Abstract
The scavenger receptor family comprises transmembrane proteins involved in the recognition of polyanionic ligands. Several studies have established that members of this family are involved both in immunity and in developmental processes. In Drosophila melanogaster, one of the best characterized scavenger receptors is Croquemort, which participates in the recognition of apoptotic cells in the embryo. Although comparative genomic studies have revealed the presence of four orthologs of this receptor in the malaria vector Anopheles gambiae, little is known about their function. We have investigated the expression pattern of the four Croquemort orthologs during the mosquito life cycle. Croquemort transcripts SCRBQ2 and SCRBQ4 are expressed at all the developmental stages, while expression of Croquemort transcripts SCRBQ1 and SCRBQ3 is more restricted. We have also investigated the expression of the four Croquemort orthologs in the different organs of the adult female. Croquemort transcript SCRBQ2 is highly expressed in the A. gambiae female midgut. SCRBQ2 midgut gene expression was up-regulated after a non-infected or a Plasmodium berghei-infected blood meal, compared to its expression in midguts of sugar-fed females. Interestingly, knockdown of SCRBQ2 expression by dsRNA injection resulted in a 62.5% inhibition of oocyst formation, suggesting that SCRBQ2 plays a role in Plasmodium–mosquito interactions.
Keywords: Croquemort, Scavenger receptor, Anopheles, CD36, RNAi
1. Introduction
The scavenger receptor (SR) family comprises transmembrane proteins with multiple domains that participate in the recognition of a broad range of polyanionic ligands, including modified or oxidized low-density lipoproteins (oxLDL), bacteria and apoptotic cells (Pluddemann et al., 2006). The SR family includes eight independent classes (A–H) based on their multidomain structure (Murphy et al., 2005; Pluddemann et al., 2007), however, there is not a common domain to all SRs. Several studies have established that members of the SR family are involved both in immunity (Gough and Gordon, 2000; Mukhopadhyay and Gordon, 2004; Peiser et al., 2002) and in developmental processes (Murphy et al., 2005).
In insects, one of the best characterized SRs is Croquemort, a member of the class B of the SR family. This class includes three members: CD36, the scavenger receptor class B type I (SR-BI) and the lysosomal integral membrane protein II (LIMP-II) (Febbraio et al., 2001; Murphy et al., 2005). Croquemort is a CD36-family homolog that was first described in Drosophila (Franc et al., 1996), where it is expressed in the plasma membrane of hemocytes. Croquemort expression is coincident with the first wave of apoptosis in the embryo, functioning as an essential receptor for phagocytosis of apoptotic corpses (Franc et al., 1999). RNAi-mediated silencing of this gene in the Drosophila S2 cell line decreased their ability to internalize Staphylococcus aureus, but not Escherichia coli, indicating that this molecule can also act as a phagocytic receptor for Gram-positive bacteria (Stuart et al., 2005). Recently, a proteomic analysis of a Drosophila cell line infected with Flock House virus indicated that Croquemort expression was up-regulated in response to the viral infection (Go et al., 2006).
There are four orthologs (SCRBQ1–4) of Drosophila’s Croquemort in Anopheles gambiae, which appear to correspond to a specific gene expansion in the mosquito (Christophides et al., 2002). Of these, Croquemort transcript SCRBQ2 (AGAP010133) was first identified among the transcripts (ESTs) expressed by the A. gambiae hemocyte-like 4A3A (Dimopoulos et al., 2000) and 4A3B (Dimopoulos et al., 2002) cell lines. Recently, a microarray assay identified the expression of three Croquemort transcripts in the midguts of fourth-instar A. gambiae larvae. In this study, the Croquemort SCRBQ2 transcript was found to be enriched in the gastric caeca and in the hindgut/Malpighian tubules versus its expression in whole larvae (Neira Oviedo et al., 2008). Changes in Croquemort SCRBQ2 mRNA abundance have also been described in the midguts of female A. gambiae mosquitoes upon Plasmodium berghei ookinete invasion (Dong et al., 2006), but the role, if any, of this molecule during parasite infection remains unknown.
Here we report on the developmental expression profile and organ expression in adult females of the four A. gambiae Croquemort orthologs and on the effect of Croquemort SCRBQ2 gene silencing on the development of P. berghei in the mosquito, as an initial approach to explore Croquemort gene function during parasite infection.
2. Materials and methods
2.1. Mosquito rearing
A. gambiae Keele strain mosquitoes were maintained on a 10% sugar solution at 27 °C and 80% humidity with a 12-h light/dark cycle according to standard rearing procedures (Benedict, 1997). dsRNA-injected, blood-fed and P. berghei-infected mosquitoes were kept at 19 °C under the same rearing conditions.
2.2. Semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR)
RT-PCR assays were used to determine the expression profile of the four Croquemort orthologs along the mosquito developmental cycle and in the different organs of the adult female. Additional RT-PCR assays, provided as Supplementary material, were also used to determine the expression profile of the four Croquemort orthologs in response to different feeding conditions as well as to assess multiple gene-knockdown in RNAi assays. 1–2 μg of DNase-treated total RNA (Turbo DNAse, Ambion, Austin, Texas, USA), isolated from A. gambiae embryos, first to fourth-instar larvae, pupae, and whole sugar-fed females and from midguts, ovaries, and carcass (whole body minus organs) of sugar-fed adult females were reverse-transcribed using SuperScript III (Invitrogen, Carlsbad, California, USA). Croquemort transcript abundance was determined using specific oligonucleotide primers corresponding to an internal region of each Croquemort sequence. All Croquemort sequences were retrieved from the A. gambiae database (http://agambiae.vectorbase.org/index.php), (Lawson et al., 2007). Gene-specific primers were: SCRBQ1-specific primers (accession no. AGAP0 10132): SCRBQ1-F, 5′-ACGAGCGTGGTATGGTATCGT-3′; SCRBQ1-R, 5′-CACTGGATTCAGCGTCGTCA-3′. SCRBQ2-specific primers (accession no. AGAP010133): SCRBQ2-F, 5′-AGTTCCGTACGAAGAGCTACAAAA-3′; SCRBQ2-R, 5′-ACACGCATTACTAACGAAACAAAA-3′. SCRB Q3-specific primers (accession no. AGAP008179): SCRBQ3-F, 5′-TGGACGTTTCGAGGTGCAA-3′; SCRBQ3-R, 5′-CATCTCGAACCGATG CTCCT-3′. SCRBQ4-specific primers (accession no. AGAP003373): SCRBQ4-F, 5′-GTGAAACGCGTCTTCCTGC-3′; SCRBQ4-R, 5′-ACTACCATTGCGATGCCACA-3′. The A. gambiae ribosomal protein (AgS7) (Salazar et al., 1993) amplification product was used as expression control. AgS7-specific primers (accession no. AGAP010592): AgS7F, 5′-TTCGTTGTGAACCCAAATAAAAATC-3′ and AgS7R, 5′-TGCGGCTTCAGATCCGAGTTC-3′. Amplification for all transcripts was performed as follows: 95 °C for 5 min, 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s (30 cycles) with a final extension at 72 °C for 7 min. All PCR products were cloned either in the pGEM-T Easy vector (Promega) or in the TOPO-4 vector (Invitrogen) and sequenced to confirm their identity. The results are representative of two independent biological replicates.
2.3. Real-time quantitative PCR
Real-time PCR was used to determine Croquemort SCRBQ2 transcript abundance in different mosquito organs and in response to different feeding conditions as well as to assess gene-knockdown in RNAi assays.
RNA samples were treated with Turbo DNase (Ambion, Austin, Texas, United States) and reverse-transcribed as above. Real-time quantification was performed using SYBR Green PCR Master Mix (Applied Biosystems) and the 7300 Real-Time PCR System (Applied Biosystems). All real-time PCR reactions were performed in triplicate. Specificity of the reactions was assessed by analysis of melting curves for each data point. Serial dilutions of a standard cDNA sample from whole A. gambiae females (six concentrations, two technical replicates each) were run along with the experimental cDNA to calculate a standard curve using the ABI Prism 7300 Sequence Detection Software (Applied Biosystems). Subsequently, cDNA templates were normalized using the ribosomal protein S7 gene. The AgS7 and SCRBQ2 Croquemort-specific primers used for all assays are the same as described above. The results are representative of at least two independent biological replicates.
2.4. RNAi gene-silencing assays
Gene-silencing assays were used to explore potential Croquemort SCRBQ2 gene function during P. berghei infection. A fragment of the SCRBQ2 transcript was amplified with specific primers flanked at their 5′ end by the T7 promoter sequence. dsRNA was produced from the PCR-amplified gene fragment using the in vitro transcription MEGAscript RNAi kit (Ambion) according to the manufacturer’s instructions. dsGFP (Green Fluorescent Protein) was used as a negative control. SCRBQ2 Croquemort-specific primers were: SCRBQ2-F, 5′-TAATACGACTCACTATAGGGAGGTTTGGCTGGTTTGTTGG-3′, SCRBQ2-R, 5′-TAATACGACTCACTATAGGGGTCCCGTTACAGCAGTTTGG-3′. GFP-specific primers were: GFP-F: 5′-TAATACGACTCACTATAGGGTGTTCCATGGCCAACACTTGTCAC-3′, GFP-R: 5′-TAATACGACTCACTATAGGGTTGGAAAGGGCAGATTGTGTGGAC-3′.
To perform the RNAi-mediated gene-silencing assays, A. gambiae females (4–6 days-old) were cold-anesthetized and inoculated with dsSCRBQ2 or dsGFP (control). About 140 nl of each dsRNA (4 μg/μl) were injected into the thorax of the mosquitoes using a nano-injector (Nanoject II; Drummond Scientific, Broomall, Pennsylvania, United States).
Gene-silencing was verified in the midguts and carcasses 48, 72 and 96 h after dsRNA injection by real-time PCR using the SCRBQ2- and AgS7-specific primers as described above. Given the sequence similarity of the four Croquemort genes, a semi-quantitative RT-PCR assay of the four genes was performed using cDNA from the midguts 72 h after dsGFP or dsSCRBQ2 injection to explore the possibility of multiple knockdowns involving one of the other Croquemort orthologs.
For Plasmodium infection assays, 3–4 days after dsRNA injection, GFP-injected and SCRBQ2-injected mosquitoes were fed on a single mouse infected with the P. berghei ANKA 2.34 strain per biological replicate. The midguts of blood-fed mosquitoes were dissected on ice 12–15 days after ingestion of infected blood. Only midguts from mosquitoes that ingested blood, as indicated by egg development, were analyzed. Mosquito midguts were stained with 0.2% mercurochrome and oocyst numbers per mosquito midgut were determined using a light microscope (Nikon). A total of 10 biological replicates (injecting 100 mosquitoes for each group in each replicate) were performed.
2.5. Statistical analysis
Averages from biological replicates are presented for the expression quantification experiments. The sum of rank test (Mann–Whitney U test) was used to test difference of continuous variables between groups. All analyses were two-tailed and p ≤ 0.05 was considered significant. In particular, to evaluate the effect of SCRBQ2 silencing, the sum of rank test was used to test the difference in the number of oocysts between dsSCRBQ2- (experimental) and dsGFP-injected (control) mosquitoes. Infection prevalence was defined as the proportion of mosquitoes with one or more oocysts. The percentage of inhibition in oocyst formation was calculated as follows: % inhibition = (control mean oocyst number – experimental mean oocyst number)/control mean oocyst number × 100. All statistical analyses were performed using the R software (R Development Core Team, 2007).
3. Results
3.1. Croquemort genes in A. gambiae
In contrast to the single Drosophila gene, there are four Croquemort genes (SCRBQ1–4) in the A. gambiae genome. Of these, the protein encoded by Croquemort transcript SCRBQ2 (AGAP010133) has the highest identity to its Drosophila counterpart (42%), while the gene products of Croquemort transcripts SCRBQ1 (AGAP010132), SCRBQ3 (AGAP008179), and SCRBQ4 (AGAP003373) have a 37%, 35% and 36% identity, respectively. Alignment of the predicted amino acid sequences using the ClustalW algorithm (Thompson et al., 1994) revealed the presence of conserved residues. In particular, several cysteines (256, 285, and 345), prolines (93, 350, 356, 374, 387, and 423) and glycines (92, 130, 185, 210, 230, 261, and 371) that were used to align the Drosophila Croquemort protein with other members of the CD36 superfamily (Franc et al., 1996) are also present in the Anopheles amino acid sequences. Likewise, a motif formed by 13 amino acids (amino acids 350–362) that has been suggested by (Franc et al., 1996) as relevant in these proteins (P I/V Y I/L S F/L P H F Y/L L A D/S) is also partially conserved in the Anopheles Croquemort proteins. In particular, the last nine amino acids are strongly conserved in the sequences (Fig. 1). The functional significance of this motif is currently under investigation (N. Franc, personal communication).
Fig. 1.
Sequence alignment of the four A. gambiae Croquemort predicted proteins (SCRBQ1–4) and Drosophila Croquemort (accession number CG4280). Identical residues are shaded black, while conserved groups are shown with a gray background. Conserved prolines, cysteines and glycines are indicated with an asterisk. A 13-amino acid motif that was originally described in Drosophila’s Croquemort and that is present in members of the CD36 superfamily is shown in a box. The last nine amino acids are strongly conserved both in Drosophila and in A. gambiae.
3.2. Developmental profile of Croquemort SCRBQ1–4 genes expression
We designed oligonucleotides to amplify a specific fragment of each mRNA to determine their expression profile at different developmental stages by RT-PCR (Fig. 2). SCRBQ1 was expressed most strongly in larval stages but its expression could not be detected in the embryo. SCRBQ2 was expressed evenly at all developmental stages. Croquemort transcript SCRBQ3 was expressed at four developmental stages: mainly in third instar larvae and weakly in first and fourth-instar larvae as well as in adult females. SCRBQ4 expression was detected in all the developmental stages, but it was strongest in third and fourth-instar larvae. These expression profiles were replicated in two independent RNA samples.
Fig. 2.
Developmental profile of the four Croquemort transcripts (SCRBQ1–4) in the A. gambiae mosquito. E, embryos; first (L1), second (L2), third (L3) and fourth (L4) instar larvae; pupae (P); ♀, females; –, negative control (no cDNA). The A. gambiae S7 ribosomal protein served as a loading control. The expression profile obtained from a second independent RNA sample reproduced these results.
3.3. Transcriptional profile of Croquemort SCRBQ1–4 genes in the different organs of A. gambiae
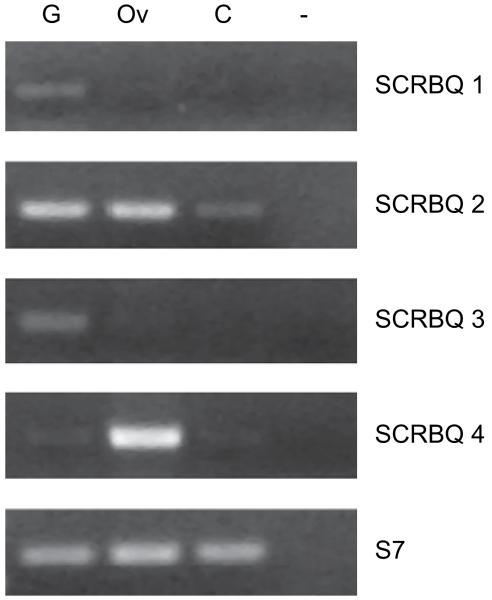
The expression profile of these genes was analyzed in the midguts, ovaries and carcasses (whole bodies minus organs) of sugar-fed mosquitoes. As shown in Fig. 3, all the transcripts were detected in the midguts, although the expression of SCRBQ2 was noticeably higher than that of the other SCRBQ genes. SCRBQ2 expression was also detected in the ovaries and carcasses, while SCRBQ4 was also expressed in the ovaries.
Fig. 3.
Transcriptional profile of the four Croquemort genes in the different A. gambiae organs. G, midguts; Ov, ovaries; C, carcass (whole body minus organs); –, negative control (no cDNA). The A. gambiae S7 ribosomal protein served as a loading control. These results were reproduced using a second independent RNA sample.
We decided to focus on Croquemort transcript SCRBQ2 (AGAP010133) since it is strongly expressed in the mosquito midgut, an organ where close interaction between the parasite and the mosquito occurs. Besides, SCRBQ2 has the highest identity to the Drosophila protein and previous studies had reported transcriptional changes for this gene upon P. berghei invasion of the mosquito midgut (Dong et al., 2006).
3.4. Croquemort SCRBQ2 mRNA is mainly expressed in the midgut of A. gambiae mosquitoes
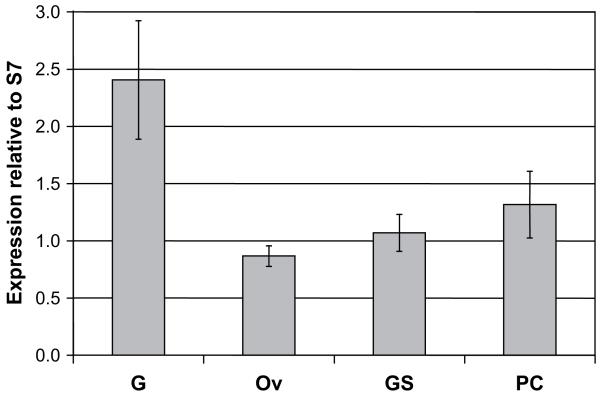
The expression profile of the Croquemort SCRBQ2 mRNA in female mosquitoes, as determined by real-time PCR assays, is shown in Fig. 4. The gene is expressed in all the organs of sugarfed A. gambiae females, but the mRNA is most abundant in the midgut. The transcript was also detected in the ovaries, salivary glands and carcass, its abundance being 36%, 44% and 55% of that observed in midguts, respectively. The differences in transcript abundance in the different organs with respect to the midguts were statistically significant (p < 0.001).
Fig. 4.
Expression profile of the Croquemort SCRBQ2 (AGAP010133) transcript in the different A. gambiae organs. Real-time quantitative PCR analysis of the SCRBQ2 Croquemort transcript. The A. gambiae S7 ribosomal protein mRNA served as a quantification standard. Bars indicate 95% confidence intervals. G, midguts; Ov, ovaries; SG, salivary glands; PC, carcass (whole body minus midgut, ovaries and salivary glands).
3.5. Croquemort SCRBQ2 mRNA expression is up-regulated in the midgut of blood-fed A. gambiae
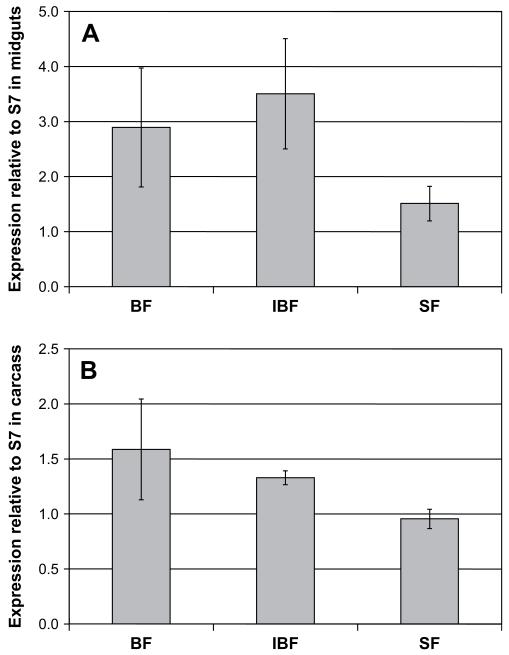
As determined by real-time PCR assays, Croquemort SCRBQ2 expression was up-regulated (1.9-fold increase, p = 0.004) in the midguts of blood-fed mosquitoes 24 h after blood feeding, compared to its expression in midguts of sugar-fed mosquitoes. Gene expression was also up-regulated (2.3-fold increase, p = 0.002) in midguts from mosquitoes 24 h after feeding on a P. berghei-infected blood meal (Fig. 5A). The expression of the gene was moderately up-regulated in the carcass of blood-fed mosquitoes (1.7-fold increase with respect to its expression in the carcass of sugar-fed females, p = 0.002) and in the carcasses of mosquitoes infected with P. berghei (1.4-fold increase, p = 0.002) (Fig. 5B). We confirmed that the differences we observed were not due to the different rearing temperatures of the mosquitoes, as no increase in the expression of SCRBQ2 was apparent by RT-PCR in sugar-fed mosquitoes that were kept for 24 h at 19 °C (data not shown). RT-PCR assays exploring the changes of the other three SCRBQ genes under different feeding conditions showed that SCRBQ2 is the only gene consistently up-regulated after blood- and infectious-blood feeding (Supplementary figure S1).
Fig. 5.
Expression profile of the Croquemort SCRBQ2 (AGAP010133) transcript in the A. gambiae mosquito under different feeding conditions. Real-time quantitative PCR analysis of the SCRBQ2 Croquemort transcript expression in response to a non-infected or infected blood meal. Transcript abundance was determined in the (A) midguts and (B) carcasses (whole body minus midgut) 24 h after a non-infectious-blood meal (BF) or a P. berghei-infected blood meal (IBF). Sugar-fed (SF) mosquitoes were used as a reference. The A. gambiae S7 ribosomal protein mRNA served as a quantification standard. Bars indicate 95% confidence intervals.
3.6. Double-stranded Croquemort SCRBQ2 RNA effectively reduces transcript abundance in female A. gambiae mosquitoes
To determine the level of RNAi-mediated gene silencing, midguts and carcasses were dissected 48, 72 and 96 h after dsRNA injection, and SCRBQ2 gene transcript abundance was determined by real-time PCR. At 48 h after the injection, Croquemort expression in the midguts of A. gambiae females injected with dsSCRBQ2 was reduced by 62% (p < 0.001) with respect to that observed at the same time point in control mosquitoes injected with dsGFP. A similar decrease in Croquemort transcript expression was observed 72 and 96 h after the injection of dsSCRBQ2 (55% reduction in both, p < 0.001). Overall, this represents a ~60% decrease in Croquemort SCRBQ2 expression in dsSCRBQ2-injected mosquitoes at all the time points examined, when compared to its expression in the midguts of dsGFP-injected (control) mosquitoes (Fig. 6A). Similarly, Croquemort expression in the carcasses of dsSCRBQ2-injected mosquitoes decreased by about 40% at the three time points (p < 0.001) (Fig. 6B).
Fig. 6.
RNAi-mediated knockdown of Croquemort transcript SCRBQ2 in the A. gambiae mosquito. Real-time quantitative PCR analysis of the SCRBQ2 Croquemort transcript expression in the (A) midguts and (B) carcasses of A. gambiae females injected with dsSCRBQ2 or dsGFP RNA. Samples were obtained 48, 72 and 96 h after dsRNA injection and analyzed by real-time RT-PCR. (C) Croquemort expression in the midguts of A. gambiae female mosquitoes injected with dsSCRBQ2 or dsGFP and fed 72 h later with P. berghei-infected blood. Samples were obtained 96 h after dsRNA injection (24 h after infectious-blood feeding). The A. gambiae S7 ribosomal protein mRNA served as a quantification standard. Bars indicate 95% confidence intervals.
The effect of dsRNA injection in Croquemort transcript levels was also determined in mosquitoes that were fed on P. berghei-infected mice 72 h after dsRNA injection. In the midguts, transcript abundance was reduced by 71% (p = 0.002) 96 h after dsRNA injection (24 h after infectious-blood feeding) (Fig. 6C). Similarly, in the carcass, SCRBQ2 expression was reduced by 33% (p = 0.002).
Possible knockdown of the other three Croquemort orthologs in the midgut by dsSCRBQ2 was explored by semi-quantitative RT-PCR 72 h after injection of dsGFP and dsSCRBQ2 (Supplementary figure S2). The expression of all three Croquemort genes in the midgut was markedly inferior to that of SCRBQ2. No differences were observed in the expression level of SCRBQ1 and SCRBQ3. However, SCRBQ4 could be weakly observed in dsGFP- but not in dsSCRBQ2-injected mosquitoes, suggesting that co-suppression by dsSCRBQ2 might occur.
3.7. Reduction of Croquemort SCRBQ2 transcript abundance affects P. berghei oocyst formation
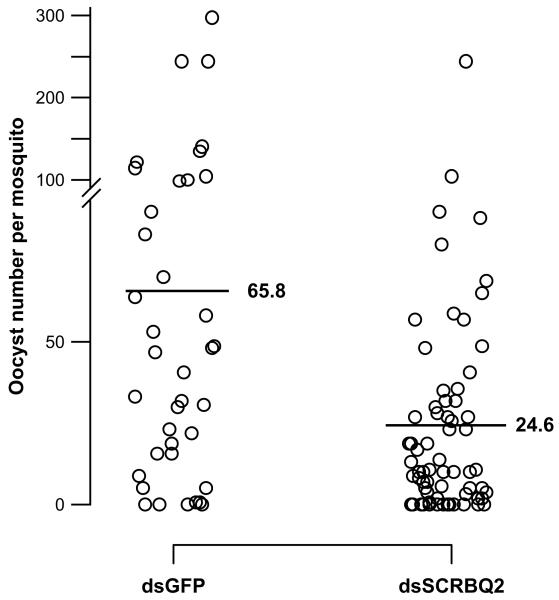
To determine whether the loss of SCRBQ2 would affect oocyst development in the mosquito, we performed RNAi-mediated gene silencing and quantified the number of P. berghei oocysts in the mosquito midgut. There was a statistically significant difference (p = 0.035, Mann–Whitney U test) in the number of oocysts observed between mosquitoes injected with dsSCRBQ2 and those injected with dsGFP. Among the ten assays performed, 4386 oocysts were observed in 151 dsGFP-injected mosquitoes (mean oocyst number per mosquito = 29.0) and 2723 oocysts were present in 162 dsSCRBQ2-injected mosquitoes (mean oocyst number per mosquito = 16.8). This corresponds to a 42.1% inhibition in oocyst formation. Given that infection levels were variable across the experiments, we performed a subgroup analysis, testing experiments separately depending on the level of infection occuring in either of both groups (cut-off: median infection 30 oocysts per midgut). Among the four experiments with higher infection levels, 2501 oocysts were observed in 38 dsGFP-injected mosquitoes (mean oocyst number per mosquito = 65.8) and 1652 oocysts were present in 67 dsSCRBQ2-injected mosquitoes (mean oocyst number per mosquito = 24.6). There was a statistically significant reduction (p < 0.001, Mann–Whitney U test) in the number of oocysts observed in dsSCRBQ2-injected mosquitoes with respect to dsGFP-injected mosquitoes (Fig. 7). This corresponds to an in vivo inhibition of 62.5% in oocyst formation. Among the six experiments with lower infection levels, there was also a marginally significant difference among the control and experimental groups (16.7 oocysts in dsGFP-injected mosquitoes versus 11.3 oocysts in dsSCRBQ2-injected mosquitoes, p = 0.05).
Fig. 7.
RNAi knockdown of Croquemort transcript SCRBQ2 decreases the number of P. berghei oocysts in midguts of A. gambiae mosquitoes. The midguts of dsSCRBQ2- and GFP-injected (control) mosquitoes were dissected 12–15 days after a P. berghei infectious-blood meal to determine the number of oocysts in each midgut. The mean number of oocysts is shown with a horizontal line. There was a statistically significant reduction of 62.5% (p < 0.001, Mann–Whitney U test, two-tailed) in the number of oocysts observed in dsSCRBQ2-injected mosquitoes with respect to dsGFP-injected mosquitoes. Data was pooled from four independent biological replicates with a median infection ≥ 30 oocysts. A break and change in the scale of the vertical axis were made for clarity purposes.
The prevalence of infection was 78.1% for dsGFP-injected mosquitoes and 69.8% for dsSCRBQ2-injected mosquitoes (p = 0.12). In the high infection group the prevalence of infection was 89.4% for dsGFP-injected mosquitoes and 79.1% for dsSCRBQ2-injected mosquitoes (p = 0.27), while in the low infection group the prevalence of infection was 74.3% for dsGFP-injected mosquitoes and 63.1% for dsSCRBQ2-injected mosquitoes (p = 0.11).
4. Discussion
Gene expansion can lead to diversification and the development of new species-specific functions (Christophides et al., 2002). Comparative genomics of D. melanogaster and A. gambiae have revealed numerous gene expansions in both genomes. In A. gambiae, the fibrinogen-domain (FBN) family (Zdobnov et al., 2002) and the carboxylesterases and cytochrome P450s families – both of them involved in insecticide resistance – (Ranson et al., 2002) are two examples. Drosophila’s Croquemort, a class B scavenger receptor, has four orthologs in the A. gambiae genome, which is another example of gene expansion (Christophides et al., 2002). Conceivably, the gene diversification of this family represents a particular adaptation to mosquito biology, since there are also four Croquemort orthologs in Aedes aegypti (Waterhouse et al., 2007).
Expression analyses evidenced differences in the developmental profiles of these four transcripts. Croquemort transcript SCRBQ2 had an almost constant expression level irrespective of the life stage, which suggests that changes in the transcriptional profile of this gene might not be developmentally regulated. However, for Croquemort transcripts SCRBQ1, SCRBQ3 and SCRBQ4, a difference in transcript abundance during the mosquito development cycle was observed. SCRBQ1 and SCRBQ4 expression was slightly up-regulated in late larval stages, while SCRBQ3 transcript was present mainly in third instar larvae and its expression in other developmental stages was more restricted, in contrast to the other Croquemort transcripts. Recently, data obtained from a microarray assay also identified the expression of Croquemort transcripts SCRBQ1, SCRBQ2, and SCRBQ3 in the midguts of fourth-instar A. gambiae larvae, however, Croquemort transcript SCRBQ4 was not detected in these assays (Neira Oviedo et al., 2008). We detected the presence of all four Croquemort transcripts in fourth-instar larvae, although we did not perform dissections of individual organs. The use of whole larvae in our assays may explain why we were able to detect the presence of transcript SCRBQ4 in fourth-instar larvae.
The precise function of the four Croquemort orthologs during the different life cycle stages of A. gambiae remains to be established. If expressed in the hemocytes, they might be involved in apoptotic cell clearance during the remodeling process of the larvae, as was previously documented in Drosophila (Franc et al., 1996; Franc et al., 1999). Additionally, they could be acting as scavenger receptors for lipoprotein complexes (Turunen, 1985) which could be used for lipid metabolism in the larval midgut. This is an interesting possibility since transcript SCRBQ1 is expressed in the anterior midgut, the only region in which transcripts involved in lipoprotein transport have been identified. Transcripts SCRBQ1 and SCRBQ3 are also expressed in the posterior midgut, a region in which lipid metabolism and absorption apparently take place (Neira Oviedo et al., 2008). Whether the different Croquemort genes have redundant or overlapping functions or whether they perform diverse roles in the mosquito awaits investigation.
Scavenger receptors bind a broad range of polyanionic ligands, in particular modified low-density lipoproteins (LDL) (McGuinness et al., 2003). In vertebrates, CD36 binds oxidized LDL, modified LDL, long chain fatty acids, and anionic phospholipids (Febbraio et al., 2001). Both in Drosophila and in Anopheles, Croquemort proteins have a CD36 domain. Therefore, the possibility exists that Croquemort could be acting as a scavenger receptor for any or several of these molecules, particularly in the ovaries, where it could be scavenging for lipids that would be useful for egg production. Croquemort might be performing the same function in the fat body, as it is also expressed in the carcass (whole body minus midgut, ovaries and salivary glands) of the mosquito.
We focused on the SCRBQ2 transcript (AGAP010133), since it has the highest identity to Drosophila’s Croquemort and importantly, it had been shown to change its expression profile in relation to P. berghei invasion (Dong et al., 2006). Besides, SCRBQ2 was the ortholog predominantly expressed in adult females (Fig. 2) and it is also the gene that has the highest expression level in the midgut (Fig. 3).
Compared to its expression in midguts of sugar-fed mosquitoes, Croquemort SCRBQ2 expression was up-regulated in the midguts of blood-fed mosquitoes. Gene expression was also up-regulated in A. gambiae midguts 24 h after receiving a blood meal infected with P. berghei. Previously, microarray data indicated that Croquemort transcript SCRBQ2 was up-regulated in the midguts of P. berghei-infected mosquitoes upon ookinete invasion (Dong et al., 2006). In contrast, our data show a similar up-regulation of the SCRBQ2 transcript in response both to normal blood feeding and blood infected with P. berghei. However, it is not uncommon for real-time PCR and microarray data to vary due to differences in technical procedures, such as data normalization and the amount of change in gene expression (Morey et al., 2006). Our results indicate that increase in SCRBQ2 transcript abundance in the mosquito midguts appears to be more a physiological response to blood feeding than a specific response to the malaria parasite crossing the midgut epithelium, which usually occurs 18–24 h after the infectious-blood meal (Beier, 1998; Dimopoulos et al., 2002).
Also, we cannot rule out the possibility that some of the Croquemort SCRBQ2 transcripts detected in the midguts could correspond to those expressed by hemocytes that remain attached to the midgut (Abraham et al., 2005; Osta et al., 2004). If expressed in A. gambiae hemocytes, this molecule could be acting as a phagocytic receptor for apoptotic cells (Franc et al., 1999) or as a phagocytic receptor for Gram-positive bacteria, as observed in a Drosophila cell line (Stuart et al., 2005). Supporting the latter hypothesis, microarray data has shown that SCRBQ2 expression is up-regulated in response to bacterial challenges (Dimopoulos et al., 2002).
During Plasmodium development in the mosquito, there is close interaction between the parasite and the midgut. RNAi-mediated gene silencing was used to assess potential functions of the Croquemort SCRBQ2 gene in vivo. Knockdown of gene expression resulted in a significant reduction (~60%, p < 0.001) in the number of P. berghei oocysts in the midgut of A. gambiae, compared to the number of oocysts in the control (GFP-injected) group, suggesting that SCRBQ2 participates in parasite development. Similarly, there were differences in the infection prevalence between the GFP- and SCRBQ2-injected mosquitoes, although they did not reach statistical significance.
Even though it is very weakly expressed in the midgut, it is possible that SCRBQ4 might have also been silenced in dsSCRBQ2-injected mosquitoes. Nevertheless, SCRBQ2 is substantially more abundant than SCRBQ4 in the midgut, which supports the fact that the results we observed are mainly due to the knockdown of SCRBQ2. The precise role that Croquemort SCRBQ2 plays in parasite development remains to be determined.
In conclusion, we present the expression profile of four Croquemort transcripts along the life cycle of A. gambiae. In particular, Croquemort transcript SCRBQ2 is highly expressed in the midguts of adult sugar-fed females, and is up-regulated after a blood meal. RNAi knockdown experiments suggest that Croquemort SCRBQ2 participates in the establishment of Plasmodium parasites in the mosquito midgut.
Supplementary Material
Acknowledgments
We wish to express our gratitude to Leticia Cortes-Martínez, Juan García-Jiménez and Juana Calderón-Amador at CINVESTAV-IPN for their excellent technical assistance. We are grateful to Kathryn Shaw for helping with P. berghei cultures and mice infections and to the Johns Hopkins Malaria Research Institute insectary for assistance with mosquito rearing. We also thank Dr. Abraham Eappen for providing the AgS7 primers and Dr. Anil Ghosh for kindly providing the GFP plasmid and primers (Johns Hopkins School of Public Health, USA). We also acknowledge the support of Dr. Fernando Navarro-García, Head of the Cell Biology Department (CINVESTAV-IPN), and we thank Dr. Juan Pedro Luna-Arias at CINVESTAV-IPN for helpful criticisms. M.G.-L. received a PhD scholarship no. 166703 from the Mexican Council of Science and Technology (CONACyT, Mexico) during the development of this work. M.G.-L. was also a Telmex Foundation Scholarship holder (Mexico). This investigation received financial support from CONACyT (grant no. 62389 to F.C.H.-H.; Apoyos Integrales para la Formación de Doctores en Ciencias & Programa de Becas Mixtas en el Extranjero para Becarios CONACyT Nacionales, both to M.G.-L.) and from the National Institutes of Health (to M.J.-L.).
Footnotes
Appendix. Supplementary material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ibmb.2009.03.008.
References
- Abraham EG, Pinto SB, Ghosh A, Vanlandingham DL, Budd A, Higgs S, Kafatos FC, Jacobs-Lorena M, Michel K. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16327–16332. doi: 10.1073/pnas.0508335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier JC. Malaria parasite development in mosquitoes. Annu. Rev. Entomol. 1998;43:519–543. doi: 10.1146/annurev.ento.43.1.519. [DOI] [PubMed] [Google Scholar]
- Benedict MQ. Care and maintenance of anopheline mosquito colonies. In: Crampton JM, Beard CB, Louis C, editors. The Molecular Biology of Insect Disease Vectors. A Methods Manual. Chapman & Hall; London: 1997. p. 579. [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Müller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Casavant TL, Chang S, Scheetz T, Roberts C, Donohue M, Schultz J, Benes V, Bork P, Ansorge W, Soares MB, Kafatos FC. Anopheles gambiae pilot gene discovery project: identification of mosquito innate immunity genes from expressed sequence tags generated from immune-competent cell lines. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Christophides GK, Meister S, Schultz J, White KP, Barillas-Mury C, Kafatos FC. Genome expression analysis of Anopheles gambiae: responses to injury, bacterial challenge, and malaria infection. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Invest. 2001;108:785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc NC, Dimarcq JL, Lagueux M, Hoffmann JA, Ezekowitz RAB. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. 1996;4:431–443. doi: 10.1016/s1074-7613(00)80410-0. [DOI] [PubMed] [Google Scholar]
- Franc NC, Heitzler P, Ezekowitz RAB, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- Go EP, Wikoff WR, Shen Z, O’Maille G, Morita H, Conrads TP, Nordstrom A, Trauger SA, Uritboonthai W, Lucas DA, Chan KC, Veenstra TD, Lewicki H, Oldstone MB, Schneemann A, Siuzdak G. Mass spectrometry reveals specific and global molecular transformations during viral infection. J. Proteome Res. 2006;5:2405–2416. doi: 10.1021/pr060215t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough PJ, Gordon S. The role of scavenger receptors in the innate immune system. Microbes Infect. 2000;2:305–311. doi: 10.1016/s1286-4579(00)00297-5. [DOI] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, Emmert D, Hammond M, Hill CA, Kennedy RC, Lobo NF, MacCallum MR, Madey G, Megy K, Redmond S, Russo S, Severson DW, Stinson EO, Topalis P, Zdobnov EM, Birney E, Gelbart WM, Kafatos FC, Louis C, Collins FH. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007;35:D503–D505. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness DH, Dehal PK, Pleass RJ. Pattern recognition molecules and innate immunity to parasites. Trends Parasitol. 2003;19:312–319. doi: 10.1016/s1471-4922(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Gordon S. The role of scavenger receptors in pathogen recognition and innate immunity. Immunobiology. 2004;209:39–49. doi: 10.1016/j.imbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Murphy JE, Tedbury PR, Homer-Vanniasinkam S, Walker JH, Ponnambalam S. Biochemistry and cell biology of mammalian scavenger receptors. Atherosclerosis. 2005;182:1–15. doi: 10.1016/j.atherosclerosis.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Neira Oviedo M, Vanekeris L, Corena-McLeod MD, Linser PJ. A microarray-based analysis of transcriptional compartmentalization in the alimentary canal of Anopheles gambiae (Diptera: Culicidae) larvae. Insect Mol. Biol. 2008;17:61–72. doi: 10.1111/j.1365-2583.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Curr. Opin. Immunol. 2002;14:123–128. doi: 10.1016/s0952-7915(01)00307-7. [DOI] [PubMed] [Google Scholar]
- Pluddemann A, Mukhopadhyay S, Gordon S. The interaction of macrophage receptors with bacterial ligands. Expert Rev. Mol. Med. 2006;8:1–25. doi: 10.1017/S1462399406000159. [DOI] [PubMed] [Google Scholar]
- Pluddemann A, Neyen C, Gordon S. Macrophage scavenger receptors and host-derived ligands. Methods. 2007;43:207–217. doi: 10.1016/j.ymeth.2007.06.004. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria: 2007. ISBN 3-900051-07-0. URL. http://www.R-project.org. [Google Scholar]
- Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, Unger MF, Collins FH, Feyereisen R. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- Salazar CE, Mills-Hamm D, Kumar V, Collins FH. Sequence of a cDNA from the mosquito Anopheles gambiae encoding a homologue of human ribosomal protein S7. Nucleic Acids Res. 1993;21:4147. doi: 10.1093/nar/21.17.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Deng J, Silver MJ, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RAB, Moore KJ. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J. Cell Biol. 2005;170:477–485. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen S. Absorption. In: Kerkut GA, Gilber LI, editors. Comprehensive Insect Physiology, Biochemistry and Pharmacology. Pergamon Press; Oxford: 1985. pp. 241–278. [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, Mueller HM, Dimopoulos G, Law JH, Wells MA, Birney E, Charlab R, Halpern AL, Kokoza E, Kraft CL, Lai Z, Lewis S, Louis C, Barillas-Mury C, Nusskern D, Rubin GM, Salzberg SL, Sutton GG, Topalis P, Wides R, Wincker P, Yandell M, Collins FH, Ribeiro J, Gelbart WM, Kafatos FC, Bork P. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.