Abstract
Malaria parasites digest hemoglobin within a food vacuole to supply amino acids, releasing the toxic product heme. During the detoxification, toxic free heme is converted into an insoluble crystalline form called hemozoin (Hz). Heme detoxification protein (HDP) in Plasmodium falciparum is one of the most potent of the hemozoin-producing enzymes. However, the reaction mechanisms of HDP are poorly understood. We identified the active site residues in HDP using a combination of Hz formation assay and spectroscopic characterization of mutant proteins. Replacement of the critical histidine residues His122, His172, His175, and His197 resulted in a reduction in the Hz formation activity to approximately 50% of the wild-type protein. Spectroscopic characterization of histidine-substituted mutants revealed that His122 binds heme and that His172 and His175 form a part of another heme-binding site. Our results show that the histidine residues could be present in the individual active sites and could be ligated to each heme. The interaction between heme and the histidine residues would serve as a molecular tether, allowing the proper positioning of two hemes to enable heme dimer formation. The heme dimer would act as a seed for the crystal growth of Hz in P. falciparum.
Malaria is one of the most common infectious diseases in the world. Plasmodium falciparum, which causes the most severe form of malaria, replicates within red blood cells. During its growth within the red blood cell, the parasite digests hemoglobin within a food vacuole to supply amino acids1,2, releasing the toxic product heme3,4,5. While free heme is detoxified by enzymatic degradation in mammals, the malaria parasite has evolved a distinct mechanism for heme detoxification6. During detoxification, free heme is converted into a crystalline form called hemozoin (Hz)7,8. Inhibition of Hz formation is one of the most important factors targeting the malaria parasite because the process of Hz formation is indispensable for the survival of Plasmodium species. Indeed, several antimalarial drugs are thought to inhibit Hz production9,10. However, resistance to currently used antimalarial drugs has led to an urgent need to develop new and effective drugs.
Spectroscopic and crystallographic studies have revealed that the Hz structure is composed of direct coordination between a heme propionate group and the iron of another heme dimer unit (Fig. 1A)11. These dimers form hydrogen bonds with their neighbors via another propionate group. The unique structure of Hz has led to the speculation that formation of the links between the monomeric heme molecules is a crucial step in Hz production. Lipids or proteins have been suggested to catalyze Hz formation12,13,14, whereas the molecular mechanism of its formation is highly controversial. Lipids fraction from P. falciparum food vacuole promote Hz formation in vitro12. On the other hand, histidine-rich protein II (HRP II) in P. falciparum was the first protein to be involved in Hz formation14. HRP II could facilitate the heme dimer formation, and lipids might then help the polymerization process of the precursor heme dimers15. However, a P. falciparum clone lacking the HRP II and III genes still forms Hz normally16, suggesting that lipids and/or other proteins may compensate for the loss of HRP II and III.
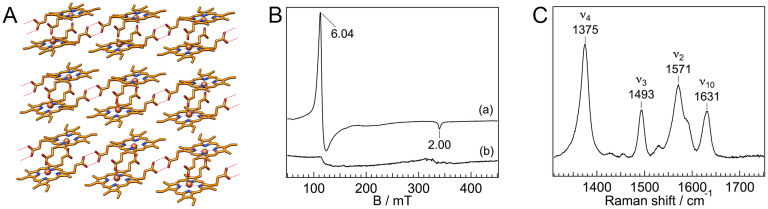
Figure 1. Heme coordination structure of the HDP–heme complex.
(A) The structure of Hz. Dotted lines represent hydrogen bonds. (B) EPR spectra of heme-bound HDP (trace a) and free heme (trace b) at 5 K at pH 5.6. EPR spectrum measurements were carried out at the X-band (9.50 GHz) microwave frequency. (C) Resonance Raman spectrum of heme-bound HDP in pH 5.6 at room temperature. The excitation wavelength was set at 410 nm.
Recently, heme detoxification protein (HDP) was identified in P. falciparum and was found to be extremely potent in converting heme into Hz17. HDP exists in the food vacuole, where it forms a complex with the hemoglobinase falcipain-2 and other proteases17,18. Moreover, HDP orthologs are found in seven other species of Plasmodium17. These results suggest that HDP is involved in the formation of Hz in Plasmodium species. P. falciparum HDP consists of 205 amino acids. Although the C-terminal region of HDP is homologous to that of fasciclin-1, there is no homology with the known heme proteins17. Therefore, the interaction between HDP and heme would be different from previously identified heme proteins. Recently, we established a method for purifying intact HDP from Escherichia coli19. The intact HDP contains two equivalent heme binding sites19. A possible Hz formation mechanism is that binding of two hemes to HDP is followed by the formation of reciprocal iron–oxygen bonds between the two heme molecules19. However, the residues essential for Hz production in HDP and other Hz-producing proteins have not been reported12,20.
In this study, we show that four of nine histidine residues in HDP are involved in Hz production. Spectroscopic characterization revealed that His122 is the most probable axial ligand of heme iron and that His172 and His175 form a part of another heme-binding site in HDP. Since the histidine-less mutant still retained its Hz-producing activity, the binding of hemes into the protein matrix is likely to be another critical factor for their activity. These results suggest that the role of histidine residues is to bring heme into the proper alignment in the active site of HDP for the efficient production of heme dimers. It is possible that these precursor heme dimers interact with lipids to achieve further growth of Hz crystals. A proposed reaction scheme for the heme dimer formation is one of the most important steps toward understanding Hz formation in malaria parasites and has significant implications for designing new drugs.
Results
Heme Coordination Structure of HDP
To investigate the interaction between HDP and heme, we measured EPR spectra for the HDP with two equivalents of heme at pH 5.6. Since HDP can produce Hz at pH 5.2 or lower, the formation of Hz does not occur in this condition. Fig. 1B shows the EPR spectra for the HDP–heme complex (trace a) and the free heme (trace b) at 5 K, where the EPR signals from high-spin species can be observed clearly. The EPR spectrum of the heme-bound HDP had a sharp signal at g = 6.04 (Fig. 1B, trace a). In contrast, the EPR signal in the free heme exhibited a broad peak around g = 6 (Fig. 1B, trace b). The EPR signal at g = 6.04 for the HDP–heme complex can be assigned to the high-spin heme21. A broad signal at approximately g = 6 arising from the free heme could be the result of the highly overlapping signals derived from the multiple configurations of free heme in the solution. These data indicate that a high-spin heme is present in the HDP–heme complex. The spin state and coordination structure of heme in HDP at room temperature was also investigated by resonance Raman spectroscopy. Fig. 1C represents the high frequency region of the resonance Raman spectrum for HDP with two equivalents of heme at pH 5.6. It is established that the lines in the high frequency region can be used as sensitive markers of the oxidation state (ν4) and spin and coordination sates (ν2, ν3, and ν10) of the heme iron22. The lines at 1375, 1493, 1571, and 1631 cm−1 observed for the HDP–heme complex can be assigned to ν4, ν3, ν2, and ν10, respectively. These frequencies indicate that the heme iron is high-spin form as found for myoglobin and horseradish peroxidase23,24. Since a mixture of high- and low-spin heme proteins exhibits two distinguishable ν3 lines25, the single band observed at 1493 cm−1 indicates that the high-spin component of heme is predominant in the HDP–heme complex at room temperature at pH 5.6. These results suggest that both the hemes in HDP have the high-spin configurations akin to that in typical hemeproteins. As reported previously, the electronic absorption spectra for the HDP-heme complex depend on pH value19. The electronic absorption spectra and EPR spectra for the HDP-heme complex at pH 5.6, 7.0, and 11.0 showed that the high-spin component of heme was increased at the low pH (Fig. S1). Since HDP can produce Hz at pH 5.2 or lower, the high-spin component of heme would play important role for its activity.
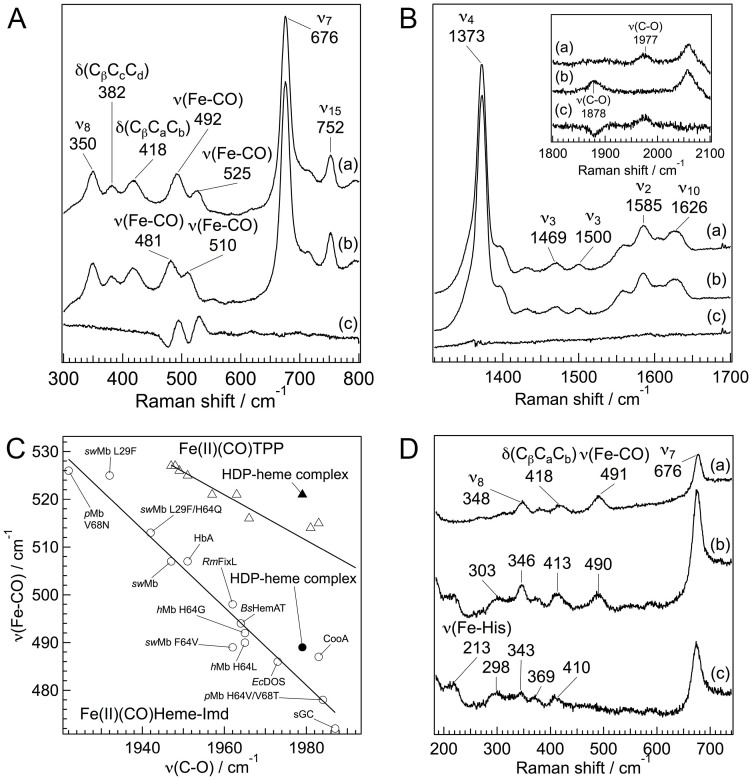
To identify the amino acid of the axial ligand, we measured the resonance Raman spectra for the CO adduct of the reduced heme in HDP. The backbonding correlation, a plot of Fe–CO stretching versus C–O stretching vibrational frequencies, is a useful tool for identifying a proximal ligand26,27,28. Fig. 2A shows the resonance Raman spectra in the Fe–CO stretching frequency region of the 12C16O and 13C18O-bound HDP–heme complex. Two lines at 492 and 525 cm−1 for the 12C16O adduct can be observed in Fig. 2A, trace a. Both lines can be assigned to the stretching mode of Fe–CO, ν(Fe–CO), since the Fe–CO stretching mode and the Fe–C–O bending mode are in general observed in the 450–550 and 560–590 cm−1 regions, respectively29. In contrast to the HDP with oxidized heme, the CO adduct of the reduced heme in HDP had at least two configurations. Multiple heme-binding configurations were confirmed by the observation of two distinct coordination-state marker lines of ν3 at 1469 and 1500 cm−1 (Fig. 2B, trace a). Because HDP binds heme with a stoichiometry of 1:2 (HDP: heme)19, two ν(Fe–CO) lines may arise from the different heme species in HDP. While the two configurations of the Fe–CO stretching mode were observed for the HDP–heme complex, there was only one Raman line at 1977 cm−1 in the C–O stretching band region (Fig. 2B, inset, trace a). The absence of two distinct lines indicates the overlap of the ν(C–O) line from the two configurations of heme in HDP.
Figure 2. Heme coordination structure of the CO adduct of the HDP–heme complex.
Resonance Raman spectra of the CO adduct of the HDP–heme complex in pH 5.6. (A and B) Resonance Raman spectra of the HDP–heme complex treated with 12C16O (trace a), 13C18O (trace b), and difference spectra (12C16O–13C18O) for the low-frequency region (A) and the high frequency region (B). The excitation wavelength was set at 410 nm. (C) Backbonding correlation plot of ν(Fe–CO) versus ν(C–O) for various histidine ligated proteins and synthetic tetraphenylporphyrin (TPP) derivatives. The black circle and triangle represent the CO adducts of the HDP–heme complex. Open circles signify proteins whose axial ligand is histidine. Open triangles are TPP that has no axial ligand. Labels HbA, hemoglobin A; Mb, Myoglobin: sGC, soluble guanylyl cyclase; Bs, Bacillus subtilis; Ec, Escherichia coli; h, human; p, pig; Rm, Rhizobium meliloti; and sw, sperm whale. (D) Resonance Raman spectra of the CO adduct of the HDP–heme complex. Spectra of the unphotolyzed (trace a), photolyzed at 100 ns after dissociation of CO (trace b), and the reduced 5-coordinate (trace c) heme of HDP. The pump and probe pulse wavelength were set at 532 and 436 nm, respectively.
The effect of the axial ligand is readily observed from the backbonding plot, which shows the ν(Fe–CO)/ν(C–O) correlations among heme proteins with different heme ligands (Fig. 2C)26,27,28. The lower line corresponds to the proteins with neutral proximal ligands that are bound to the heme iron through a histidine residue. The upper line corresponds to heme–CO complexes without amino acid axial ligands for heme iron. The two sets of ν(Fe–CO)/ν(C–O) frequencies for the CO-bound HDP–heme complex fall on the upper line (Fig. 2C, filled triangle) and the lower line (Fig. 2C, filled circle), indicating that HDP has two distinct heme coordination structures in the CO-bound reduced form of heme. The backbonding plot shows that a histidine residue forms a bond with heme in HDP, whereas there is a distinct heme–CO complex without an amino acid ligand for the heme iron. Because HDP binds two equivalents of heme per molecule of protein19, these two configurations of heme could be derived from the two independent heme-binding sites. Another possibility is that the reduction of heme iron and/or the binding of CO to heme may break the bond between an amino acid residue and the heme iron. The EPR and resonance Raman spectra indicate that both the hemes coordinate with amino acid residues in the oxidized form, one of which may lose an axial ligand following reduction of the heme iron.
The histidine ligation of reduced heme in a protein can be directly probed via the stretching mode between the heme iron and the proximal histidine, ν(Fe–His). A reduced five-coordinate heme with a histidine residue as an axial ligand confers a ν(Fe–His) Raman band in the 200–250 cm−1 region22. However, due to instability of the complex of HDP with reduced heme during the measurements, we were unable to measure the resonance Raman spectrum for the reduced heme in HDP. To obtain the spectrum for the reduced five-coordinate heme in HDP, a resonance Raman spectrum of the photolyzed CO adducts of the reduced heme was obtained at 100 ns after dissociation of CO (Fig. 2D). The observed time-resolved resonance Raman spectrum included contributions from both the photolyzed and CO-bound forms (Fig. 2D, trace b). To obtain the spectrum for the photolyzed form, the contribution of unphotolyzed species (Fig. 2D, trace a) was subtracted (Fig. 2D, trace c). An intense band assignable to ν(Fe–His) was observed at 213 cm−1, confirming ligation of a histidine residue to the heme iron.
Role of Histidine Residues in Hz formation
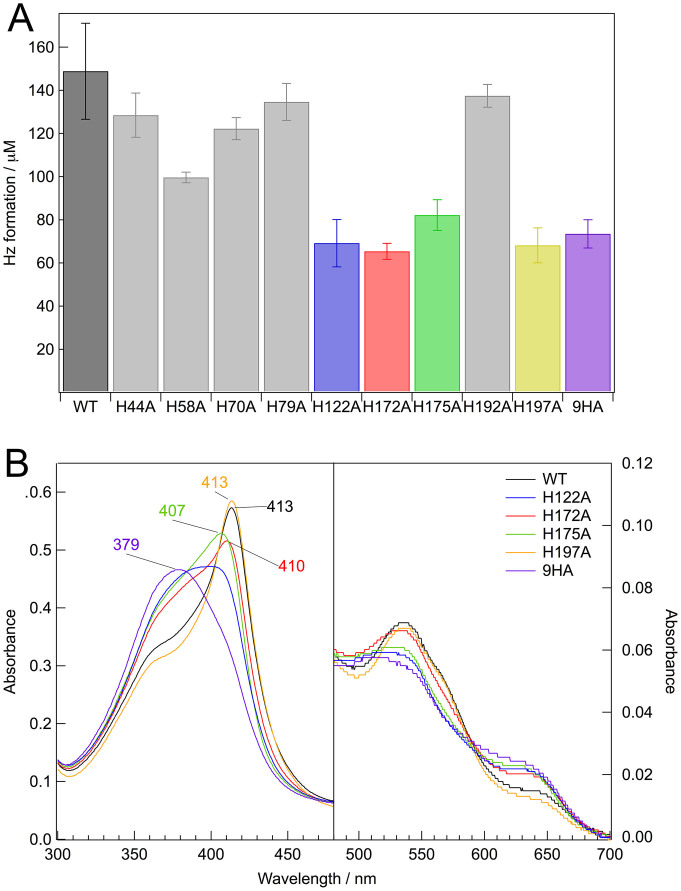
Wild-type HDP contains nine histidine residues17. To investigate the role of histidine residues in Hz formation, we constructed nine HDP mutants, H44A, H58A, H70A, H79A, H122A, H172A, H175A, H192A, and H197A. In addition to the single mutant proteins, we constructed a histidine-less mutant, in which all nine histidine residues were replaced by alanine, and we named the resulting mutant 9HA. We then assessed the Hz formation activity of the recombinant wild-type and mutant proteins (Fig. 3A). In a Hz formation assay, where the concentration of wild-type HDP and heme was 5 μM and 600 μM, respectively, heme was converted at approximately a 25% yield into Hz over 15 min (Fig. 3A, black bar). As reported previously17,18, the recombinant HDP exhibited the concentration dependent Hz production (Fig. S2). On the other hand, there was no activity detected in the absence of HDP. The formation of Hz was further confirmed by FTIR spectroscopy, as described previously19. The crystalline material produced by HDP exhibited two strong absorbance peaks at 1210 and 1663 cm−1 in the FTIR spectrum, indicating the formation of reciprocal iron-oxygen bonds between two heme molecules19. In contrast to wild-type HDP, the efficiency of Hz production by the 9HA mutant was reduced to less than half of that of the wild-type HDP (Fig. 3A, purple bar), suggesting that the histidine residues play a critical role in the formation of Hz by HDP. We assessed Hz formation activity of the single mutant proteins to identify the specific histidine residues involved in its enzyme activity. We found that four of the nine single mutations, H122A (Fig. 3A, blue bar), H172A (Fig. 3A, red bar), H175A (Fig. 3A, green bar), and H197A (Fig. 3A, yellow bar), reduced Hz formation activity to approximately 50% of the wild-type HDP. The H172A mutant showed the greatest reduction in Hz formation activity to 44% of the wild-type. These results suggest that these four histidine residues are located in the active site of HDP.
Figure 3. Hz formation activity and electronic absorption spectra for HDP and its mutant proteins.
(A) Hz formation assay for wild-type HDP (WT) and mutant proteins. The assays of Hz formation were carried out for 15 min at 37°C as described in the Methods section. Data are expressed as the mean and standard deviation of at least four independent experiments. (B) Electronic absorption spectra of heme-bound wild-type HDP (WT) and mutant proteins. The sample concentration was approximately 40 μM in 50 mM MOPS–NaOH, pH 7.0, at room temperature. The path-length of the cell was 0.2 cm.
To explore the role of individual histidine residues in HDP, electronic absorption spectra for wild-type HDP and the mutant proteins with heme were measured at pH 7.0 (Fig. 3B). Since the electronic absorption spectrum for the HDP-heme complex in pH 5.6 was found to have a rather broad peak (Fig. S1), it was difficult to distinguish free heme from ligated heme. Therefore, we carried out the electronic absorption measurements at neutral pH. With the exception of the 9HA mutant, the heme titration to HDP proteins showed that the other mutant proteins and wild-type HDP had two independent heme-binding sites (Fig. S3). In the case of the 9HA mutant, we were unable to determine the exact number of heme-binding sites because of relatively broad Soret absorption. To obtain the electronic absorption spectra, the wild-type HDP and all mutant proteins were incubated with two equivalents of heme and passed through a gel filtration column to remove unbound heme. The wild-type HDP–heme complex exhibited a Soret band at 413 nm, which was identical to that observed previously (Fig. 3B, black line)19. HDP showed an electronic absorption spectrum typical of heme-binding proteins. The most drastic change was observed in the 9HA mutant with heme, which exhibited a relatively broad Soret maximum at 379 nm (Fig. 3B, purple line), very similar to that of the free heme30. This result confirmed that the histidine residues in the wild-type HDP act as an axial ligand for the heme iron. Since unbound heme was removed by gel filtration, the 9HA mutant must also interact with heme. The structural properties of the heme-binding environment in the 9HA mutant were examined by measurement of the EPR spectrum. The EPR spectrum of the 9HA mutant with heme exhibited a sharp signal at g = 6.03 (Fig. S4), which is very similar to the wild-type HDP–heme complex (Fig. 1B, trace a). Since the free heme in the solution exhibited a broad signal at approximately g = 6 (Fig. 1B, trace b), the 9HA mutant could incorporate heme in the protein matrix. Therefore, the 9HA mutant retains its heme-binding capability without the histidine residues. At present, however, it is difficult to determine the coordination structure of heme in the 9HA mutant.
Similar to the 9HA mutant, the H122A mutant with heme also exhibited a rather broad Soret band maximum at approximately 390 nm (Fig. 3B, blue line), which is approximately 15 nm blue-shifted compared with that of the wild-type HDP–heme complex. The spectral changes of the Soret band in the H122A mutant indicate that His122 is the most probable axial ligand of heme iron in HDP, whereas the position and shape of the Soret band for the H122A mutant was different from that for the 9HA mutant. The HDP–heme complex can bind two equivalents of heme per monomer. Thus, there would be another heme-binding site that remains bound to the heme iron. Both H172A (Fig. 3B, red line) and H175A (Fig. 3B, green line) exhibited a broad Soret band with a peak at 410 and 407 nm, respectively. The peak positions of the Soret band for both mutants were close to that for the wild-type HDP, although the shoulders of the Soret band at 380 nm for mutant proteins were very similar to that for the 9HA mutant that has no axial ligands for heme iron. The most likely explanation for the spectral changes in the H172A and H175A mutants is that H122 acts as a heme-binding site and that H172 and H175 exist in another heme-binding site. Resonance Raman results indicate that both hemes in HDP have an amino acid residue for its axial ligand in the oxidized form. The secondary heme-binding site is most likely to have H172 or H175 as an axial ligand. From the present data, it is difficult to determine which histidine residue binds heme. These two histidine residues would be located close to each other and would act as an alternative axial heme ligand in their mutant proteins. In contrast, the spectrum of the H197A mutant with heme was very similar to that of the wild-type HDP–heme complex, although H197A exhibited a reduction in Hz formation activity to approximately 50% of the wild-type HDP. Taken together, these results suggest that three histidine residues (H122 and H172/H175) are candidates for the axial ligands for the heme iron in HDP.
Discussion
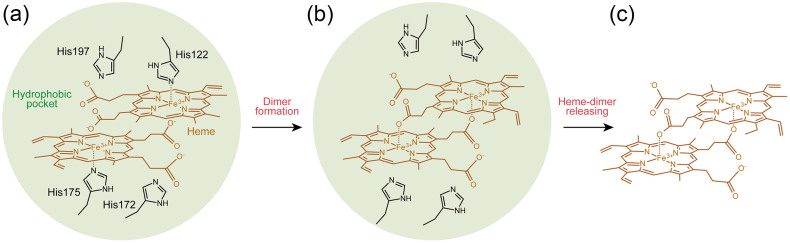
To address the question of the Hz formation mechanism, we examined the heme coordination structure and spin state of the complex of HDP with heme. A sharp high-spin signal at g = 6.04 in the EPR spectrum of the HDP–heme complex was clearly distinct from that of free heme (Fig. 1B), indicating the existence of an interaction between HDP and heme. The EPR signal around g = 6 is most often associated with the histidine-ligated hemeproteins where the most common another axial ligand is either water or hydroxide. The high-spin component of heme was also predominant in the HDP–heme complex at room temperature (Fig. 1C). These results reveal that both hemes in HDP have high-spin configurations in the oxidized form. The backbonding plot for the ν(Fe–CO)/ν(C–O) correlations and the observation of a ν(Fe–His) band at 213 cm−1 demonstrated ligation of the histidine residue to the heme (Fig. 2C and 2D). The high-spin heme in HDP could help explain the molecular mechanism of Hz formation. The formation of the links between the monomeric hemes seems to be a crucial step in the nucleation process of Hz formation. The heme sixth-ligand of either water or hydroxide in HDP could be easily replaced by a heme propionate group from another heme in HDP (Fig. 4).
Figure 4. A proposed reaction mechanism for Hz formation in HDP.
(a) HDP captures heme into a hydrophobic pocket. His122 and His175 are important for anchoring heme. His 172 and His197 also bring heme into proper alignment for the reaction. (b) A coordination bond between a heme propionate group and the iron of another heme dimer unit is formed. (c) HDP releases the heme dimer, serving as a seed for the crystal growth of Hz.
In the present study, the role of the individual histidine residues of HDP in Hz production was investigated using a combination of a Hz formation assay and spectroscopic characterization of histidine-substituted HDP mutants. The most drastic changes in the electronic absorption spectrum were observed in the H122A mutant HDP that showed a reduction in Hz formation activity to 46% of the wild-type activity (Fig. 3A, blue bar and 3B, blue line). His122 is a conserved amino acid residue in the Plasmodium species HDP17, indicating that this conserved histidine residue plays an important role in Hz formation. Mutation of His175, another conserved histidine residue in HDP, also resulted in spectral changes and decreased Hz formation activity (Fig. 3A, green bar and 3B, green line). Therefore, His122 and His175 are the most probable axial ligands of heme iron in HDP. Since the resonance Raman measurement for the HDP–heme complex indicates a high-spin form with an axial ligand, His122 and His175 could be present in the individual active sites and could be ligated to each heme.
In contrast, although the substitution of His172 resulted in decreased Hz formation activity and spectral changes in the electronic absorption spectrum (Fig. 3A, red bar and 3B, red line), the histidine residue is only conserved in P. falciparum and P. reichenowi17. Other Plasmodium species have an asparagine residue in the corresponding position. Residues acting as heme axial ligands are mostly His, Cys, Met, Lys, or Tyr. However, since cytochrome c in Rhodobacter sphaeroides has an asparagine residue for the heme axial ligand31, the asparagine residue in other Plasmodium species may functionally compensate for the histidine residue in HDP of P. falciparum. The substitution of His197 resulted in decreased Hz formation activity, whereas the electronic absorption spectrum for H197A HDP with heme was almost identical to that for wild-type HDP with heme (Fig. 3A, yellow bar and 3B, yellow line). These results suggest that His197 is not an axial ligand for the heme iron but plays a crucial role in stabilizing the Hz intermediate. The hydrogen-bonding network between the histidine residue and the heme propionate is important for maintaining the position of the heme in myoglobin32. Similarly, the role of His197 in HDP may be to anchor the heme molecule in the correct position for the reaction.
The involvement of histidine residues has also been reported in other Hz-producing proteins. HRP II in P. falciparum forms a complex with heme, which involves a bis-histidine coordinate heme33,34. Hz formation activity of α-glucosidase in Rhodnius prolixus is inhibited by diethlpyrocarbonate that modifies histidine residues20. However, the specific heme-binding sites in histidine-rich protein 2 and α-glucosidase have not yet been identified. We here identified important histidine residues involved in Hz formation, during which they act as an axial ligand for the heme iron. Our results indicate that the role of histidine residues is to bring heme into the proper alignment in the active site of HDP for efficient production of Hz (Fig. 4).
Our present study demonstrates that the histidine residues in HDP regulate Hz formation. However, the histidine-less mutant, 9HA, retained 49% of its Hz-producing activity. Hz production by HDP proceeded even with no histidine residues in its sequence. In the case of α-glucosidase in R. prolixus, modification of the histidine residues reduces Hz production but does not completely inhibit its activity20. These results indicate the involvement of histidine residues in Hz formation, whereas the binding of heme into the protein matrix would be another critical factor for activity. The primary factor in the production of Hz seems to be the binding of heme within the hydrophobic core of HDP. Although heme proteins generally have amino acid residues for the axial ligand binding the heme iron, the heme-degradation protein MhuD in Mycobacterium tuberculosis contains two hemes, one of which has no specific amino acid residue for the axial ligand35. The histidine substituted mutant HmuO can also bind heme without a specific amino acid residue for heme36. Because of the hydrophobic nature of heme, the hydrophobic core of the protein can incorporate heme even without an axial ligand. The 9HA mutant protein could thus also bind heme in its hydrophobic pocket. It should be noted here that the previous study of the lipid-mediated formation of Hz pointed out the importance of a hydrophobic environment for heme dimer formation7. Molecular dynamics simulations indicate that removal of the water molecule that binds to the heme iron facilitates heme dimer formation37. In the presence of water molecules, coordination between a heme propionate group and the iron of another heme were found to be unstable37. The binding of heme by HDP seems to provide a hydrophobic environment and facilitate the formation of a heme dimer.
These results conclusively demonstrate that HDP in P. falciparum provides for the formation of precursor heme dimers to initiate Hz nucleation. The interaction between heme and the histidine residues in the hydrophobic pocket would serve as a molecular tether, allowing the proper positioning of the heme to form a covalent bond with a heme propionate group from another heme. Hydrophobic environments would increase the rate of formation of the heme dimer in HDP. The heme dimers released from HDP could interact with other dimers through hydrogen bonds between the propionate side chains. However, the polymerization mechanism of these heme dimers has remained obscure. The dimeric form of HDP may have a significance in the polymerization mechanism. HDP was present predominantly as a dimer in the trophozoite-infected red blood cells and found as both the monomeric and dimeric form in the food vacuole preparation17. The dimeric form of HDP could be enabled to polymerize the heme dimers, because the multiple active sites present in the dimeric HDP serve as an increase in the local concentration of the heme dimers. Hz crystal is found within neutral lipids in the food vacuole of the parasite38, suggesting that the synergistic effect on HDP and lipids would also be important to Hz formation in vivo. Since the heme dimer is thought to be unstable in a water37, the lipid nanospheres with a hydrophobic environment may help the stacking of the heme dimers for further extension. Although the Hz production activities of HDP is much higher than that of the neutral lipids in vitro17, the addition of lipids extracted from the lysed parasites or mono-oleoyl glycerol (MOG) exhibits an additive effect on HDP-mediated Hz formation18. The molecular mechanisms of the heme dimers polymerization will be the subject of further research.
In conclusion, the Hz formation assay showed that four of nine single mutations, H122A, H172A, H175A, and H197A, reduced Hz formation activity to approximately 50% of the wild-type HDP, indicating that these histidine residues are involved in Hz production. The residual activity of the histidine-less mutant protein suggests that the binding of heme in the hydrophobic core of HDP is another critical factor in Hz formation. Specific heme binding to H122 and H172/H175 was also revealed by spectroscopic characterization of histidine-substituted HDP mutants. These results will provide valuable information for designing antimalarial drugs to specifically inhibit Hz formation in malaria parasites.
Methods
Protein expression and purification
Expression and purification of P. falciparum HDP and mutant proteins were performed as described previously17,19. In brief, BL21(DE3) cells transformed with the pCold HDP plasmid were grown at 37°C in 2 × YT containing 100 µg/mL ampicillin. Expression was induced with 1 mM IPTG at 15°C overnight. The recombinant protein was mainly localized in the inclusion bodies and refolded as described previously. HDP protein was purified with a HiTrap Q HP anion exchange column (GE Healthcare). The colorless apo-HDP was obtained. The protein concentration was determined using BCA protein assay reagent (Thermo Scientific). HDP has an estimated extinction coefficient of 36.0 mM−1cm−1 at 280 nm. The purified HDP, dissolved in an appropriate buffer, was incubated with two equivalents of heme in N, N-dimethylformamide and passed through a Sephadex G25 column (GE Healthcare) to remove the unbound heme. The heme concentration in stock solution was determined in 0.1 M NaOH using an extinction coefficient of 58.4 mM−1cm−1 at 385 nm. The PrimeSTAR max mutagenesis kit (Takara Bio) was used to prepare the mutants.
Hz formation assay
The Hz formation assay was performed as described with some modification14,17,19. A 10-mM fresh stock solution of hemin in N,N-dimethylformamide was prepared. The hemozoin formation assays were performed at 37°C for 15 min in 0.5 M sodium acetate pH 5.2 containing 5 μM HDP and 600 μM hemin. After incubation, the reaction mixture was centrifuged at 15,000 × g for 10 min at room temperature. The precipitates were washed three times with 1 mL 0.1 M NaHCO3, 2.5% SDS, pH 9.1, followed by three washes with deionized water. The final adducts were solubilized in 0.1 M NaOH, and the amount of heme was determined by the absorption spectra. Hemin concentration was determined in 0.1 M NaOH using an extinction coefficient of 58.4 mM−1cm−1 at 385 nm.
Spectroscopy
Electronic absorption spectra were obtained using a Shimadzu UV-3150 spectrometer. The path-length of the cell was 0.2 cm. The sample concentration was approximately 40 μM in 50 mM MOPS–NaOH, pH 7.0. Resonance Raman measurements were performed with a nanosecond pulse laser operating at 1 kHz39,40. Probe pulses at 410 nm were second harmonics of the output of an Nd:YLF-pumped Ti:sapphire laser (Photonics Industries, TU-L). For the nanosecond time-resolved resonance measurements, probe pulses at 436 nm were second harmonics of the output of an Nd:YLF-pumped Ti:sapphire laser (Photonics Industries, TU-L), and pump pulses at 532 nm were generated with a diode-pumped Nd:YAG laser (Megaopto, LR-SHG). Pulse widths of the pump and probe pulses were 20 and 30 ns, respectively. Scattered light was detected with a liquid nitrogen-cooled CCD camera (Roper Scientific, Spec-10:400B/LN) attached to a custom-made prism prefilter (Bunko Keiki) equipped with a single spectrograph (HORIBA, iHR550). Raman shifts were calibrated with Raman bands of cyclohexane, carbon tetrachloride, indene, and ferrocyanide. The sample concentration was approximately 100 μM in 50 mM MES–NaOH, pH 5.6. Sample solutions for the resonance Raman measurements were contained in an airtight 10-mm ϕ NMR tube and spun with a spinning cell device rotating at 2000 rpm at room temperature. EPR spectra were measured on a Bruker EMX Plus Premium. Measurements were carried out at the X-band (9.50 GHz) microwave frequency. The sample concentration was approximately 500 μM in 50 mM MES–NaOH, pH 5.6.
Author Contributions
H.I., S.A. and Y.M. designed the experiments. K.N. and H.I. prepared samples and performed experiments. K.N. performed spectral analysis. H.I. and Y.M. prepared the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by the Human Frontier Science Program CDA-00046/2009 (to H.I.); Grants-in-Aid for Scientific Research for Young Scientist (B) 22770141 and 24770124 (to H.I.), Scientific Research on the Priority Areas (Molecular Science for Supra Functional Systems) 19056013 (to Y.M.), and Scientific Research (B) 2350084 (to S.A.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan; and by the Join Studies Program (2011–2012) of the Institute for Molecular Science.
References
- Bonilla J. et al. Effects on growth, hemoglobin metabolism and paralogous gene expression resulting from disruption of genes encoding the digestive vacuole plasmepsins of Plasmodium falciparum. Int. J. Parasitol. 37, 317–327 (2007). [DOI] [PubMed] [Google Scholar]
- Goldberg D., Slater A., Cerami A. & Henderson G. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc. Natl. Acad. Sci. USA 87, 2931–2935 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou A. & Fitch C. Mechanism of hemolysis induced by ferriprotoporphyrin IX. J. Clin. Invest. 68, 672–677 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Tekwani B., Singh R. & Chauhan V. Artemisinin, an endoperoxide antimalarial, disrupts the hemoglobin catabolism and heme detoxification systems in malarial parasite. J. Biol. Chem. 274, 19383–19388 (1999). [DOI] [PubMed] [Google Scholar]
- Vincent S. H. Oxidative effects of heme and porphyrins on proteins and lipids. Semin. Hematol. 26, 105–113 (1989). [PubMed] [Google Scholar]
- Sigala P. et al. Direct tests of enzymatic heme degradation by the malaria parasite Plasmodium falciparum. J. Biol. Chem. 287, 37793–37807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan T. Recent advances in understanding the mechanism of hemozoin (malaria pigment) formation. J. Inorg. Biochem. 102, 1288–1299 (2008). [DOI] [PubMed] [Google Scholar]
- Egan T. Haemozoin formation. Mol. Biochem. Parasitol. 157, 127–136 (2008). [DOI] [PubMed] [Google Scholar]
- Meshnick S. et al. Second-generation antimalarial endoperoxides. Parasitol. Today 12, 79–82 (1996). [DOI] [PubMed] [Google Scholar]
- Sullivan D., Gluzman I., Russell D. & Goldberg D. On the molecular mechanism of chloroquine's antimalarial action. Proc. Natl. Acad. Sci. USA 93, 11865–11870 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagola S. et al. The structure of malaria pigment β-haematin. Nature 404, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- Bendrat K., Berger B. & Cerami A. Haem polymerization in malaria. Nature 378, 138–139 (1995). [DOI] [PubMed] [Google Scholar]
- Fitch C., Cai G., Chen Y. & Shoemaker J. Involvement of lipids in ferriprotoporphyrin IX polymerization in malaria. Biochim. Biophys. Acta 1454, 31–37 (1999). [DOI] [PubMed] [Google Scholar]
- Sullivan D., Gluzman I. & Goldberg D. Plasmodium hemozoin formation mediated by histidine-rich proteins. Science 271, 219–222 (1996). [DOI] [PubMed] [Google Scholar]
- Pandey A. et al. Hemozoin formation in malaria: a two-step process involving histidine-rich proteins and lipids. Biochem. Biophys. Res. Commun. 308, 736–743 (2003). [DOI] [PubMed] [Google Scholar]
- Sullivan D. Theories on malarial pigment formation and quinoline action. Int. J. Parasitol. 32, 1645–1653 (2002). [DOI] [PubMed] [Google Scholar]
- Jani D. et al. HDP-a novel heme detoxification protein from the malaria parasite. PLoS pathog. 4, e1000053 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh M. et al. Protein complex directs hemoglobin-to-hemozoin formation in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 110, 5392–5397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani K., Ishikawa H., Aono S. & Mizutani Y. Heme-binding properties of heme detoxification protein from Plasmodium falciparum. Biochem. Biophys. Res. Commun. 439, 477–480 (2013). [DOI] [PubMed] [Google Scholar]
- Mury F. et al. Alpha-glucosidase promotes hemozoin formation in a blood-sucking bug: an evolutionary history. PloS One 4, e6966 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani T., Iizuka T. & Waterman M. Studies on modified hemoglobins. 3. Spin states of ferric hemoglobin, semi-hemoglobin, and isolated subunit chains. J. Biol. Chem. 246, 7683–7689 (1971). [PubMed] [Google Scholar]
- Spiro T. G. Resonance Raman spectra of heme proteins and other metalloproteins. Wiley (1988). [Google Scholar]
- Rakshit G., Spiro T. G. Resonance Raman spectra of horseradish peroxidase: evidence for anomalous heme structure. Biochemistry 13, 5317–5323 (1974). [DOI] [PubMed] [Google Scholar]
- Hu S., Smith K. & Spiro T. G. Assignment of protoheme resonance Raman spectrum by heme labeling in myoglobin. J. Am. Chem. Soc. 118, 12638–12646 (1996). [Google Scholar]
- Ishikawa H. et al. Unusual heme binding in the bacterial iron response regulator protein: spectral characterization of heme binding to the heme regulatory motif. Biochemistry 50, 1016–1022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray G. et al. How far can proteins bend the FeCO unit? Distal polar and steric effects in heme proteins and models. J. Am. Chem. Soc. 116, 162–176 (1994). [Google Scholar]
- Smulevich G., Evangelista-Kirkup R., English A. & Spiro T. G. Raman and infrared spectra of cytochrome c peroxidase-carbon monoxide adducts in alternative conformational states. Biochemistry 25, 4426–4430 (1986). [DOI] [PubMed] [Google Scholar]
- Spiro T. G. & Wasbotten I. CO as a vibrational probe of heme protein active sites. J. Inorg. Biochem. 99, 34–44 (2005). [DOI] [PubMed] [Google Scholar]
- Rousseau D. L. et al. Ligand-Protein Interactions in Mammalian Nitric Oxide Synthase. The Smallest Biomolecules: Diatomics and their Interactions with Heme Proteins Ghosh, A. (ed.). Elsevier, Amsterdam, (2008). [Google Scholar]
- Ishikawa H. et al. Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol. Cell 19, 171–181 (2005). [DOI] [PubMed] [Google Scholar]
- Leys D. et al. Crystal structures of an oxygen-binding cytochrome c from Rhodobacter sphaeroides. J. Biol. Chem. 275, 16050–16056 (2000). [DOI] [PubMed] [Google Scholar]
- Peterson E., Friedman J., Chien E. & Sligar S. G. Functional implications of the proximal hydrogen-bonding network in myoglobin: a resonance Raman and kinetic study of Leu89, Ser92, His97, and F-helix swap mutants. Biochemistry 37, 12301–12319 (1998). [DOI] [PubMed] [Google Scholar]
- Choi C. et al. Spectroscopic characterization of the heme-binding sites in Plasmodium falciparum histidine-rich protein 2. Biochemistry 38, 16916–16924 (1999). [DOI] [PubMed] [Google Scholar]
- Schneider E. & Marletta M. Heme binding to the histidine-rich protein II from Plasmodium falciparum. Biochemistry 44, 979–986 (2005). [DOI] [PubMed] [Google Scholar]
- Chim N., Iniguez A., Nguyen T. & Goulding C. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J. Mol. Biol. 395, 595–608 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G. et al. Histidine 20, the crucial proximal axial heme ligand of bacterial heme oxygenase Hmu O from Corynebacterium diphtheriae. J. Biol. Chem. 275, 17494–17500 (2000). [DOI] [PubMed] [Google Scholar]
- de Villiers K., Kaschula C., Egan T. & Marques H. Speciation and structure of ferriprotoporphyrin IX in aqueous solution: spectroscopic and diffusion measurements demonstrate dimerization, but not μ-oxo dimer formation. J. Biol. Inorg. Chem. 12, 101–117 (2007). [DOI] [PubMed] [Google Scholar]
- Pisciotta J. et al. The role of neutral lipid nanospheres in Plasmodium falciparum haem crystallization. Biochem. J. 402, 197–204 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Ishikawa H. & Mizutani Y. Protein dynamics of isolated chains of recombinant human hemoglobin elucidated by time-resolved resonance Raman spectroscopy. J. Phys. Chem. B 116, 1992–1998 (2012). [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Ishikawa H., Aono S. & Mizutani Y. Structural dynamics of proximal heme pocket in HemAT-Bs associated with oxygen dissociation. Biochim. Biophys. Acta 1824, 866–872 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information