Abstract
Most rare-variant association tests for complex traits are applicable only to population-based or case-control resequencing studies. There are fewer rare-variant association tests for family-based resequencing studies, which is unfortunate since pedigrees possess many attractive characteristics for such analyses. Family-based studies can be more powerful than their population-based counterparts due to increased genetic load and further enable the implementation of rare-variant association tests that, by design, are robust to confounding due to population stratification. With this in mind, we propose a rare-variant association test for quantitative traits in families; this test integrates the QTDT approach of Abecasis et al. [Abecasis, et al. 2000a] into the kernel-based SNP association test KMFAM of Schifano et al. [Schifano, et al. 2012]. The resulting within-family test enjoys the many benefits of the kernel framework for rare-variant association testing, including rapid evaluation of p-values and preservation of power when a region harbors rare causal variation that acts in different directions on phenotype. Additionally, by design, this within-family test is robust to confounding due to population stratification. While within-family association tests are generally less powerful than their counterparts that use all genetic information, we show that we can recover much of this power (while still ensuring robustness to population stratification) using a straightforward screening procedure. Our method accommodates covariates and allows for missing parental genotype data, and we have written software implementing the approach in R for public use.
Keywords: Sequencing, rare variants, quantitative traits, family studies
Introduction
The emergence of next-generation sequencing technology, along with the development of the exome chip, has led many investigators to study the role of rare genetic variation in complex human traits. Rather than analyze rare variants individually, many statistical approaches for rare-variant association mapping employ grouping strategies that aggregate rare variants in a gene or region for analysis to improve power. These approaches can be broadly categorized as either burden tests that collapse grouped rare variants into a single aggregate variable that is then regressed on phenotype [Li and Leal 2008; Madsen and Browning 2009; Morris and Zeggini 2010; Zawistowski, et al. 2010], kernel tests that relate phenotype to rare variants in a region as a function of a variance component (SKAT, [Wu, et al. 2011]), and unified tests that combine burden and kernel tests together (SKAT-O, [Lee, et al. 2012]). Burden tests are preferred when a region harbors rare causal variants that all act in the same direction on phenotype (all protective or all deleterious) whereas kernel tests are optimal when a region harbors rare causal variants that act in different directions on phenotype [Wu, et al. 2011].
Although these rare-variant methods generally have improved power compared to tests of individual rare variants, almost all of these tests are restricted to case-control or population-based study designs and cannot be used in family-based studies. Family-based designs have several advantages over population-based designs in that they enable the use of statistics that, by design, are robust to confounding due to population stratification. Family designs also can solve genetic problems that are hard to answer in population-based studies. For example, sequencing the parents of affected subjects can identify de novo mutations and also allow the study of rare homozygous genotypes, which are difficult to find in population-based designs [Do, et al. 2012]. Families are also attractive to study because they often provide increased genetic load for a disease or trait: while carriers of a minor risk allele will be hard to sample in the general population, they are more likely to be found in families of probands [Zollner 2012]. Finally, family studies allow the study of the segregation pattern of complex disease [Ott, et al. 2011]. Because of these appealing features and the fact that there are many familial samples from past linkage studies, family-based resequencing studies are gaining in popularity. Several recent studies have identified disease-associated rare variants through family-based designs, including rare variants associated with multiple sclerosis [Ramagopalan, et al. 2011], simplex autism [Krumm, et al. 2013], dilated cardiomyopathy [Norton, et al. 2011], and Alzheimer’s disease [Cruchaga, et al. 2012].
Recently, a few methods have been proposed for rare-variant association testing in families. Schaid et al. [Schaid, et al. 2013] developed a method for complex traits that accounts for relatedness among study subjects. Their method took a retrospective view of the sample, which assumes that the outcome is fixed while the genotype is random, and is particularly appealing for the analysis of datasets that are collected under non-random ascertainment (such as those collected for linkage studies). Chen et al. [Chen, et al. 2013] developed a rare-variant test for quantitative traits in families by extending kernel-machine methods [Kwee, et al. 2008; Wu, et al. 2011] to pedigree analysis by inserting a random familial effect due to shared polygenes within the modeling framework; a similar idea was employed by Schifano et al. [Schifano, et al. 2012] and Oualkacha et al [Oualkacha, et al. 2013]. Jiang and McPeek [Jiang and McPeek 2014] adopted a similar strategy to extend the SKAT-O [Lee, et al. 2012] method to family studies of quantitative traits. Although the methods of both groups adjust for kinship in family studies, they do not consider potential bias caused by population stratification. Population stratification can lead to substantially inflated false-positive rates in sequencing studies of rare variants [Epstein, et al. 2012; Jiang, et al. 2013; Liu, et al. 2013], and standard GWAS approaches to correct for such stratification (such as principal components or EMMAX [Kang, et al. 2010]) may not be effective when applied to rare variants [Mathieson and McVean 2012]. Therefore, a rare-variant association test that maintains validity in the presence of such stratification is needed. Ionita-Laza et al. [Ionita-Laza, et al. 2013] proposed such a method based on the family-based association test (FBAT) framework. Although this method is robust to population stratification, it ignores between-family information that could perhaps be exploited to boost power. Fang et al. [Fang, et al. 2012; Fang, et al. 2013] used between-family information for this purpose in an adaptive rare-variant association test for quantitative traits; however, the procedure requires computationally intensive permutations for inference, so it is unclear whether the approach is scalable to large-scale resequencing efforts.
In this paper, we propose a novel two-stage method for rare-variant analysis of quantitative traits in trios and nuclear families. The approach is based on the QTDT (quantitative transmission disequilibrium test) framework of Abecasis et al. [Abecasis, et al. 2000a] for SNP association mapping. The QTDT framework decomposes the observed individual genotypes into between-family and within-family components. The within-family component is robust to population stratification, while the between-family component is sensitive to the phenomenon. In this paper, we calculate the within-family component for each rare variant in a region, and then integrate these components within the kernel procedure KMFAM of Schifano et al. [Schifano, et al. 2012], which was previously developed for SNP-set association testing of quantitative traits in families. Specifically, within KMFAM, we create a kernel matrix based on the within-family component, and then use this kernel matrix to test for association with phenotype using a modified score statistic. By using the within-family component only, our rare-variant association test for quantitative traits is robust to confounding due to population stratification. Also, the approach calculates p-values analytically rather than via resampling and is thus scalable to exome sequencing and whole-genome resequencing studies. Because the approach relies on a kernel framework, it also preserves power when a region contains a mixture of trait-increasing and trait-decreasing variation. The approach also allows for covariates and, for nuclear families, can be implemented when phenotype and genotype data on parents are missing, so it can be applied in the study of quantitative traits related to late-onset diseases.
A potential drawback of using only within-family information for analysis is that power is reduced by ignoring the (sensitive) between-family information within the analysis [Ionita-Laza, et al. 2013]. However, borrowing ideas from Purcell et al. [Purcell, et al. 2005] and Van Steen et al. [Van Steen, et al. 2005], we propose using between-family information as a screening tool to identify the most interesting regions (based on the magnitude of the p-value for the region) that merit further investigation. We then apply our within-family test to only these top regions, thereby reducing the multiple-testing burden (compared to within-family testing of all regions) and potentially gaining power. We note that the first stage of the analysis (using the between-family information) is independent of the second stage (which uses orthogonal within-family information). We also note that, by using within-family information in the second stage, our approach is still robust to confounding due to population stratification.
In subsequent sections, we first describe the KMFAM procedure and then, for rare variants, discuss how we integrate the QTDT framework into the model to make the method robust to population stratification. We next describe our screening procedure to improve power. We then evaluate our approaches using simulated sequence data in trios and nuclear families and show how screening can improve power of within-family testing while maintaining an appropriate type I error rate, even under population stratification. Finally, we summarize our method and discuss potential extensions.
Materials and Methods
Notation and KMFAM Model
We initially present the KMFAM model of Schifano et al. ([Schifano, et al. 2012] (also used by Chen et al. [Chen, et al. 2013]), and then show how to modify the framework to develop a within-family association test of rare variation for quantitative traits. As in KMFAM, we assume a sample of N nuclear families that are genotyped for s rare-variants in a gene or region of interest. Let Yij denote the quantitative outcome for the jth individual in the ith family, where i=1,2,3…N and j=1,2,..ni. We define Xij as a c×1 vector that represents the covariates for the jth individual in the ith family and further define Gij as an s×1 vector that represents the genotypes of the s rare variants for each subject (where each rare-variant genotype is coded as the number of copies of the rare allele the subject possesses at each site). We assume that the outcome, Yij, follows a multivariate normal distribution with mean and variance defined through the model:
| (1) |
where α is a c×1 vector of coefficients for Xij and β is a sx1 vector of coefficients for Gij. While we assume the coefficients in α are fixed effects, we instead assume the coefficients for the genotype effects β are random and follow an arbitrary distribution with variance τ. With this assumption, we can test for association between rare variants and phenotype by considering the hypothesis τ = 0 rather than an s degree of freedom fixed-effects test: β1 = β2 = β3 = ..... = βs = 0, which will have low power.
To complete the formulation of model (1) for pedigree data, we let fij denote the random effect to account for within-family correlation due to shared polygenes. We assume the effect within a family follows a multivariate normal distribution: , where Φi is the kinship matrix for family i and is the variance due to the effect of polygenes. We also define as the random error term. From model (1), we calculate the variance of outcome as
| (2) |
where K=GIGT is the kernel matrix, and G is a matrix composed of the vectors Gij such that each row is GijT for a single individual. Note that here we use a linear kernel, but if previous information is available for the rare variants in the gene, the use of other kernels, such as a linear weighted kernel, can increase power (Wu et al., 2011); in this case I can be replaced with a weighting matrix Z, where elements in Z represent the weight. There are several methods to specify the weight, based on the belief of the variant’s contribution to the outcome. One common method is to calculate weight as a function of the minor allele frequency (MAF); Wu et al. [Wu, et al.] considered such a weight that modeled MAF using a Beta distribution, but other weights are possible, as well.
To test whether the rare variants in the gene are associated with the outcome, we construct a variance component score test derived from model (1) [Lin 1997; Zhang and Lin 2003]. The null hypothesis is H0: τ = 0, and the test statistic takes the form:
| (3) |
where all parameters are estimated under the null hypothesis. We define V̂0 and α̂0 as the estimates of V in (1) and α under the null. Further, we define a projection matrix , such that PV0P = P. Thus, under the null, we have
| (4) |
where λi are eigenvalues of , here . As are independently and identically distributed random variables, Q is distributed as an asymptotic mixture of chi-square distributions, and the p-values can be calculated using the Davies method [Davies 1980].
Robust Rare-Variant Association Test
One issue with the KMFAM framework described above is that the resulting score tests from model (1) are sensitive to population stratification. To resolve this issue, we integrate the QTDT [Abecasis, et al. 2000a] framework into our model. The QTDT framework decomposes the observed genotype Gij into a between-family component (which we denote by Bij) and an orthogonal within-family component (which we denote by Wij). The between-family component takes the following value:
Once we obtain the between-family component, we then construct the within-family component, Wij, by subtracting the between-family component from the observed genotype such that Wij=Gij−Bij.
By design, association analyses of complex traits that base inference on the within-family component Wij, are robust to population stratification. Based on this observation, we can construct a robust rare-variant association test for trios and nuclear families by replacing the observed genotypes G in the kernel matrix K described in (2) with their corresponding within-family components W. We then construct the score statistic Q in (3) as before to derive our robust family-based association test.
Screening Procedure
Although the QTDT framework ensures the robustness of our proposed score test to potential confounding due to population stratification, the discarding of between-family information when confounding due to population stratification is not an issue can lead to sizable power loss compared to use of the observed genotype. In attempts to restore the power of our within-family association test to levels anticipated when using observed-genotype information, we suggest a two-stage screening approach that uses both the within- and between-family rare-variant information. In the first stage, we use between-family information to screen and identify the top regions for follow up. If parental phenotype and genotype information are available, we carry out the first stage by performing the SKAT [Wu, et al. 2011] test on parents only, and then select a subset of regions for follow-up investigation based on smallest p-values. If parental information is unavailable, we instead conduct the first-stage screening by applying KMFAM to the outcomes and between-family components of the offspring. In the second stage, we construct the robust test (using the within-family components calculated for the offspring) only on those top regions selected from the first stage. By only testing a reduced number of regions in the second stage using the within-family component, we reduce the number of robust tests that are conducted thereby reducing the multiple-testing burden and increasing power. As discussed in Abecasis et al. [Abecasis, et al.], the between-family and within-family components are orthogonal to each other, such that the first-stage and second-stage tests are independent.
Type I Error Simulations
We evaluated the type I error and power of our approach using simulated sequencing data. We used cosi [Schaffner, et al. 2005] to simulate sequence data for a pool of 5000 European and 5000 African haplotypes, each of length 30 kb. Rare variants were defined as variants with a minor-allele frequency greater than 0% and less than 3% in the region. To simulate family data, we randomly paired subjects within each population and simulated offspring by sampling one haplotype from each parent. When considering nuclear families with 2 or more offspring, we performed simulations for the situation where all parental information is available, as well as where 20–100% parental information is missing.
Using this concept, we first performed type 1 error rate simulations to verify that our method is robust to population stratification. We simulated the outcome through the null model:
| (5) |
where γ is the mean trait difference between European and African subjects, IAfrican, ij is an indicator variable that is 1 if the subject is African and 0 otherwise, and all other terms are the same as defined in model (1). We specified fij and eij such that the overall trait heritability was 0.35. To induce confounding due to population stratification in our simulations, we first assumed our sample consisted of a mixture of European and African families, with the percentage of European families ranging from 25% to 75%. We then assumed a value of γ in model (5) that ranged from 0 (no confounding due to population stratification) to 3 (extreme confounding due to population stratification).
Power Simulations
To estimate power, we simulated a region of 300 kb, divided into 10 non-overlapping regions of 30 kb each, and selected one region at random as causal (the other 9 regions are assumed to be independent of outcome). To generate trait data for each subject based on the causal region, we used the idea of Wu et al. [Wu, et al. 2011] and assumed a certain percentage (5% or 15%) of rare variants (defined as variants with a minor allele frequency less than 3%) in the region influenced the outcome, with the effect size of a causal variant defined as β = c× |log10 MAF|, where we varied the constant c among values between 0.4 and 0.6. We then included these effects due to rare variants within model (5) to simulate the outcome. To keep power at a reasonable range for the 300kb region, we fixed γ at 0.25 for power simulations under stratification. As with the null simulations, we assumed the trait heritability was 0.35.
Results
Type I Error
We first performed type I error rate simulations on parent-offspring trios to demonstrate that population stratification can lead to spurious rare-variant association with quantitative traits in families. Figure 1 presents type I error results for two methods: our robust rare-variant approach that uses within-family information from the offspring only and a SKAT test of rare-variant association that uses the observed offspring genotype (constituting both the within- and between-family components). For these simulations, simulated datasets consisted of 500 trios where 50% are of European descent and the remaining 50% are of African descent. When the mean trait difference between European and African populations is 0 (such that there is no confounding due to population stratification), both the within-family test and observed-genotype test had appropriate type I error. However, when we induced confounding due to population stratification by assuming a non-zero mean trait difference between Africans and Europeans, we found the standard SKAT test using the observed genotype had inflated type I error. Our robust rare-variant association test, in contrast, maintained the proper type I error rate under confounding.
Figure 1.
Empirical type 1 error rates of rare-variant association tests applied to 30-kb sequenced regions in parent-offspring trios. Simulated datasets consisted of 500 parent-offspring trios (50% of European ancestry, 50% of African ancestry). The mean trait difference between European and African subjects varies from 0 (no stratification) to 3 (extreme stratification). Total trait heritability is 0.35. We analyzed each simulated trio dataset twice: once using SKAT to analyze the observed offspring genotypes (“Observed genotype,” blue line) and once using our proposed kernel test that used only the within-family component of the observed offspring genotypes (“Within component,” red line). Each result is based on 1000 replicates.
We next performed another set of type I error simulations, where we assumed datasets consisting of 500 nuclear families each with two children. We varied the proportion of nuclear families that were of European origin between 25% and 75% and assumed the mean trait difference between African and European samples to be 2 (thereby inducing confounding due to population stratification). We further assumed the proportion of nuclear families within each dataset that was missing parental genotype information ranged from 0% to 100%. In our first set of simulations (shown in Figure 2), we studied the type I error rates of methods assuming examination of the 30-kb region in its entirety. We compared the type I error rates using the observed genotype information in the offspring only, accounting for kinship (which corresponds to the KMFAM test of Schifano et al. as well as the test of Chen et al. [Chen, et al. 2013]), as well as using our robust rare-variant association test that relies only on the within-family information in the offspring. Our results indicated that rare-variant association tests using observed genotype information led to considerable inflation in type I error rates across different simulation models, whereas our robust within-family association test remained valid in all situations. The validity of the robust rare-variant association test was confirmed both when parental genotype information was available on all participants, as well as when such genotype information was completely absent in the dataset. Thus, for late-onset diseases in which parental information might not be available, our method is still robust to population stratification.
Figure 2.
Empirical type 1 error rates of rare-variant association tests applied to 30-kb sequenced regions for nuclear families with 2 children each (total heritability is 0.35). Simulated datasets consisted of 500 nuclear familes each with 2 children. Percentage of European varies from 25% to 75%. Percentage of missing parents varies from 0% to 100%. The mean trait difference between European and African subjects is 2. For each simulated dataset, we used KMFAM to analyze the observed offspring genotypes (“Observed genotype,” brown bars) and used our proposed kernel test that used only the within-family component of observed offspring. For within-family results, we present findings assuming percentage of missing parents was 0%, 20%, 60%, and 100%. Each result is based on 1000 replicates.
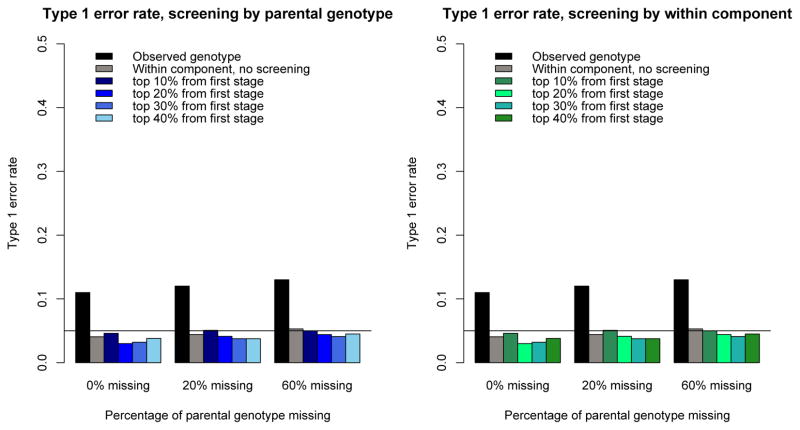
We performed a final set of type I error simulations for nuclear families of size two under our proposed screening scheme where, in this instance, we split the 300-kb region into 10 non-overlapping regions, each of size 30 kb. Using between-family information, we identified a subset of regions for follow up (based on p-value) that we then investigated further using the within-family component. Our results are shown in Figure 3. Overall, our results show that our screening procedure (conducted using either parental information or between-family information in siblings, if parents are not available) preserved type I error across models, with differing missing parental information as well as different proportions of regions that were then followed up using within-family information. These results demonstrate that our screening procedure maintains appropriate type I error, even when there is confounding due to population stratification, due to the fact that the between-family component and within-family component of the offspring genotype are orthogonal to one another.
Figure 3.
Empirical type 1 error rates of rare-variant association tests applied to ten 30-kb sequenced regions for nuclear families with 2 children each (total heritability is 0.35). Simulated datasets consisted of 500 nuclear families each with 2 children. The mean trait difference between European and African subjects is 0.25. For each simulated dataset, we first used KMFAM to analyze the observed offspring genotypes (“Observed genotype,” black bars); we then used our proposed kernel test to analyze the within-family component of offspring without screening, and then applied screening procedures. We applied two screening processes: screening by parental information (blue bars) and screening by the between-family component (green bars). Left: screen by parental genotype. Right: screen by within-family component. Top 10% to 40% of regions with smallest p-value were selected through the screening process and analyzed in the second stage. Each result is based on 1000 replicates.
Power
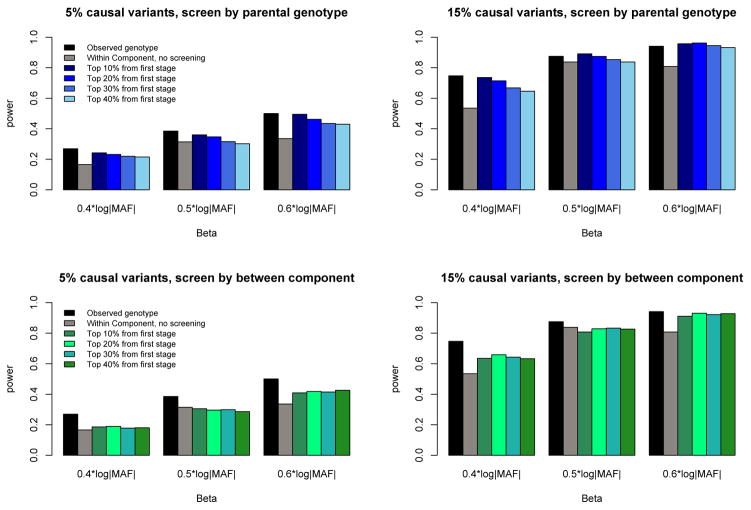
In the previous section, we showed that our robust rare-variant association test that uses the within-family component remains valid in the presence of population stratification. We next studied the power of our proposed robust test to detect association with a trait under various trait-influencing models. We assumed either 5% or 15% of rare variants in a region were causal and assumed the effect size of such causal variants was β = c×|log10 MAF|, where c ranged from 0.4 to 0.6. We first compared the power of our robust within-family association test to the standard observed-genotype test considered by Chen et al. and Schifano et al. under models with no population stratification (to ensure the power of the observed-genotype test was valid). We generated sequence and trait data on 500 nuclear families each with two offspring. We first analyzed the observed rare-variant genotypes in the family using the kernel test of Chen et al., and then repeated the analysis using our robust within-family association test. As shown in Figure 4, the power of the kernel test using observed genotype information (shown as black bars) is, as expected, more powerful than the same test using within-family information alone (shown in gray bars) across different simulation models. In attempts to see whether we could restore some power to the robust test, we then applied our screening procedure to these simulated datasets using between-family information. For each dataset, we tested the between-family components of each of the 10 regions, and then subsequently considered only the top 10%, 20%, 30%, or 40% (based on minimum p-value) of these regions using our within-family test. The results show that, when screening is performed using parental genotype and trait information, our screening procedure restores power to levels similar to those using the observed-genotype information (see top panels of Figure 4). If screening is instead performed using between-family information, the robust within-family association test also shows a power increase, although it is not as notable as using parental information (see bottom panels of Figure 4). Thus, it appears that our initial screening step improves the power of the within-family association test, while preserving appropriate type I error under the null.
Figure 4.
Empirical power of rare-variant association tests applied to ten 30-kb sequenced regions for nuclear families without stratification. Simulated datasets consisted of 500 European families each with 2 children. Three effect sizes were used: 0.4×|log10 MAF| 0.5×|log10 MAF|, and 0.6×|log10 MAF|. As in Figure 3, for each dataset we used KMFAM to test the observed genotype; then we used our method to test the within-family component without screening, and then applied two screening methods. Top panel: screen by parental genotype. Bottom panel: screen by between-family component. Each result is based on 1000 replicates.
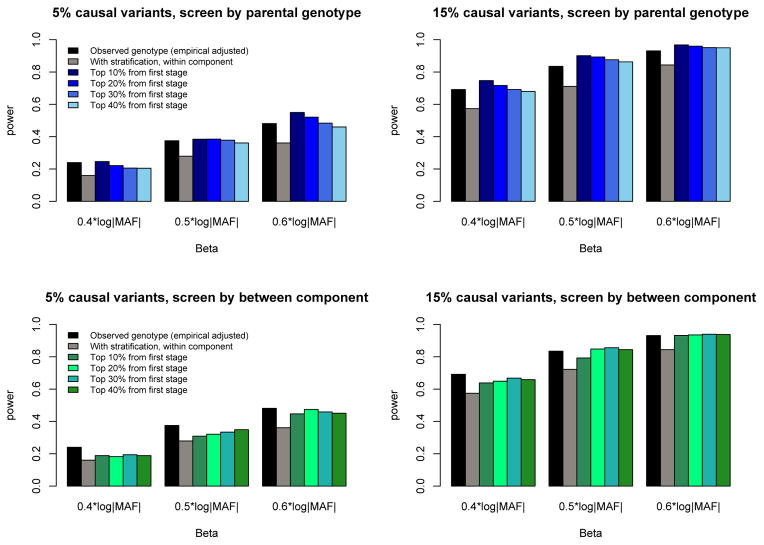
While we obtained our results in Figure 4 under simulation models that assumed no confounding due to population stratification, we also observed similar trends in simulation models that were generated with confounding due to population stratification. Figure 5 presents power results under confounding due to population stratification that assumed a mean trait difference between African and European samples. For the observed-genotype analyses, we report empirical adjusted power accounting for population stratification (black bars) rather than the naïve power that does not account for population stratification (which is invalid). To obtain the empirical adjusted power, we simulated and analyzed null datasets generated with the same amount of confounding as in the power datasets and used the empirical distribution of the null tests to determine an appropriate threshold to declare significance. We then evaluated the observed genotype’s power based on this empirical threshold. The remaining bars denote the power of the robust within-family association test, along with variations that screen using parental or between-family information. The results show that screening can improve power of the robust rare-variant test, particularly as the percentage of causal variants and the magnitudes of their effect increase. The results in Figure 5 were for simulated datasets consisting of nuclear families with two offspring each; we saw similar trends when analyzing parent-offspring trios, as well as when a region harbored variants that acted in different directions on outcome (see Supplemental Figures 1 and 2).
Figure 5.
Empirical power of rare-variant association tests applied to ten 30-kb sequenced regions for nuclear families with/without stratification. Simulations were performed under population structure such that 25% of families are European, and the mean trait difference between European and African subjects is 0.25. Black bars denote empirical power of observed genotypes adjusted for population stratification. Three effect sizes were used: 0.4×|log10 MAF|, 0.5×|log10 MAF|, and 0.6×|log10 MAF|. Top panel: screen by parental genotype. Bottom panel: screen by between-family component. Each result is based on 1000 replicates.
Discussion
In this paper, we proposed a kernel method for analyzing rare-variant sequencing studies in trios and nuclear families that is robust to confounding due to population stratification. We also introduced a screening procedure using parental or between-family information to improve the power of this robust test and showed that this procedure can increase power to levels similar to those of the observed-genotype test when confounding due to stratification is not an issue. In addition to robustness, our approach has many other practical features. The method easily allows for covariates and permits rapid calculation of p-values using analytic procedures. We have implemented our procedure in R software, which is available from our website (see Web Resources). Our approach is computationally efficient, as the analysis of a 30-kb region for 500 nuclear families each of size two takes on average 53.08 seconds on a 768 processor running Linux OS with 2.6 gigahertz of RAM. Based on the computational speed, we believe the approach can be scaled reasonably to whole-exome or whole-genome resequencing studies on a multi-node cluster.
Our approach currently considers either parent-offspring trios or nuclear families with an arbitrary number of offspring. In future work, we intend to extend the approach to consider pedigrees of any arbitrary size or structure. Saad and Wijsman [Saad and Wijsman 2014] have highlighted the value of using such large families for rare-variant analysis by showing that sequencing a small proportion of subjects while genotyping the remaining subjects using a sparser set of markers, and then subsequently using pedigree information to impute the missing variants among genotyped subjects, will lead to greater power compared to analyzing the sequenced individuals alone. Given such appealing features of such large pedigrees, we will expand our framework to handle such pedigrees by leveraging the work of Abecasis et al [Abecasis, et al. 2000b]. In that work, if a person is a genotyped founder, the within-family component is defined as 0; if a person is genotyped but not a founder, the within-family component is defined as genotype minus between-family component or sufficient statistics; if a person is not genotyped, then the within-family component is left as undefined. We can then incorporate the within-family information for each subject within our kernel test for inference.
Family-based genetic studies of complex traits occasionally have information available from additional unrelated singletons. While we cannot use these individuals within our robust within-family association test of rare variation, the information from such singletons can be used in our screening step (treating them in the same way as the parental information) to identify the most interesting regions for follow up using the robust test. Such information could be helpful in screening and should not affect the validity of the second-stage robust test, even if there is confounding due to population stratification and/or coverage differences between the family and unrelated arms of the study.
In this paper, we focused on familial studies of quantitative traits. To extent our method to binary traits, we can look into applying estimating-equation procedures similar to those proposed by Wang et al. [Wang, et al. 2013] for analyzing observed genotypes. A few additional methods have discussed familial rare-variant analysis of binary traits: Preston and Dudbridge [Preston and Dudbridge 2014] compared the power of several family-based designs for binary traits, and found that using cases from affected families and unrelated controls often has optimal power. The use of unrelated controls may raise concerns of bias caused by population stratification; to avoid this problem, one could implement an idea similar to Mirea et al. [Mirea, et al. 2012], who adopted a weighting strategy where the between-family and within-family contributions to a test statistic are weighted by a test of population-stratification bias. We will explore these ideas in future work.
Acknowledgments
This work was supported by National Institutes of Health grant HG007508. We thank Cheryl Strauss for her editorial assistance. Dr. Epstein is a paid consultant for Amnion Laboratories
Footnotes
Web Resources
Cosi simulation package, http://www.broadinstitute.org/~sfs/cosi
Epstein Software, http://www.genetics.emory.edu/labs/epstein/software
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000a;66(1):279–92. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WOC, Cardon LR. Pedigree tests of transmission disequilibrium. European Journal of Human Genetics. 2000b;8(7) doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet Epidemiol. 2013;37(2):196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Haller G, Chakraverty S, Mayo K, Vallania FL, Mitra RD, Faber K, Williamson J, Bird T, Diaz-Arrastia R, et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PLoS One. 2012;7(2):e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RB. Algorithm AS 155: The Distribution of a Linear Combination of χ2 Random Variables. Journal of the Royal Statistical Society Series C (Applied Statistics) 1980;29(3):323–333. [Google Scholar]
- Do R, Kathiresan S, Abecasis GR. Exome sequencing and complex disease: practical aspects of rare variant association studies. Hum Mol Genet. 2012;21(R1):R1–9. doi: 10.1093/hmg/dds387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein MP, Duncan R, Jiang Y, Conneely KN, Allen AS, Satten GA. A permutation procedure to correct for confounders in case-control studies, including tests of rare variation. Am J Hum Genet. 2012;91(2):215–23. doi: 10.1016/j.ajhg.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Sha Q, Zhang S. Two adaptive weighting methods to test for rare variant associations in family-based designs. Genet Epidemiol. 2012;36(5):499–507. doi: 10.1002/gepi.21646. [DOI] [PubMed] [Google Scholar]
- Fang S, Zhang S, Sha Q. Detecting association of rare variants by testing an optimally weighted combination of variants for quantitative traits in general families. Annals of Human Genetics. 2013;77(6):524–534. doi: 10.1111/ahg.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Family-based association tests for sequence data, and comparisons with population-based association tests. Eur J Hum Genet. 2013;21(10):1158–62. doi: 10.1038/ejhg.2012.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, McPeek MS. Robust Rare Variant Association Testing for Quantitative Traits in Samples With Related Individuals. Genetic epidemiology. 2014;38(1):10–20. doi: 10.1002/gepi.21775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Epstein MP, Conneely KN. Assessing the impact of population stratification on association studies of rare variation. Hum Hered. 2013;76(1):28–35. doi: 10.1159/000353270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, O’Roak BJ, Karakoc E, Mohajeri K, Nelson B, Vives L, Jacquemont S, Munson J, Bernier R, Eichler EE. Transmission disequilibrium of small CNVs in simplex autism. Am J Hum Genet. 2013;93(4):595–606. doi: 10.1016/j.ajhg.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. Am J Hum Genet. 2008;82(2):386–97. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224–37. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Variance component testing in generalized linear models with random effects. Biometrika. 1997;(84):309–326. [Google Scholar]
- Liu Q, Nicolae DL, Chen LS. Marbled inflation from population structure in gene-based association studies with rare variants. Genet Epidemiol. 2013;37(3):286–92. doi: 10.1002/gepi.21714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS genetics. 2009;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44(3):243–6. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirea L, Infante-Rivard C, Sun L, Bull SB. Strategies for genetic association analyses combining unrelated case-control individuals and family trios. American journal of epidemiology. 2012;176(1):70–79. doi: 10.1093/aje/kwr494. [DOI] [PubMed] [Google Scholar]
- Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34(2):188–93. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton N, Li D, Rieder MJ, Siegfried JD, Rampersaud E, Zuchner S, Mangos S, Gonzalez-Quintana J, Wang L, McGee S, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88(3):273–82. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nat Rev Genet. 2011;12(7):465–74. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- Oualkacha K, Dastani Z, Li R, Cingolani PE, Spector TD, Hammond CJ, Richards JB, Ciampi A, Greenwood CM. Adjusted sequence kernel association test for rare variants controlling for cryptic and family relatedness. Genet Epidemiol. 2013;37(4):366–76. doi: 10.1002/gepi.21725. [DOI] [PubMed] [Google Scholar]
- Preston MD, Dudbridge F. Utilising Family - Based Designs for Detecting Rare Variant Disease Associations. Annals of human genetics. 2014;78(2):129–140. doi: 10.1111/ahg.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Sham P, Daly MJ. Parental phenotypes in family-based association analysis. Am J Hum Genet. 2005;76(2):249–59. doi: 10.1086/427886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Dyment DA, Cader MZ, Morrison KM, Disanto G, Morahan JM, Berlanga-Taylor AJ, Handel A, De Luca GC, Sadovnick AD, et al. Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Annals of Neurology. 2011;70(6):881–886. doi: 10.1002/ana.22678. [DOI] [PubMed] [Google Scholar]
- Saad M, Wijsman EM. Power of Family - Based Association Designs to Detect Rare Variants in Large Pedigrees Using Imputed Genotypes. Genetic epidemiology. 2014;38(1):1–9. doi: 10.1002/gepi.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15(11):1576–83. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Sinnwell JP, Thibodeau SN. Multiple genetic variant association testing by collapsing and kernel methods with pedigree or population structured data. Genet Epidemiol. 2013;37(5):409–18. doi: 10.1002/gepi.21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifano ED, Epstein MP, Bielak LF, Jhun MA, Kardia SLR, Peyser PA, Lin X. SNP set association analysis for familial data. Genetic Epidemiology. 2012;36(8):797–810. doi: 10.1002/gepi.21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steen K, McQueen MB, Herbert A, Raby B, Lyon H, DeMeo DL, Murphy A, Su J, Datta S, Rosenow C. Genomic screening and replication using the same data set in family-based association testing. Nature genetics. 2005;37(7):683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- Wang X, Lee S, Zhu X, Redline S, Lin X. GEE - Based SNP Set Association Test for Continuous and Discrete Traits in Family - Based Association Studies. Genetic epidemiology. 2013;37(8):778–786. doi: 10.1002/gepi.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawistowski M, Gopalakrishnan S, Ding J, Li Y, Grimm S, Zöllner S. Extending rare-variant testing strategies: analysis of noncoding sequence and imputed genotypes. The American Journal of Human Genetics. 2010;87(5):604–617. doi: 10.1016/j.ajhg.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Lin X. Hypothesis testing in semiparametric additive mixed models. Biostatistics. 2003;4(1):57–74. doi: 10.1093/biostatistics/4.1.57. [DOI] [PubMed] [Google Scholar]
- Zollner S. Sampling strategies for rare variant tests in case-control studies. Eur J Hum Genet. 2012;20(10):1085–91. doi: 10.1038/ejhg.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]