Our understanding of genome biology, genomics, and disease, and even human history, has advanced tremendously with the completion of the Human Genome Project. Technological advances coupled with significant cost reductions in genomic research have yielded novel insights into disease etiology, diagnosis, and therapy for some of the world’s most intractable and devastating diseases—including malaria, HIV/AIDS, tuberculosis, cancer, and diabetes. Yet, despite the burden of infectious diseases and, more recently, noncommunicable diseases (NCDs) in Africa, Africans have only participated minimally in genomics research. Of the thousands of genome-wide association studies (GWASs) that have been conducted globally, only seven (for HIV susceptibility, malaria, tuberculosis, and podoconiosis) have been conducted exclusively on African participants; four others (for prostate cancer, obsessive compulsive disorder, and anthropometry) included some African participants (www.genome.gov/gwastudies/). As discussed in 2011 (www.h3africa.org), if the dearth of genomics research involving Africans persists, the potential health and economic benefits emanating from genomic science may elude an entire continent.
The lack of large-scale genomics studies in Africa is the result of many deep-seated issues, including a shortage of African scientists with genomic research expertise, lack of biomedical research infrastructure, limited computational expertise and resources, lack of adequate support for biomedical research by African governments, and the participation of many African scientists in collaborative research at no more than the level of sample collection. Overcoming these limitations will, in part, depend on African scientists acquiring the expertise and facilities necessary to lead high-quality genomics research aimed at understanding health problems relevant to African populations and to become internationally competitive in genomic science and its applications.
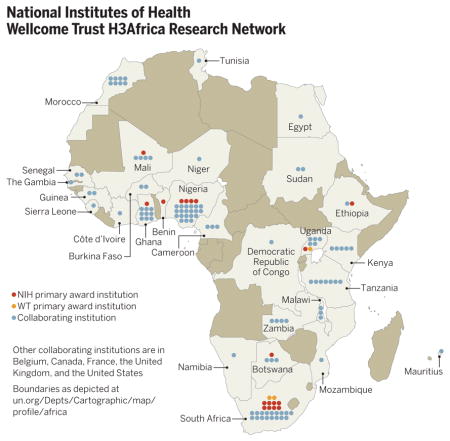
In June 2010, the U.S. National Institutes of Health (NIH) and UK-based Wellcome Trust, in partnership with the African Society of Human Genetics, announced a plan to enhance the ability of African scientists to apply genomic and epidemiological approaches to shed light on the determinants of chronic and infectious diseases in Africa (1). The Human Heredity and Health in Africa (H3Africa) initiative, now funded at $76 million over 5 years, is focused on capacity building, as well as specific scientific goals. H3Africa research grants are awarded directly to African institutions where principal investigators are based (table S1), which allows African scientists to develop and direct their independent research agendas. The program encourages formation of intra-continental collaborations and development of specific infrastructural elements, i.e., African-based biorepositories and a pan-African bioinformatics network (H3ABio-Net). H3Africa also includes training programs aimed at retaining African scientists on the continent to help build a sustainable critical mass of researchers. Open calls for research proposals have emphasized collaborations within Africa, plus accessible biorepositories and a bioinformatics network with nodes across the continent (table S2). The footprint of H3Africa extends across Africa (see the map), comprising 21 grants (table S1). It is anticipated that, together, H3Africa projects will analyze samples from 50,000 to 75,000 participants.
H3Africa is predicated on the belief that diseases and nonmedical issues relevant to Africans can be best explored in partnership with inhabitants of Africa (both researchers and research participants) who can provide a rich context and deep knowledge of the continent’s past and present environment. African genomes and the unique genetic structure of African populations harbor many clues to understanding human evolutionary history which, in turn, can help shed light on disease etiology. For example, recent genomic studies showed that African Americans (AA) with chronic kidney disease (CKD) who harbor risk variants of the apolipoprotein L1 gene (APOL1) have a risk for accelerated CKD progression and the development of end-stage renal disease, that is two to five times normal, respectively (2, 3). These variants also confer 29 times the risk of HIV-associated nephropathy (HIVAN) (4). Despite these renal outcomes, the prevalence of the risk genotype is 13% among AA and virtually absent among those of non-African ancestry. The prevailing hypothesis is that APOL1 renal risk variants evolved in sub-Saharan Africa about 10,000 years ago to confer protection against the regionally endemic trypanosome parasite, the cause of African sleeping sickness. Recent studies led by African scientists showed that the frequency of the risk variants, as well as the prevalence of CKD and HIVAN in carriers of the risk variant, are much higher in West Africa (Yoruba, 28%; Igbo, 23%; the major ancestral populations of AA) where the trypanosome parasite is endemic as compared with the non-endemic region of Ethiopia (~1%) (5–7). The association between CKD and APOL1 [a component of high-density lipoprotein (HDL) cholesterol] is shedding light on the complicated protective relation between HDL cholesterol and CKD in global populations (8). In another example, African scientists participating in H3Africa have used genomic tools to understand how genes interact with life style (barefoot farming practices) to increase susceptibility to podoconiosis, a neglected tropical disease in Ethiopia and Cameroon (9).
A key challenge to building critical mass for genomic research in Africa is the retention of scientific leadership capable of developing and maintaining sustainable research programs. The dearth of research-intensive institutions on the continent, coupled with a shortage of funded positions, continues to drive Africa’s talented scientists to countries where they have better opportunities to develop their potential and pursue their interests. Furthermore, the African continent lacks a strong history of collaborative scientific endeavor (10), as African researchers have turned to their well-resourced counterparts from Europe, North America, and Asia, rather than to their neighbors, to achieve scientific excellence and strong publication records. Consequently, African scientists have not adequately developed the necessary infrastructure and large-scale biomedical research culture required to promote research in Africa. H3Africa has begun building a strong foundation for genomic research based on collaboration among African scientists. Perhaps more important, H3Africa is facilitating the implementation of the norms and standards for project oversight, goal orientation, and timely dissemination of discoveries and training of the next generation of biomedical researchers across Africa. The consortium is also addressing the use of standardized protocols with detailed attention to community engagement and ethics approval (see below), protocols and policies for sharing biospecimens and data, and publication policies for large collaborative groups.
Approaches to these issues are facilitated by frequent interactions among consortium members to share experiences in developing genomic research programs, to support and promote interactions among the collaborative projects, and to jointly tackle ethical and policy concerns. An important example is data harmonization. By standardizing phenotype measurement and how collected responses are coded to facilitate data merging, statistical power for discovery of genetic variants and for modeling gene-by-environment interactions can be greatly increased.
Implementation of multinational and multiinstitutional genomics research projects in Africa faces additional challenges. Many local ethics review committees have little experience in genomic studies that require broad consent for long-term storage and sharing of biospecimens and data, and some have balked at the concept of global sharing of biospecimens and data with no immediate local benefit, viewing it as another form of exploitation. Several African countries have restrictive legislative policies that hamper sharing across national boundaries. Cultural beliefs and practices regarding donating any body part, including blood, need to be addressed. The growing international debate about return of individual genomic results is also an issue in Africa (11). Finally, there are huge disparities across Africa that span rural communities adhering to long-established cultural beliefs and practices on the one hand to sophisticated “citizens of the world” residing in major cities on the other. These communities share genetic heritage, but require different approaches to engagement and informed consent. Thus, H3Africa includes a grant program that supports empirical research on innovative approaches to informed consent; community engagement; and the ethical, legal, social, and cultural factors unique to the African research environment.
The H3Africa Consortium has developed an approach that attempts to balance (i) protection of the ability of African scientists to be the first to analyze and publish findings about their main research questions, given their limited resources and capacity to deal with data as quickly as scientists in developed countries with (ii) the benefit of global access to H3Africa data and biospecimens. To reach these not completely compatible ends, the H3Africa Consortium has agreed that data will be made initially available to the consortium members via H3ABioNet until submission to the European Genome-phenome Archive, from which they will be publicly accessible (through an independent Data and Biospecimen Access Committee). As is common in genomics, there will be a short lag (12 months) between data submission and publication; this is somewhat longer than the norm (6 to 9 months) to provide resource-challenged African investigators a bit more time to analyze and submit their manuscripts for peer review.
Similar considerations went into development of the policy for the release of biospecimens collected in H3Africa. The biospecimens will be stored in an African biorepository (with backup elsewhere on the continent), and from there shared globally for further research. Data and biospecimen sharing does, however, raise the often contentious issues of ownership and commercialization rights. The H3Africa Consortium is addressing this issue while embracing an ethos that promotes research for the global common good. Resources generated by H3Africa are expected to be useful in future genomic research not only in Africa but also globally.
H3ABioNet has also embarked on a program of training and accreditation of its bio-informatics nodes to carry out specific data analysis techniques, i.e., of GWAS or next-generation sequencing data. Part of the training involves a series of workshops, often held at the nodes, to prepare for an accreditation exercise. The accreditation involves giving the nodes raw data sets to analyze, with their results being assessed by an international accreditation committee. One of the major challenges in holding training courses or even just joining working-group Skype calls, is poor Internet connectivity. H3ABioNet is using creative approaches to overcome these issues by seeking low latency alternatives and using portable devices that host data and tools and run independently of the network.
There are several criteria for success that have been defined to assess the accomplishments of the H3Africa initiative (see the table). Each of the component grants has a set of specific, yearly milestones, progress toward which is assessed on an annual basis by the funders (with input from an Independent Experts Committee of outside scientists. Both Wellcome Trust and the NIH will also critically evaluate the progress of H3Africa through peer review toward the end of the initial funding period. Accomplishments of both individual grants and the overall program will be considered in each funder decision process to determine whether continued support is justified.
The efforts of the African scientific community and their international colleagues will not in themselves be sufficient. It is essential that national governments and regional political and economic organizations support sustained funding of all research fields, including genomics and research infrastructure development. In fact, H3Africa has been useful in leveraging additional funding from local sources, as demonstrated by support from the South African Department of Science and Technology to enhance data collection in an H3Africa project of cardiometabolic disease genomics, an early promise of potential long-term success.
Supplementary Material
Measures of success for the 5-year H3Africa program.
Publication in high-impact journals with African lead and senior authors
Increased availability of funding for biomedical and genomics research in Africa
Effective operation of a pan-African bioinformatics network
Regular data release
Establishment of one or more full-scale biorepositories
Effective release of samples within and outside of the African continent
Contribution to the ongoing efforts to reverse African “brain drain”
Extension of funding for a second 5 years
Footnotes
References
- 1.Nature. 2010;465:990. [Google Scholar]
- 2.Genovese G, et al. Science. 2010;329:841. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsa A, et al. N Engl J Med. 2013;369:2183. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopp JB, et al. J Am Soc Nephrol. 2011;22:2129. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behar DM, et al. Am J Nephrol. 2011;34:452. doi: 10.1159/000332378. [DOI] [PubMed] [Google Scholar]
- 6.Ulasi II, et al. Nephron Clin Pract. 2013;123:123. doi: 10.1159/000353223. [DOI] [PubMed] [Google Scholar]
- 7.Tayo BO, et al. Int Urol Nephrol. 2013;45:485. doi: 10.1007/s11255-012-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley AR, et al. Int J Nephrol. 2012;2012:748984. doi: 10.1155/2012/748984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tekola Ayele F, et al. N Engl J Med. 2012;366:1200. doi: 10.1056/NEJMoa1108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vré R, et al. McKinsey Quarterly Report. McKinsey & Co; Washington, DC: Jun, 2010. Closing the R&D gap in African health care. [Google Scholar]
- 11.Rotimi CN, Marshall PA. Genome Med. 2010;2:20. doi: 10.1186/gm141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


