells predominantly by human equilibrative nucleoside transporters[79]. A deficiency of activity of one of them, hENT1, conferred high-level resistance to the toxicity of gemcitabine[80-83], and patients with PDAC that have detectable hENT1 or high hENT1 gene expression have significantly prolonged survival after gemcitabine chemotherapy when given as adjuvant treatment after resection[84,85]. After transport, Gemcitabine must be sequentially converted into mono- di- or triphosphorylated forms to exert its full cytotoxic activity. In this cascade, the first two steps of phosphorylation are rate-limiting. In mammalian cells, gemcitabine conversion into gemcitabine monophosphate is performed by the deoxycytidine kinase (DCK)[80-81]. Then, the Uridylate monophosphate kinase yields gemcitabine diphosphate[81]. Gemcitabine derivatives exhibit antitumor activity either by interfering with intracellular nucleotide pools, or through direct inhibition of DNA synthesis. Resistance to gemcitabine has been reported to involve a deficiency in DCK enzyme, a decrease in nucleoside transport and an overexpression of ribonucleotide reductase. After cellular entry, gemcitabine must be phosphorylated by deoxycytidine kinase (dCK), which is a rate limiting step. We previously demonstrated that down-regulation of dCK specifically enhanced acquired resistance to gemcitabine in pancreatic cancer cells, whereas transfection of wild-type dCK restored sensitivity to the drug[83]. Conversely, active metabolites of gemcitabine are reduced by 5′-nucleotidase, and gemcitabine itself is inactivated by cytidine deaminase (CDA). High levels of these catabolic enzymes are associated with resistance to the drug. Ribonucleic Reductase (RR) is a dimeric enzyme composed of regulatory subunit M1 and catalytic subunit M2. PDAC patients with high levels of RRM1 expression had poor survival rates after gemcitabine treatment. Moreover, RRM2 gene silencing by RNA interference is an effective therapeutic adjunct to gemcitabine treatment. These data suggest that the genes encoding proteins involved in the transport and metabolism of gemcitabine and in the metabolism of targets can be potential candidates to predict sensitivity to gemcitabine.
Fujita et al[84] investigated 70 patients with PDAC. Of the 70 patients, 40 received gemcitabine-based adjuvant chemotherapy. They measured hENT1, dCK, CDA, RRM1, and RRM2 mRNA levels by quantitative real-time reverse transcription-polymerase chain reaction in endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) cytological specimens. High dCK, low RRM2 groups had a significantly longer disease-free survival in the gemcitabine-treated group[85]. Itoi et al[86] assessed 35 PDAC biopsy specimens for RRM2 expression levels. In the low RRM2 expression group, a complete response was observed in one patient, and a partial response was observed in eight patients. In contrast, in the high RRM2 expression group, complete response was not observed. In the work from Ashida et al[87] mRNAs were extracted from 35 unresectable pancreatic adenocarcinoma tissues obtained by EUS-FNA before GEM-treatment. Among these GEM sensitivity-related genes, dCK alone showed a significant correlation with Gemcitabine efficacy. Among all molecules that are crucial for Gemcitabine intracellular transport and metabolism, hENT1, dCK and RRM2 appear important. If their expression has been studied at the mRNA levels on EUS-FNA, immunohistochemistry on these materials remains to be validated.
CONCLUSION
Both clinician and scientist take benefit from cellular and tissue material sampled under EUS-FNA in PDAC patients. The progress of molecular biology authorizes now extraction of DNA, mRNA and miRNA. After amplification identification of genetic abnormalities and quantification of biomarkers for improvement of diagnosis, prognosis and hopefully treatment together with a better knowledge of pancreatic carcinogenesis especially in locally advanced pancreatic adenocarcinoma. Among these markers, Kras oncogene assay appears now the most robust for improvement of positive and differential diagnosis of pancreatic cancer especially when FNA are inconclusive or doubtful. Clinical implication or miRNA, Mucins and markers of invasion is still debated. Future molecular developments may open windows towards personalized treatments after molecular characterization of a single patient.
Footnotes
P- Reviewer: Kawa S S- Editor: Ma YJ L- Editor: A E- Editor: Liu XM
References
- 1.Hidalgo M, Maitra A. The hedgehog pathway and pancreatic cancer. N Engl J Med. 2009;361:2094–2096. doi: 10.1056/NEJMcibr0905857. [DOI] [PubMed] [Google Scholar]
- 2.Buscail L, Faure P, Bournet B, Selves J, Escourrou J. Interventional endoscopic ultrasound in pancreatic diseases. Pancreatology. 2006;6:7–16. doi: 10.1159/000090022. [DOI] [PubMed] [Google Scholar]
- 3.Savides TJ, Donohue M, Hunt G, Al-Haddad M, Aslanian H, Ben-Menachem T, Chen VK, Coyle W, Deutsch J, DeWitt J, et al. EUS-guided FNA diagnostic yield of malignancy in solid pancreatic masses: a benchmark for quality performance measurement. Gastrointest Endosc. 2007;66:277–282. doi: 10.1016/j.gie.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Al-Haddad M, Eloubeidi MA. Interventional EUS for the diagnosis and treatment of locally advanced pancreatic cancer. JOP. 2010;11:1–7. [PubMed] [Google Scholar]
- 5.Yoshinaga S, Suzuki H, Oda I, Saito Y. Role of endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) for diagnosis of solid pancreatic masses. Dig Endosc. 2011;23 Suppl 1:29–33. doi: 10.1111/j.1443-1661.2011.01112.x. [DOI] [PubMed] [Google Scholar]
- 6.Kato K, Kamada H, Fujimori T, Aritomo Y, Ono M, Masaki T. Molecular Biologic Approach to the Diagnosis of Pancreatic Carcinoma Using Specimens Obtained by EUS-Guided Fine Needle Aspiration. Gastroenterol Res Pract. 2012;2012:243524. doi: 10.1155/2012/243524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddiqui AA, Brown LJ, Hong SK, Draganova-Tacheva RA, Korenblit J, Loren DE, Kowalski TE, Solomides C. Relationship of pancreatic mass size and diagnostic yield of endoscopic ultrasound-guided fine needle aspiration. Dig Dis Sci. 2011;56:3370–3375. doi: 10.1007/s10620-011-1782-z. [DOI] [PubMed] [Google Scholar]
- 8.Bang JY, Hebert-Magee S, Trevino J, Ramesh J, Varadarajulu S. Randomized trial comparing the 22-gauge aspiration and 22-gauge biopsy needles for EUS-guided sampling of solid pancreatic mass lesions. Gastrointest Endosc. 2012;76:321–327. doi: 10.1016/j.gie.2012.03.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janssen J, Schlörer E, Greiner L. EUS elastography of the pancreas: feasibility and pattern description of the normal pancreas, chronic pancreatitis, and focal pancreatic lesions. Gastrointest Endosc. 2007;65:971–978. doi: 10.1016/j.gie.2006.12.057. [DOI] [PubMed] [Google Scholar]
- 10.Kitano M, Sakamoto H, Matsui U, Ito Y, Maekawa K, von Schrenck T, Kudo M. A novel perfusion imaging technique of the pancreas: contrast-enhanced harmonic EUS (with video) Gastrointest Endosc. 2008;67:141–150. doi: 10.1016/j.gie.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 11.Hirche TO, Ignee A, Barreiros AP, Schreiber-Dietrich D, Jungblut S, Ott M, Hirche H, Dietrich CF. Indications and limitations of endoscopic ultrasound elastography for evaluation of focal pancreatic lesions. Endoscopy. 2008;40:910–917. doi: 10.1055/s-2008-1077726. [DOI] [PubMed] [Google Scholar]
- 12.Ryozawa S, Iwano H, Taba K, Sen-yo M, Uekitani T. Genetic diagnosis of pancreatic cancer using specimens obtained by EUS-FNA. Dig Endosc. 2011;23 Suppl 1:43–45. doi: 10.1111/j.1443-1661.2011.01117.x. [DOI] [PubMed] [Google Scholar]
- 13.Khalid A, Nodit L, Zahid M, Bauer K, Brody D, Finkelstein SD, McGrath KM. Endoscopic ultrasound fine needle aspirate DNA analysis to differentiate malignant and benign pancreatic masses. Am J Gastroenterol. 2006;101:2493–2500. doi: 10.1111/j.1572-0241.2006.00740.x. [DOI] [PubMed] [Google Scholar]
- 14.Brais RJ, Davies SE, O’Donovan M, Simpson BW, Cook N, Darbonne WC, Chilcott S, Lolkema MP, Neesse A, Lockley M, et al. Direct histological processing of EUS biopsies enables rapid molecular biomarker analysis for interventional pancreatic cancer trials. Pancreatology. 2012;12:8–15. doi: 10.1016/j.pan.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Varadarajulu S, Hasan MK, Bang JY, Hebert-Magee S, Hawes RH. Endoscopic ultrasound-guided tissue acquisition. Dig Endosc. 2014;26 Suppl 1:62–69. doi: 10.1111/den.12146. [DOI] [PubMed] [Google Scholar]
- 16.Hamada S, Shimosegawa T. Biomarkers of pancreatic cancer. Pancreatology. 2011;11 Suppl 2:14–19. doi: 10.1159/000323479. [DOI] [PubMed] [Google Scholar]
- 17.Mishra G. DNA analysis of cells obtained from endoscopic ultrasound-fine needle aspiration in pancreatic adenocarcinoma: Fool’s Gold, Pandora’s Box, or Holy Grail? Am J Gastroenterol. 2006;101:2501–2503. doi: 10.1111/j.1572-0241.2006.00929.x. [DOI] [PubMed] [Google Scholar]
- 18.Bournet B, Pointreau A, Delpu Y, Selves J, Torrisani J, Buscail L, Cordelier P. Molecular endoscopic ultrasound for diagnosis of pancreatic cancer. Cancers (Basel) 2011;3:872–882. doi: 10.3390/cancers3010872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Laethem JL, Vertongen P, Deviere J, Van Rampelbergh J, Rickaert F, Cremer M, Robberecht P. Detection of c-Ki-ras gene codon 12 mutations from pancreatic duct brushings in the diagnosis of pancreatic tumours. Gut. 1995;36:781–787. doi: 10.1136/gut.36.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berthélemy P, Bouisson M, Escourrou J, Vaysse N, Rumeau JL, Pradayrol L. Identification of K-ras mutations in pancreatic juice in the early diagnosis of pancreatic cancer. Ann Intern Med. 1995;123:188–191. doi: 10.7326/0003-4819-123-3-199508010-00005. [DOI] [PubMed] [Google Scholar]
- 21.Costentin L, Pagès P, Bouisson M, Berthelémy P, Buscail L, Escourrou J, Pradayrol L, Vaysse N. Frequent deletions of tumor suppressor genes in pure pancreatic juice from patients with tumoral or nontumoral pancreatic diseases. Pancreatology. 2002;2:17–25. doi: 10.1159/000049443. [DOI] [PubMed] [Google Scholar]
- 22.Hanoun N, Delpu Y, Suriawinata AA, Bournet B, Bureau C, Selves J, Tsongalis GJ, Dufresne M, Buscail L, Cordelier P, Torrisani J. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010;56:1107–1118. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- 23.Bournet B, Pointreau A, Souque A, Oumouhou N, Muscari F, Lepage B, Senesse P, Barthet M, Lesavre N, Hammel P, et al. Gene expression signature of advanced pancreatic ductal adenocarcinoma using low density array on endoscopic ultrasound-guided fine needle aspiration samples. Pancreatology. 2012;12:27–34. doi: 10.1016/j.pan.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Szafranska AE, Doleshal M, Edmunds HS, Gordon S, Luttges J, Munding JB, Barth RJ, Gutmann EJ, Suriawinata AA, Marc Pipas J, et al. Analysis of microRNAs in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clin Chem. 2008;54:1716–1724. doi: 10.1373/clinchem.2008.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn SA, Schmiegel WH. Recent discoveries in cancer genetics of exocrine pancreatic neoplasia. Digestion. 1998;59:493–501. doi: 10.1159/000007526. [DOI] [PubMed] [Google Scholar]
- 26.Koorstra JB, Hustinx SR, Offerhaus GJ, Maitra A. Pancreatic carcinogenesis. Pancreatology. 2008;8:110–125. doi: 10.1159/000123838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Gao J, Ren Y, Gu J, Du Y, Chen J, Jin Z, Zhan X, Li Z, Huang H, et al. Detection of KRAS gene mutations in endoscopic ultrasound-guided fine-needle aspiration biopsy for improving pancreatic cancer diagnosis. Am J Gastroenterol. 2011;106:2104–2111. doi: 10.1038/ajg.2011.281. [DOI] [PubMed] [Google Scholar]
- 28.Bournet B, Souque A, Senesse P, Assenat E, Barthet M, Lesavre N, Aubert A, O’Toole D, Hammel P, Levy P, et al. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with KRAS mutation assay to distinguish pancreatic cancer from pseudotumoral chronic pancreatitis. Endoscopy. 2009;41:552–557. doi: 10.1055/s-0029-1214717. [DOI] [PubMed] [Google Scholar]
- 29.Ogura T, Yamao K, Sawaki A, Mizuno N, Hara K, Hijioka S, Niwa Y, Tajika M, Kondo S, Shimizu Y, et al. Clinical impact of K-ras mutation analysis in EUS-guided FNA specimens from pancreatic masses. Gastrointest Endosc. 2012;75:769–774. doi: 10.1016/j.gie.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Reicher S, Boyar FZ, Albitar M, Sulcova V, Agersborg S, Nga V, Zhou Y, Li G, Venegas R, French SW, et al. Fluorescence in situ hybridization and K-ras analyses improve diagnostic yield of endoscopic ultrasound-guided fine-needle aspiration of solid pancreatic masses. Pancreas. 2011;40:1057–1062. doi: 10.1097/MPA.0b013e3182200201. [DOI] [PubMed] [Google Scholar]
- 31.Tada M, Komatsu Y, Kawabe T, Sasahira N, Isayama H, Toda N, Shiratori Y, Omata M. Quantitative analysis of K-ras gene mutation in pancreatic tissue obtained by endoscopic ultrasonography-guided fine needle aspiration: clinical utility for diagnosis of pancreatic tumor. Am J Gastroenterol. 2002;97:2263–2270. doi: 10.1111/j.1572-0241.2002.05980.x. [DOI] [PubMed] [Google Scholar]
- 32.Ginestà MM, Mora J, Mayor R, Farré A, Peinado MA, Busquets J, Serrano T, Capellá G, Fabregat J. Genetic and epigenetic markers in the evaluation of pancreatic masses. J Clin Pathol. 2013;66:192–197. doi: 10.1136/jclinpath-2012-201123. [DOI] [PubMed] [Google Scholar]
- 33.Maluf-Filho F, Kumar A, Gerhardt R, Kubrusly M, Sakai P, Hondo F, Matuguma SE, Artifon E, Monteiro da Cunha JE, César Machado MC, et al. Kras mutation analysis of fine needle aspirate under EUS guidance facilitates risk stratification of patients with pancreatic mass. J Clin Gastroenterol. 2007;41:906–910. doi: 10.1097/MCG.0b013e31805905e9. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi K, Yamao K, Okubo K, Sawaki A, Mizuno N, Ashida R, Koshikawa T, Ueyama Y, Kasugai K, Hase S, et al. Differential diagnosis of pancreatic cancer and focal pancreatitis by using EUS-guided FNA. Gastrointest Endosc. 2005;61:76–79. doi: 10.1016/s0016-5107(04)02224-2. [DOI] [PubMed] [Google Scholar]
- 35.Pellisé M, Castells A, Ginès A, Solé M, Mora J, Castellví-Bel S, Rodríguez-Moranta F, Fernàndez-Esparrach G, Llach J, Bordas JM, et al. Clinical usefulness of KRAS mutational analysis in the diagnosis of pancreatic adenocarcinoma by means of endosonography-guided fine-needle aspiration biopsy. Aliment Pharmacol Ther. 2003;17:1299–1307. doi: 10.1046/j.1365-2036.2003.01579.x. [DOI] [PubMed] [Google Scholar]
- 36.Khalid A, Dewitt J, Ohori NP, Chen JH, Fasanella KE, Sanders M, McGrath KM, Nikiforova M. EUS-FNA mutational analysis in differentiating autoimmune pancreatitis and pancreatic cancer. Pancreatology. 2011;11:482–486. doi: 10.1159/000331505. [DOI] [PubMed] [Google Scholar]
- 37.Ogura T, Yamao K, Hara K, Mizuno N, Hijioka S, Imaoka H, Sawaki A, Niwa Y, Tajika M, Kondo S, et al. Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J Gastroenterol. 2013;48:640–646. doi: 10.1007/s00535-012-0664-2. [DOI] [PubMed] [Google Scholar]
- 38.Didelot A, Le Corre D, Luscan A, Cazes A, Pallier K, Emile JF, Laurent-Puig P, Blons H. Competitive allele specific TaqMan PCR for KRAS, BRAF and EGFR mutation detection in clinical formalin fixed paraffin embedded samples. Exp Mol Pathol. 2012;92:275–280. doi: 10.1016/j.yexmp.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Bournet B, Selves J, Grand D, Danjoux M, Hanoun N, Cordelier P, Buscail L. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with a KRAS mutation assay using allelic discrimination improves the diagnosis of pancreatic cancer. J Clin Gastroenterol. 2014:In press. doi: 10.1097/MCG.0000000000000053. [DOI] [PubMed] [Google Scholar]
- 40.Oliveira-Cunha M, Hadfield KD, Siriwardena AK, Newman W. EGFR and KRAS mutational analysis and their correlation to survival in pancreatic and periampullary cancer. Pancreas. 2012;41:428–434. doi: 10.1097/MPA.0b013e3182327a03. [DOI] [PubMed] [Google Scholar]
- 41.da Cunha Santos G, Dhani N, Tu D, Chin K, Ludkovski O, Kamel-Reid S, Squire J, Parulekar W, Moore MJ, Tsao MS. Molecular predictors of outcome in a phase 3 study of gemcitabine and erlotinib therapy in patients with advanced pancreatic cancer: National Cancer Institute of Canada Clinical Trials Group Study PA.3. Cancer. 2010;116:5599–5607. doi: 10.1002/cncr.25393. [DOI] [PubMed] [Google Scholar]
- 42.Kullmann F, Hartmann A, Stöhr R, Messmann H, Dollinger MM, Trojan J, Fuchs M, Hollerbach S, Harder J, Troppmann M, et al. KRAS mutation in metastatic pancreatic ductal adenocarcinoma: results of a multicenter phase II study evaluating efficacy of cetuximab plus gemcitabine/oxaliplatin (GEMOXCET) in first-line therapy. Oncology. 2011;81:3–8. doi: 10.1159/000330194. [DOI] [PubMed] [Google Scholar]
- 43.Bournet B, Muscari F, Guimbaud R, Cordelier P, Buscail L. KRAS mutations and their correlation with survival of patients with advanced pancreatic cancer. Pancreas. 2013;42:543–544. doi: 10.1097/MPA.0b013e31826b388b. [DOI] [PubMed] [Google Scholar]
- 44.Kim ST, Lim do H, Jang KT, Lim T, Lee J, Choi YL, Jang HL, Yi JH, Baek KK, Park SH, et al. Impact of KRAS mutations on clinical outcomes in pancreatic cancer patients treated with first-line gemcitabine-based chemotherapy. Mol Cancer Ther. 2011;10:1993–1999. doi: 10.1158/1535-7163.MCT-11-0269. [DOI] [PubMed] [Google Scholar]
- 45.Franko J, Krasinskas AM, Nikiforova MN, Zarnescu NO, Lee KK, Hughes SJ, Bartlett DL, Zeh HJ, Moser AJ. Loss of heterozygosity predicts poor survival after resection of pancreatic adenocarcinoma. J Gastrointest Surg. 2008;12:1664–1672; discussion 1672-1673. doi: 10.1007/s11605-008-0577-9. [DOI] [PubMed] [Google Scholar]
- 46.Lee J, Jang KT, Ki CS, Lim T, Park YS, Lim HY, Choi DW, Kang WK, Park K, Park JO. Impact of epidermal growth factor receptor (EGFR) kinase mutations, EGFR gene amplifications, and KRAS mutations on survival of pancreatic adenocarcinoma. Cancer. 2007;109:1561–1569. doi: 10.1002/cncr.22559. [DOI] [PubMed] [Google Scholar]
- 47.Itoi T, Takei K, Sofuni A, Itokawa F, Tsuchiya T, Kurihara T, Nakamura K, Moriyasu F, Tsuchida A, Kasuya K. Immunohistochemical analysis of p53 and MIB-1 in tissue specimens obtained from endoscopic ultrasonography-guided fine needle aspiration biopsy for the diagnosis of solid pancreatic masses. Oncol Rep. 2005;13:229–234. [PubMed] [Google Scholar]
- 48.Jahng AW, Reicher S, Chung D, Varela D, Chhablani R, Dev A, Pham B, Nieto J, Venegas RJ, French SW, et al. Staining for p53 and Ki-67 increases the sensitivity of EUS-FNA to detect pancreatic malignancy. World J Gastrointest Endosc. 2010;2:362–368. doi: 10.4253/wjge.v2.i11.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salek C, Benesova L, Zavoral M, Nosek V, Kasperova L, Ryska M, Strnad R, Traboulsi E, Minarik M. Evaluation of clinical relevance of examining K-ras, p16 and p53 mutations along with allelic losses at 9p and 18q in EUS-guided fine needle aspiration samples of patients with chronic pancreatitis and pancreatic cancer. World J Gastroenterol. 2007;13:3714–3720. doi: 10.3748/wjg.v13.i27.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gheonea DI, Ciurea ME, Săftoiu A, Ioana M. Quantitative RT-PCR analysis of MMR genes on EUS-guided FNA samples from focal pancreatic lesions. Hepatogastroenterology. 2012;59:916–920. doi: 10.5754/hge11463. [DOI] [PubMed] [Google Scholar]
- 51.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 52.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 53.Redis RS, Berindan-Neagoe I, Pop VI, Calin GA. Non-coding RNAs as theranostics in human cancers. J Cell Biochem. 2012;113:1451–1459. doi: 10.1002/jcb.24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M, Marin-Muller C, Bharadwaj U, Chow KH, Yao Q, Chen C. MicroRNAs: control and loss of control in human physiology and disease. World J Surg. 2009;33:667–684. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, Tsongalis GJ, Suriawinata AA, Carrère N, Buscail L, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56:603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 56.Choi MR, In YH, Park J, Park T, Jung KH, Chai JC, Chung MK, Lee YS, Chai YG. Genome-scale DNA methylation pattern profiling of human bone marrow mesenchymal stem cells in long-term culture. Exp Mol Med. 2012;44:503–512. doi: 10.3858/emm.2012.44.8.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torrisani J, Bournet B, du Rieu MC, Bouisson M, Souque A, Escourrou J, Buscail L, Cordelier P. let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther. 2009;20:831–844. doi: 10.1089/hum.2008.134. [DOI] [PubMed] [Google Scholar]
- 59.Delpu Y, Lulka H, Sicard F, Saint-Laurent N, Lopez F, Hanoun N, Buscail L, Cordelier P, Torrisani J. The rescue of miR-148a expression in pancreatic cancer: an inappropriate therapeutic tool. PLoS One. 2013;8:e55513. doi: 10.1371/journal.pone.0055513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–346. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui SY, Wang R, Chen LB. MicroRNAs: key players of taxane resistance and their therapeutic potential in human cancers. J Cell Mol Med. 2013;17:1207–1217. doi: 10.1111/jcmm.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 64.Yu J, Ohuchida K, Mizumoto K, Sato N, Kayashima T, Fujita H, Nakata K, Tanaka M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol Cancer. 2010;9:169. doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steele CW, Oien KA, McKay CJ, Jamieson NB. Clinical potential of microRNAs in pancreatic ductal adenocarcinoma. Pancreas. 2011;40:1165–1171. doi: 10.1097/MPA.0b013e3182218ffb. [DOI] [PubMed] [Google Scholar]
- 66.Szafranska-Schwarzbach AE, Adai AT, Lee LS, Conwell DL, Andruss BF. Development of a miRNA-based diagnostic assay for pancreatic ductal adenocarcinoma. Expert Rev Mol Diagn. 2011;11:249–257. doi: 10.1586/erm.11.10. [DOI] [PubMed] [Google Scholar]
- 67.Preis M, Gardner TB, Gordon SR, Pipas JM, Mackenzie TA, Klein EE, Longnecker DS, Gutmann EJ, Sempere LF, Korc M. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2011;17:5812–5821. doi: 10.1158/1078-0432.CCR-11-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagata K, Horinouchi M, Saitou M, Higashi M, Nomoto M, Goto M, Yonezawa S. Mucin expression profile in pancreatic cancer and the precursor lesions. J Hepatobiliary Pancreat Surg. 2007;14:243–254. doi: 10.1007/s00534-006-1169-2. [DOI] [PubMed] [Google Scholar]
- 69.Carrara S, Cangi MG, Arcidiacono PG, Perri F, Petrone MC, Mezzi G, Boemo C, Talarico A, Cin ED, Grassini G, et al. Mucin expression pattern in pancreatic diseases: findings from EUS-guided fine-needle aspiration biopsies. Am J Gastroenterol. 2011;106:1359–1363. doi: 10.1038/ajg.2011.22. [DOI] [PubMed] [Google Scholar]
- 70.Giorgadze TA, Peterman H, Baloch ZW, Furth EE, Pasha T, Shiina N, Zhang PJ, Gupta PK. Diagnostic utility of mucin profile in fine-needle aspiration specimens of the pancreas: an immunohistochemical study with surgical pathology correlation. Cancer. 2006;108:186–197. doi: 10.1002/cncr.21913. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Gao J, Li Z, Jin Z, Gong Y, Man X. Diagnostic value of mucins (MUC1, MUC2 and MUC5AC) expression profile in endoscopic ultrasound-guided fine-needle aspiration specimens of the pancreas. Int J Cancer. 2007;121:2716–2722. doi: 10.1002/ijc.22997. [DOI] [PubMed] [Google Scholar]
- 72.Laurell H, Bouisson M, Berthelemy P, Rochaix P, Dejean S, Besse P, Susini C, Pradayrol L, Vaysse N, Buscail L. Identification of biomarkers of human pancreatic adenocarcinomas by expression profiling and validation with gene expression analysis in endoscopic ultrasound-guided fine needle aspiration samples. World J Gastroenterol. 2006;12:3344–3351. doi: 10.3748/wjg.v12.i21.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zihao G, Jie Z, Yan L, Jing Z, Jing C, Xue L, Jing Z, Heng LW, Ru G, Jianyu H. Analyzing S100A6 expression in endoscopic ultrasonography-guided fine-needle aspiration specimens: a promising diagnostic method of pancreatic cancer. J Clin Gastroenterol. 2013;47:69–75. doi: 10.1097/MCG.0b013e3182601752. [DOI] [PubMed] [Google Scholar]
- 74.Deng H, Shi J, Wilkerson M, Meschter S, Dupree W, Lin F. Usefulness of S100P in diagnosis of adenocarcinoma of pancreas on fine-needle aspiration biopsy specimens. Am J Clin Pathol. 2008;129:81–88. doi: 10.1309/5D76NDE81LE8G545. [DOI] [PubMed] [Google Scholar]
- 75.Kosarac O, Takei H, Zhai QJ, Schwartz MR, Mody DR. S100P and XIAP expression in pancreatic ductal adenocarcinoma: potential novel biomarkers as a diagnostic adjunct to fine needle aspiration cytology. Acta Cytol. 2011;55:142–148. doi: 10.1159/000320913. [DOI] [PubMed] [Google Scholar]
- 76.Ohuchida K, Mizumoto K, Ishikawa N, Fujii K, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. The role of S100A6 in pancreatic cancer development and its clinical implication as a diagnostic marker and therapeutic target. Clin Cancer Res. 2005;11:7785–7793. doi: 10.1158/1078-0432.CCR-05-0714. [DOI] [PubMed] [Google Scholar]
- 77.Chen Y, Zheng B, Robbins DH, Lewin DN, Mikhitarian K, Graham A, Rumpp L, Glenn T, Gillanders WE, Cole DJ, et al. Accurate discrimination of pancreatic ductal adenocarcinoma and chronic pancreatitis using multimarker expression data and samples obtained by minimally invasive fine needle aspiration. Int J Cancer. 2007;120:1511–1517. doi: 10.1002/ijc.22487. [DOI] [PubMed] [Google Scholar]
- 78.Dim DC, Jiang F, Qiu Q, Li T, Darwin P, Rodgers WH, Peng HQ. The usefulness of S100P, mesothelin, fascin, prostate stem cell antigen, and 14-3-3 sigma in diagnosing pancreatic adenocarcinoma in cytological specimens obtained by endoscopic ultrasound guided fine-needle aspiration. Diagn Cytopathol. 2014;42:193–199. doi: 10.1002/dc.21684. [DOI] [PubMed] [Google Scholar]
- 79.Rauchwerger DR, Firby PS, Hedley DW, Moore MJ. Equilibrative-sensitive nucleoside transporter and its role in gemcitabine sensitivity. Cancer Res. 2000;60:6075–6079. [PubMed] [Google Scholar]
- 80.Hapke DM, Stegmann AP, Mitchell BS. Retroviral transfer of deoxycytidine kinase into tumor cell lines enhances nucleoside toxicity. Cancer Res. 1996;56:2343–2347. [PubMed] [Google Scholar]
- 81.Vernejoul F, Ghénassia L, Souque A, Lulka H, Drocourt D, Cordelier P, Pradayrol L, Pyronnet S, Buscail L, Tiraby G. Gene therapy based on gemcitabine chemosensitization suppresses pancreatic tumor growth. Mol Ther. 2006;14:758–767. doi: 10.1016/j.ymthe.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 82.Maréchal R, Bachet JB, Mackey JR, Dalban C, Demetter P, Graham K, Couvelard A, Svrcek M, Bardier-Dupas A, Hammel P, et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology. 2012;143:664–674.e1-6. doi: 10.1053/j.gastro.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 83.Giovannetti E, Del Tacca M, Mey V, Funel N, Nannizzi S, Ricci S, Orlandini C, Boggi U, Campani D, Del Chiaro M, et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66:3928–3935. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- 84.Fujita H, Ohuchida K, Mizumoto K, Itaba S, Ito T, Nakata K, Yu J, Kayashima T, Souzaki R, Tajiri T, et al. Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia. 2010;12:807–817. doi: 10.1593/neo.10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanaka M, Javle M, Dong X, Eng C, Abbruzzese JL, Li D. Gemcitabine metabolic and transporter gene polymorphisms are associated with drug toxicity and efficacy in patients with locally advanced pancreatic cancer. Cancer. 2010;116:5325–5335. doi: 10.1002/cncr.25282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Itoi T, Sofuni A, Fukushima N, Itokawa F, Tsuchiya T, Kurihara T, Moriyasu F, Tsuchida A, Kasuya K. Ribonucleotide reductase subunit M2 mRNA expression in pretreatment biopsies obtained from unresectable pancreatic carcinomas. J Gastroenterol. 2007;42:389–394. doi: 10.1007/s00535-007-2017-0. [DOI] [PubMed] [Google Scholar]
- 87.Ashida R, Nakata B, Shigekawa M, Mizuno N, Sawaki A, Hirakawa K, Arakawa T, Yamao K. Gemcitabine sensitivity-related mRNA expression in endoscopic ultrasound-guided fine-needle aspiration biopsy of unresectable pancreatic cancer. J Exp Clin Cancer Res. 2009;28:83. doi: 10.1186/1756-9966-28-83. [DOI] [PMC free article] [PubMed] [Google Scholar]


