Abstract
Enzymes that regulate histone lysine methylation play important roles in neuronal differentiation, but little is known about their contributions to activity-regulated gene transcription in differentiated neurons. We characterized activity-regulated expression of lysine demethylases and lysine methyltransferases in the hippocampus of adult male mice following pilocarpine-induced seizure. Pilocarpine drove a 20-fold increase in mRNA encoding the histone H3 lysine27-specific demethylase Kdm6b selectively in granule neurons of the dentate gyrus, and this induction was recapitulated in cultured hippocampal neurons by bicuculline and 4-aminopyridine (Bic+4AP) stimulation of synaptic activity. Because activity-regulated gene expression is highly correlated with neuronal survival, we tested the requirement for Kdm6b expression in Bic+4AP induced preconditioning of neuronal survival. Prior exposure to Bic+4AP promoted neuronal survival in control neurons upon growth factor withdrawal, however this effect was ablated when we knocked down Kdm6b expression. Loss of Kdm6b did not disrupt activity-induced expression of most genes, including that of a gene set previously established to promote neuronal survival in this assay. However using bioinformatic analysis of RNA sequencing data, we discovered that Kdm6b knockdown neurons showed impaired inducibility of a discrete set of genes annotated for their function in inflammation. These data reveal a novel function for Kdm6b in activity-regulated neuronal survival, and they suggest that activity- and Kdm6b-dependent regulation of inflammatory gene pathways may serve as an adaptive pro-survival response to increased neuronal activity.
Introduction
Stimulus-regulated changes in gene transcription play an essential role in coordinating cellular adaptations to the environment. Given the broad relevance of neuronal activity-regulated transcription to many of the adaptive processes that mediate brain plasticity - including axon outgrowth, synapse development, excitation/inhibition balance, and network homeostasis (West and Greenberg, 2011) - it is important to identify the transcriptional regulators that underlie activity-regulated changes in neuronal gene transcription.
In addition to regulating the expression and/or activity of sequence-specific DNA binding transcription factors (Lyons and West, 2011), neuronal activity also modulates gene transcription by regulating histone modifying enzymes (Graff and Tsai, 2013). The acetylation, methylation, or phosphorylation of specific amino acid residues in the N-terminal tails of the histone proteins alters chromatin structure and function to either activate or repress transcription of genes (Bonasio et al., 2010). The activity-dependent regulation of histone acetylation and phosphorylation in neurons has been well documented (Guan et al., 2002; Huang et al., 2002; Chwang et al., 2006). Multiple activity-dependent mechanisms converge to regulate these histone modifications in neurons, via the activation of Erk1/2 (Huang et al., 2002; Ciccarelli et al., 2013), the recruitment of phosphorylated CBP to CREB (Chrivia et al., 1993; Kwok et al., 1994; Chawla et al., 1998; Hu et al., 1999; Impey et al., 2002), and the nuclear export of class II histone deacetylases (Chawla et al., 2003; Nott et al., 2008; Guan et al., 2009).
Histone methylation is a complex and information-rich histone modification, however comparatively little is known regarding neuronal activity-dependent regulation of this mark or its mediators. Histones H3 and H4 can be mono-, di-, or tri-methylated at multiple lysine and arginine residues resulting in transcriptional activation or repression. Histone methylation status is controlled by members of the lysine demethylases (KDM) and lysine methyltransferases (KMT) families, each of which has specificity for the modification of histones H3 and H4 at specific amino acid residues (Black et al., 2012; Greer and Shi, 2012). Neurological disease-associated mutations have been identified in a number of these enzymes including JMJD1C (autism), JARID2 (schizophrenia), KDM5C (X-linked mental retardation), KDM5A and KDM6B (intellectual disability), suggesting the functional importance of regulating lysine methylation in the brain (Castermans et al., 2007; Iwase et al., 2007; Pedrosa et al., 2007; Najmabadi et al., 2011). Emerging evidence also links KDMs and KMTs with at least some aspects of activity-dependent brain plasticity. For example, inhibition of the histone methyltransferase Ehmt2 (a.k.a. G9a/Glp) in the entorhinal cortex results in enhancement of fear conditioning and dysregulation of gene transcription and hippocampal synaptic plasticity (Gupta-Agarwal et al., 2012). However the mechanisms that couple neuronal activity with the function of KDMs and KMTs have remained unknown.
To gain insight into the potential roles of KDMs and KMTs in activity-dependent brain plasticity, here we characterize the activity-regulated mRNA expression of these enzymes in the mouse hippocampus following pilocarpine-induced status epilepticus, an in vivo model of elevated neuronal activity. We identify multiple activity-regulated KDMs and KMTs, and we characterize the anatomical distribution and functional consequences of induction of the H3K27-specific histone demethylase Kdm6b in hippocampal neurons. These data identify a novel program of activity-regulated and Kdm6b-dependent genes that are annotated for their function in inflammation, and we show for the first time that Kdm6b is required for the activity-dependent preconditioning of hippocampal neuronal survival.
Materials and Methods
Pilocarpine-induced seizure
Adult (8–12 week old) male C57BL6/J mice (Charles River Labs) were weighed and injected with 1 mg/kg methyl scopolamine nitrate (i.p.). 30 min later mice were injected i.p. with either 337 mg/kg pilocarpine HCl or saline (control mice). Stock solutions were prepared in 0.9% injectable NaCl and each animal received 0.1 cc of drug. Following pilocarpine injection, animals were monitored for the onset of status epilepticus, which was defined using behavioral criteria as a continuous limbic motor seizure of stage 2 or higher (Liu et al., 1999). During the seizure, mice were provided with food and water placed in a dish at the bottom of the cage. Status epilepticus was allowed to proceed for 3 hrs and then was terminated by administration of diazepam (10mg/kg, i.p.). Pilocarpine-treated animals that failed to develop status epilepticus were excluded from the study. 1, 3, or 6 hrs following the onset of status epilepticus, mice were deeply anesthetized with isofluorane and decapitated for brain harvesting. Brains of control mice were harvested 3 hrs after the saline injection. 5–6 mice were used for each time point. For RNA expression studies, the hippocampus was rapidly dissected bilaterally, flash frozen on dry ice/ethanol, and stored at −80°C prior to RNA harvesting. For in situ studies, whole brains were flash frozen in OCT on a dry ice/ethanol bath and stored at −80°C. All experiments were conducted in accordance with an animal protocol approved by the Duke University Institutional Animal Care and Use Committee.
RNA harvesting and quantitative PCR
RNA was harvested using the Absolutely RNA kit (Agilent). 800ng of RNA was used for reverse transcription with oligo dT primers and Superscript II (Invitrogen). Quantitative SYBR green PCR was performed on an ABI 7300 real-time PCR machine (Applied Biosystems) using the intron-spanning primers (IDT) listed in Table S1. Each sample was measured in triplicate, and all data were normalized to the expression of the housekeeping gene Gapdh as a control for RNA quantity and sample processing.
In situ hybridization and immunostaining of brain sections and cultured neurons
Fresh frozen brains were cryostat sectioned coronally at 18μm, then fixed in 3% paraformaldehyde, and rinsed with 1x phosphate-buffer saline (PBS). For radioactive probe detection, slides were dehydrated in ethanol, air dried, and hybridized at 65°C with 1×106 cpm per slide of either antisense or sense 35S-UTP-labeled riboprobes for the gene of interest. We generated riboprobes from cloned cDNA of mouse Kdm6b exon III (NM_001017426) and mouse Bdnf exon IX (NM_001017426). Slides were processed for autoradiography on film (Kodak) to detect the in situ signal, and counterstained with cresyl-violet for identification of anatomical landmarks. For fluorescent in situ hybridization, digoxigenin (DIG)-labeled antisense riboprobes targeting mouse Kdm6b exon III were used. Hybridized riboprobes were visualized by detection with peroxidase-conjugated anti-DIG Fab fragments (Roche) and developed using TSA (Perkin Elmer). For immunohistochemistry, slides were blocked in 14% NGS and permeabilized in 0.3% Triton X-100 prior to antibody incubation. Hoechst dye was used to label nuclei for identification of anatomical landmarks. Images were captured on a Leica DMI4000 inverted fluorescence microscope at 20× magnification. Primary antibodies used in this study were mouse α-human Ki67 (BD Pharmingen, 1:100), rabbit anti-Histone H3 pS10/K14Ac (Millipore 07-081), mouse anti-histone H3K27me3 (Abcam 2006), rabbit anti-histone H3K27me3 (Active Motif 39155), rabbit anti-histone H3K27me3 (Millipore 07-499), mouse anti-glial-fibrillary acid protein (GFAP; Abcam 10062) and chicken anti-microtubule associated protein 2 (MAP2, Millipore AB5543).
Hippocampal neuronal culture and lentiviral RNA interference
Hippocampal cultures were prepared from P0 CD1 mouse pups (Charles River Laboratories). Cells were plated on poly-D-lysine (PDL) and laminin (Invitrogen)-coated coverslips for cell survival assay or plates for RNA analysis in Neurobasal-A medium (Invitrogen) containing B27 supplements (Invitrogen), 1% fetal bovine serum and penicillin and streptomycin (Sigma). 5μM AraC was added on the fourth day in vitro (DIV) to block the proliferation of glial cells. For RNA interference, we purchased two independent shRNAs targeting non-overlapping sequences in mouse Kdm6b that were cloned in the lentiviral vector pLKO.1 (Thermo Scientific). Kdm6b shRNA1 (TRC#0000095265): CTGTTCTTGAGGGACAAACTC. Kdm6b shRNA2 (TRC#0000095266): GTTCGTTACAAGTGAGAACTC. Empty pLKO.1 vector was used as the control. Viral shRNAs were packaged as lentivirus in HEK293T cells (ATCC). Neurons were infected with virus at a multiplicity of infection of 1 on DIV4.
Western blot analysis
The Kdm6b expression plasmid containing FLAG-tagged Kdm6b (base pairs 3513-5336) in the vector pcDNA3 was a kind gift of Dr. Mary E. Donohoe (Burke Research Institute of Weill Cornell Medical School)(Kamikawa and Donohoe, 2014). HEK-293T cells were transfected by calcium phosphate precipitation and extracts were harvested two days later for expression analysis. Nuclear extracts were run for SDS-PAGE and transferred to PVDF for western blotting. Bands were visualized with fluorescent secondary antibodies using the Odyssey imaging system (LI-COR Bioscience). Actin was used as a loading control. Primary antibodies used in this study were mouse anti-FLAG (Sigma F3165), mouse anti-Actin (Millipore MAB1501), and chicken anti-GFP (Chemicon AB16901).
Neuronal survival assay
The neuronal survival assay was carried out in accordance with previously published protocols (Zhang et al., 2007). DIV11 neurons were either left untreated or treated with the addition of 50μM bicuculline (Sigma) and 2.5 mM 4-aminopyridine (Sigma) (Bic+4AP). After 16 hrs of Bic+4AP treatment, media was switched to minimal medium lacking serum and growth factors - salt-glucose-glycine (SGG) solution [10mM Hepes (pH7.4), 114mM NaCl, 26.1mM NaHCO3, 5.3mM KCI, 1mM MgCl2, 2mM CaCl2, 30mM glucose, 1mM glycine, 0.5mM sodium pyruvate, and 0.001% phenol red] and phosphate-free Eagle’s minimum essential medium (MEM) (99:1 vol:vol) or complete media – above media supplemented with insulin (7.5pg/mL), transferrin (7.5g/mL), sodium selenite (7.5ng/mL), penicillin G (50U/mL), and streptomycin sulfate (50pg/mL) (Bading et al., 1993). Neuronal firing was terminated by addition of 1μM TTX (Tocris). Cells were then fixed in 4% paraformaldehyde and nuclei were visualized by Hoechst 33258 staining and fluorescent microscopy. Cells undergoing apoptosis were identified by their condensed nuclear morphology (Bonni et al., 1999) and confirmed by immunoreactivity with an antibody against cleaved-caspase-3 (Cell Signaling 9661, 1:100). Neurons were identified by immunoreactivity with MAP2 (Millipore AB5543, 1:2000). Neuronal death was quantified by counting the number of apoptotic neuronal nuclei as a percentage of the total neuronal nuclei.
RNA-Seq
For gene expression studies, mouse hippocampal neurons were plated, cultured, and infected as described above for the neuronal survival assay. DIV11 neurons were either left untreated or treated with the addition of 50μM bicuculline (Sigma) and 2.5 mM 4-aminopyridine (Sigma) (Bic+4AP) for 3 hrs prior to RNA harvesting. For each treatment condition (pLKO vector/control, pLKO vector/Bic+4AP, Kdm6b shRNA1/control, Kdm6b shRNA1/Bic+4AP), triplicate replicates were pooled. Each pool was then used to generate the four treatment condition libraries for sequencing. RNA-Seq libraries were generated by the Duke University sequencing core facility and subjected to 50bp paired-end Illumina Hi-Seq sequencing. Following quality score-based trimming and adapter filtering, sequences were aligned to the mouse UCSC gene reference transcriptome (mm9, July 2007) using Tophat v2.0.8b with options -T, -x 4, and -n 2 (Trapnell et al., 2009). Cufflinks v2.0.2 with option –u for multi-read correction was used to extract normalized read counts collapsed to a single value of Fragments Per Kilobase of exon per Million fragments mapped (FPKM) for each gene (Trapnell et al., 2010). Gene sets were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov) (Huang et al., 2008). We used default parameters with the following modifications: KEGG and Panther were used for pathways analysis and we focused our search on biological pathways by choosing GO_TERM_BP_FAT for GO analysis. We considered as significant those pathways with Benjamini-corrected p values less than 0.05 and FDR less than 0.2. Fully independent replicate samples with both Kdm6b shRNAs were used for quantitative PCR validation of sequencing results.
Astrocyte cultures
Astrocyte cultures were generated by plating papain-dissociated cells from postnatal day 0–2 mouse brains onto poly-D-lysine coated flasks in DMEM with 10% FBS and N2 supplements (Invitrogen). 3 days after plating, flasks were shaken vigorously to remove any neurons that may have attached. Flasks were cultured until astrocytes were confluent, then cells were split with trypsin, replated, and grown until confluent again. For the experiments in which we assessed the regulation of gene expression in astrocytes, Bic+4AP was first added to DIV 11 hippocampal neuronal cultures in the neuronal culture medium for three hours as described in the RNA-Seq section above. Conditioned medium from control or Bic+4AP-treated neurons was then removed and placed on astrocyte cultures for 3 hours prior to astrocyte RNA harvesting.
Statistical analyses
Unless otherwise indicated, all data presented are the average of at least three measurements from each of at least two independent experiments. Quantitative PCR data following pilocarpine injection were analyzed by one-way ANOVA followed by Bonferroni corrected post-hoc comparisons for changes in gene expression over time after onset of seizure. All other data were analyzed by a Student’s unpaired t-test. Bar graphs show mean values and error bars show S.E.M. In all cases a p<0.05 was considered significant.
Results
Pilocarpine-induced seizure regulates expression of multiple KDMs in the hippocampus
To identify KDMs that may contribute to activity-dependent changes in gene transcription, we quantified expression of mRNAs in extracts from adult male mouse hippocampus either following a control saline injection or 1, 3, or 6 hrs after pilocarpine-induced status epilepticus, an in vivo experimental model of elevated synaptic activity (Curia et al., 2008). We quantified expression of all 21 mouse KDMs that have experimentally validated enzymatic activities as histone demethylases as well as 5 additional neuronally-expressed jumonji C (JmjC) domain containing proteins whose enzymatic activities as histone demethylases are either absent or uncharacterized (Black et al., 2012; Greer and Shi, 2012). We detected all 26 of these mRNA transcripts in hippocampal extracts (Table 1).
Table 1. Expression level of KDMs in the hippocampus.
Quantitative PCR was used to determine expression of KDMs relative to expression of the housekeeping gene Gapdh in the control hippocampus of adult male mice. One-way ANOVA was used to assess for change in expression over time following pilocarpine-induced seizure.
| Gene symbol | Synonyms | Histone substrate | % of Gapdh | F (3,19) | p-value |
|---|---|---|---|---|---|
| Kdm1a | Lsd1/Aof2 | H3K4, H3K9 | 0.469 | 2.658 | 0.079 |
| Kdm1b | Lsd2/Aof1 | H3K4 | 0.2969 | 1.668 | 0.22 |
| Kdm5a | Jarid1a/Rbbp2/Rbp2 | H3K4 | 1.8674 | 0.187 | 0.9 |
| Kdm5b | Jarid1b/Plu1 | H3K4 | 2.1733 | 2.297 | 0.116 |
| Kdm5c | Jarid1c/Smcx | H3K4 | 0.8874 | 2.113 | 0.139 |
| Kdm5d | Jarid1d/Smcy | H3K4 | 0.3059 | 1.106 | 0.38 |
| Kdm2a | Jhdm1a/Fbxl11 | H3K36 | 2.0218 | 5.883 | 0.007* |
| Kdm2b | Jhdm1b/Fbxl10 | H3K4, H3K36 | 0.1603 | 1.599 | 0.23 |
| No66 | H3K4, H3K36 | 0.3865 | 2.683 | 0.084 | |
| Kdm4a | Jmjd2A/JHDM3A | H3K9, H3K36, H1.4K26 | 0.8227 | 3.467 | 0.041* |
| Kdm4b | Jmjd2b/JHDM3B | H3K9, H3K36, H1.4K26 | 1.5448 | 16.49 | <0.0001* |
| Kdm4c | Jmjd2c/JHDM3C/Gasc1 | H3K9, H3K36, H1.4K26 | 0.4319 | 10.23 | 0.0005* |
| Kdm4d | Jmjd2d/JHDM3D | H3K9, H3K36, H1.4K26 | 0.0141 | 5.731 | 0.007* |
| Kdm3a | Jmjd1a/JHDM2A/TSGA | H3K9 | 0.3668 | 0.93 | 0.446 |
| Kdm3b | Jmjd1b/JHDM2B | H3K9 | 1.1403 | 0.81 | 0.51 |
| Jmjd1c | Kdm3c/TRIP8 | H3K9 | 2.2607 | 26.84 | <0.0001* |
| Phf2 | JHDM1E | H3K9 | 3.5025 | 1.509 | 0.25 |
| Phf8 | JHDM1F | H3K9, H4K20 | 0.9482 | 1.081 | 0.39 |
| Jhdm1d | Kdm7a/mKIAA1718 | H3K9, H3K27 | 4.9903 | 5.404 | 0.009* |
| Kdm6a | Utx | H3K27 | 0.1513 | 4.293 | 0.019* |
| Kdm6b | Jmjd3 | H3K27 | 0.8094 | 12.71 | 0.0002* |
| Uty | - | 0.476 | 3.741 | 0.032* | |
| Hr | - | 0.1584 | 1.631 | 0.222 | |
| Hif1an | - | 0.3803 | 5.158 | 0.011* | |
| Jarid2 | - | 0.7204 | 24.31 | <0.0001* | |
| Jmjd6 | PTDSR | H3R2, H4R3 | 0.2784 | 6.509 | 0.004* |
p<0.05.
Light grey shading denotes KDMs that decreased in expression over time after pilocarpine treatment, and dark grey highlighting denotes the KDMs that increased in expression. Hif1an was the only KDM to show both increased and decreased expression after pilocarpine at different time points.
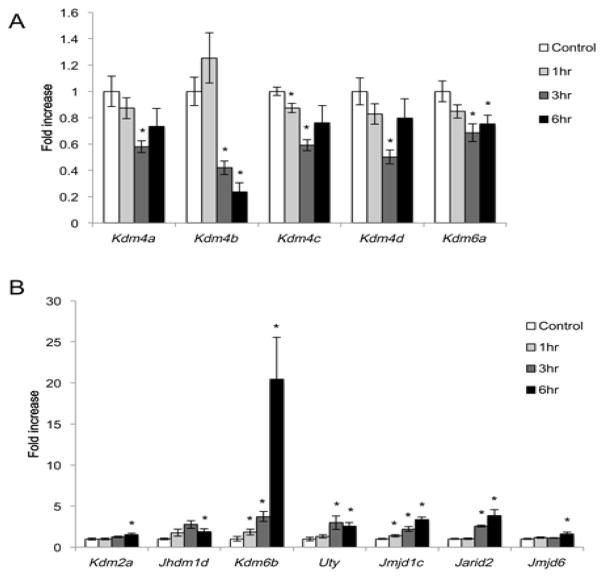
One-way ANOVA revealed significant pilocarpine-induced, time-dependent increases or decreases in the expression of 12 KDM family genes (Table 1). 5 KDMs showed decreased expression over time after pilocarpine (Fig 1A). These included the H3K27 demethylase Kdm6a and the mixed specificity (H3K9/36) demethylases Kdm4a, Kdm4b, Kdm4c, and Kdm4d. Overall these reductions were small in magnitude, with only Kdm4b mRNA levels falling more than 2-fold during the 6hr period. 7 KDMs showed significantly increased mRNA expression after seizure (Fig. 1B). These included the H3K9 demethylase Jmjd1c, the H3K27 demethylase Kdm6b, the H3K36 demethylase Kdm2a, the mixed specificity (H3K9/27) demethylase Jhdm1d, and the JmjC-domain containing genes Jmjd6, Jarid2, and Uty. These inductions were of larger magnitude than the activity-induced decreases in KDM expression, with all but Kdm2a and Jmjd6 rising more than 2-fold during the 6hr period. Most strikingly, Kdm6b showed a 20-fold increase in mRNA expression by 6hrs after pilocarpine injection. This induction is similar in magnitude to the induction of the canonical activity-regulated gene encoding Brain-Derived Neurotrophic Factor, Bdnf (fold increase Bdnf after pilocarpine compared to control: 1hr, 9.25±1.54; 3hr, 22.96±1.7; 6hr, 17.51±1.95) (Castren et al., 1993; West, 2008). These data reveal that multiple KDMs undergo synaptic activity-dependent regulation of their expression in the hippocampus in vivo, and furthermore our findings suggest that among the KDMs, Kdm6b may be particularly relevant for synaptic activity-dependent transcriptional regulation.
Figure 1.
Regulated expression of KDMs in the hippocampus following pilocarpine-induced seizure. RNA isolated from the hippocampi of saline injected animals (control) or animals that underwent pilocarpine-induced status epilepticus for 1, 3, or 6 hrs was used for quantitative PCR. (A) 5 KDMs show decreased expression following pilocarpine. (B) 7 KDMs show increased expression following pilocarpine. Data are normalized to Gapdh expression and expressed as fold induction over control. *p < 0.05 compared with control, n=5–6/group.
Pilocarpine induces Kdm6b mRNA expression in granule neurons of the dentate gyrus
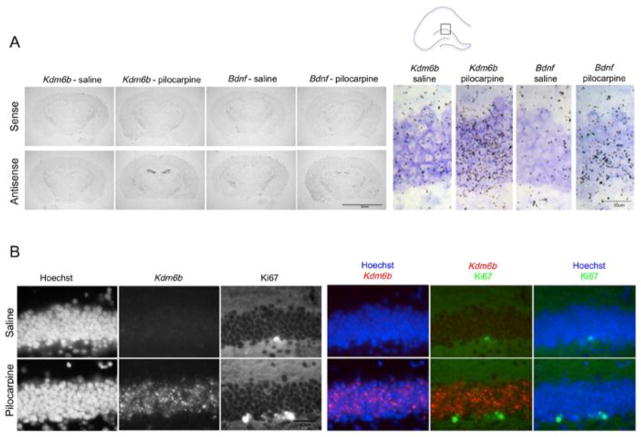
Very little is known about the expression or functions of Kdm6b in the adult CNS. Thus to investigate potential functions for Kdm6b in the hippocampus we set out to determine in which cells Kdm6b mRNA was induced following seizure. We first localized Kdm6b mRNA expression within the subregions of the hippocampus by performing in situ hybridization on coronal brain sections of adult mice after either saline or pilocarpine injection. A robust increase of Kdm6b mRNA was seen selectively over the dentate gyrus (DG) of the hippocampus in pilocarpine-treated mice. These data again mimic the magnitude and pattern of pilocarpine-induced Bdnf (Fig. 2A). Minimal induction of either Kdm6b or Bdnf was seen in the CA1 or CA3 regions of the hippocampus, which is consistent with previous studies showing predominant Bdnf induction in DG compared with CA1 or CA3 at 3–6 hrs after pilocarpine treatment (Schmidt-Kastner et al., 1996). No significant hybridization was observed with sense probes against either target in control or pilocarpine-treated mice validating the specificity of the hybridization signal.
Figure 2.
Pilocarpine induces Kdm6b expression in the granule neurons of the dentate gyrus (DG). (A) Autoradiography on coronal slices through hippocampus taken from animals 3 hr after saline or pilocarpine injection using sense (control) and antisense probes against Kdm6b or Bdnf. Both antisense probes reveal a similar selective pattern of signal induction over the DG following pilocarpine. Box shows region enlarged on right. Cresyl violet counterstain (purple) used to visualize all cells. Scale bar left 5mm, scale bar right 30μm. (B) Fluorescent in situ hybridization using antisense riboprobes against Kdm6b 3hr following saline or pilocarpine shows dense labeling in granule neurons but no colocalization with Ki67+ SGZ progenitors cells. The DNA binding dye Hoeschst is used to label all nuclei. For colocalization, Kdm6b mRNA (red), Ki67 (green), and Hoechst (blue). Orientation and region as shown in the diagram in A. Scale bar 40μm
The subgranular zone (SGZ) of the dentate gyrus in the hippocampus is one of only two places in the adult rodent brain that have ongoing neurogenesis (Alvarez-Buylla and Garcia-Verdugo, 2002) and pilocarpine-induced seizure is known to induce hippocampal neurogenesis (Parent et al., 1997). Previous studies have shown transient upregulation of Kdm6b expression at times of fate transition in differentiating neurons (Jepsen et al., 2007; Burgold et al., 2008); thus to determine whether pilocarpine-induced Kdm6b expression might be occurring selectively in neural precursors or newborn neurons of the hippocampus, we used fluorescent in situ (FISH) to achieve cellular resolution of the Kdm6b mRNA signal. Neural progenitor cells are located in the SGZ just under the hilar side of the granule cell layer (Lledo et al., 2006). We identified these precursors by immunolabeling with the Ki67 antibody, which reacts with an antigen in dividing cells. Sparse Ki67+ precursors were apparent, however Kdm6b mRNA was not detected in Ki67+ cells either in control mice or after pilocarpine-induced seizure (Fig. 2B). By contrast, after pilocarpine, Kdm6b mRNA was robustly and widely induced in an even distribution across the full extent of the densely packed granule neuron layer of the DG (Fig. 2B). This pattern is distinct from the localization of newborn neurons, which can be identified by their expression of Doublecortin and are found along the hilar side of the DG (Ming and Song, 2005). It is also distinct from the distribution of non-neuronal cells such as astrocytes and microglia, which are sparsely and evenly distributed both within and around the granule cell layer (Williamson et al., 2012). Although we cannot rule out Kdm6b expression in additional cell types, our data showing dense pilocarpine-induced Kdm6b expression enriched within the granule neuron layer of the DG are most consistent with pilocarpine selectively inducing expression of Kdm6b in granule neurons of the adult DG. These data provide the first evidence that neuronal Kdm6b expression is regulated by synaptic activity in neurons in vivo and they raise the possibility that Kdm6b may contribute to activity-dependent aspects of plasticity in differentiated neurons.
Upregulation of Kdm6b in the DG is not accompanied by global demethylation of H3K27
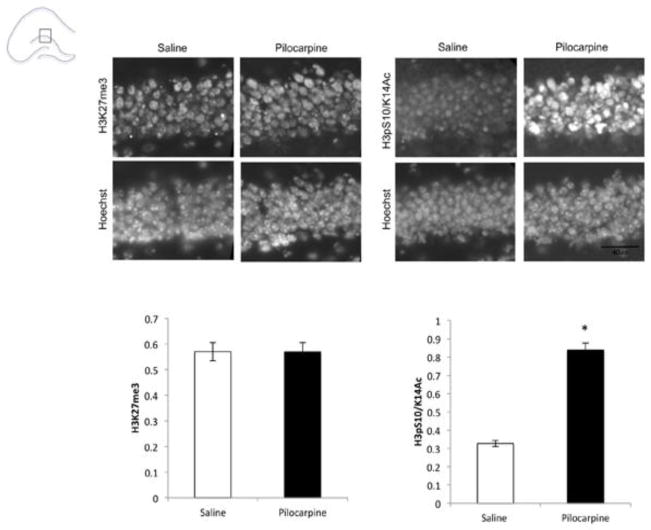
As a histone lysine demethylase, Kdm6b is part of a small subfamily of just two enzymatically active members (Kdm6b/Jmjd3 and Kdm6a/Utx) that have specificity for di- and trimethylated H3K27 (Greer and Shi, 2012). Because we saw increases in expression of Kdm6b in dentate granule neurons of the hippocampus, we wanted to determine whether seizure would result in concomitant decreases in H3K27 methylation. To test for global changes in this histone modification, we used antibodies raised against the trimethylated form of H3K27 (H3K27me3) and stained nuclei of hippocampal sections of control mice or mice that had received pilocarpine (Fig. 3). We saw no differences in the intensity of H3K27me3 immunoreactivity with any of three different antibodies raised against this modification (Fig. 3A and data not shown). By contrast, immunoreactivity with an antibody raised against a different histone H3 modification (Ser10 phosphorylation/K14 acetylation) that has previously been shown to be neuronal activity-regulated in the hippocampus (Levenson et al., 2004; Chwang et al., 2006) was robustly enhanced following pilocarpine treatment in the same brains (Fig. 3B).
Figure 3.
No change in global H3K27me3 levels in the DG following pilocarpine. Mouse brains were frozen 3hrs after injection with saline (control) or pilocarpine. Immunostaining was carried out using antibodies specific for the following posttranslationally modified histones: trimethylated histone H3 lysine 27 (H3K27me3), serine 10 phosphorylated and lysine 14 acetylated histone H3 (H3pS10/K14Ac). Nuclei were labeled with Hoechst, and Hoechst intensity was used to normalize immunostaining intensity in each sample. Data shown are using H3K27me3 antibody Active Motif 39155 (1:500), similar results were seen with two additional antibodies selective for this mark. n=2/treatment group, *p<0.05 pilocarpine compared with saline. Scale bar 40μm.
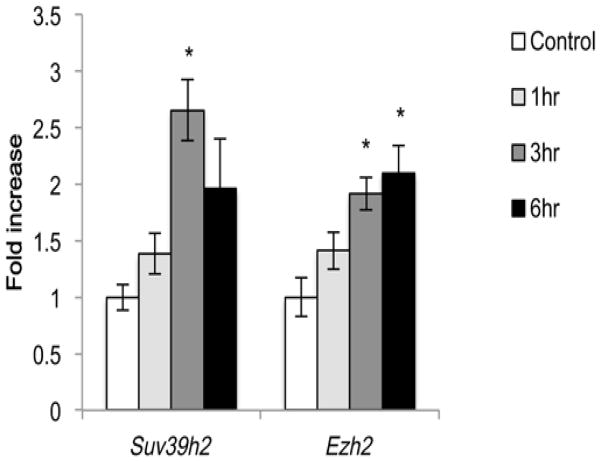
Our data showing no global change in H3K27me3 in the hippocampus after pilocarpine-induced seizure do not rule out the possibility that changes in H3K27me3 may be localized to specific regions of the genome. However since histone methylation is a dynamic process, we also considered whether activity-induced increases in KDMs might be offset by activity-induced expression of KMTs, leading to a globally balanced steady-state distribution of these histone modifications. Because many of the activity-regulated KDMs we identified in the hippocampus have specificity for histones methylated at K9 or K27, we analyzed the hippocampal mRNA expression of 10 KMTs with specificity for methylation at these sites (Black et al., 2012). All 10 KMTs were detected in the same hippocampal lysates used for analysis of KDM expression (Table 2). ANOVA revealed significant time-dependent increases in expression of two of these KDMs following pilocarpine-induced seizure: the H3K9 methyltransferase Suv39h2 and the H3K27 methyltransferase Ezh2 (Fig. 4) both of which showed ~2-fold increases in mRNA expression. These data raise the possibility that balanced regulation of both KDMs and KMTs may act to maintain global chromatin state following pilocarpine-induced increases in hippocampal neuronal activity.
Table 2. Expression levels of KMTs in the hippocampus.
Quantitative PCR was used to determine expression of H3K9 and H3K27 KMTs relative to expression of the housekeeping gene Gapdh in the control hippocampus of adult male mice. One-way ANOVA was used to assess for change in expression over time following pilocarpine-induced seizure.
| Gene symbol | Synonyms | Histone substrate | % of Gapdh | F(3,19) | p-value |
|---|---|---|---|---|---|
| Suv39h1 | KMT1A | H3K9 | 0.6265 | 0.562 | 0.65 |
| Suv39h2 | KMT1B | H3K9 | 0.1516 | 6.317 | 0.019* |
| Ehmt1 | KMT1D/GLP | H3K9, H1.4K26 | 0.5717 | 1.549 | 0.24 |
| Ehmt2 | KMT1C/G9a | H3K9, H1.4K26 | 6.1235 | 1.012 | 0.41 |
| Setdb1 | KMTIE | H3K9 | 0.6010 | 2.134 | 0.14 |
| Kmt2a | Mll1 | H3K4 | 0.8498 | 0.772 | 0.53 |
| Kmt2b | Mll2 | H3K4 | 0.2941 | 0.839 | 0.49 |
| Kmt2c | Mll3 | H3K4 | 0.2755 | 1.341 | 0.3 |
| Ezh1 | KMT6B | H3K27 | 0.7335 | 0.976 | 0.43 |
| Ezh2 | KMT6A | H3K27, H1.4K26 | 0.2959 | 7.468 | 0.04* |
p<0.05.
Dark grey highlighting denotes two KMTs that increased in expression after pilocarpine treatment.
Figure 4.
Regulated expression of H3K9 and H3K27 KMTs in the hippocampus following pilocarpine-induced seizure. RNA isolated from the hippocampi of saline injected animals (control) or animals that underwent pilocarpine-induced status epilepticus for 1, 3, or 6 hrs was used for quantitative PCR. Data are normalized to Gapdh expression and expressed as fold induction over control. *p < 0.05 compared with control, n=5–6/group.
Kdm6b is required for activity-dependent priming of neuronal survival
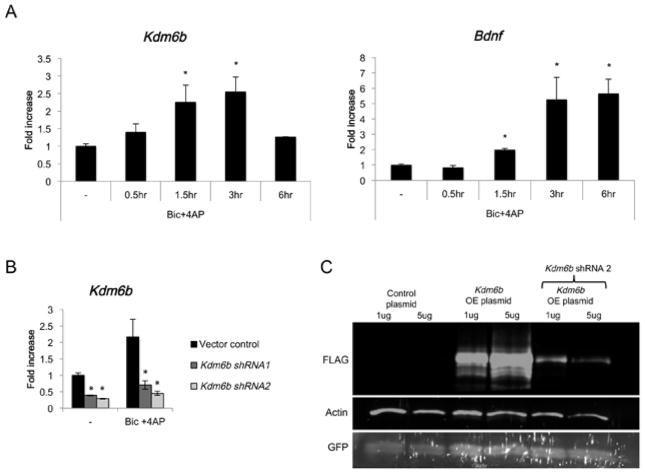
DG granule neurons are relatively resistant to cell death following seizure, and the induction of synaptic activity-dependent gene transcription in the DG is thought to contribute to this survival process (Clifford et al., 1987; Ploski et al., 2006). Given the induction of Kdm6b expression in the DG following pilocarpine treatment and the overlap of this expression pattern with the pro-survival neurotrophin Bdnf, we hypothesized that Kdm6b may contribute to the activity-dependent promotion of hippocampal neuronal survival. This ability of synaptic activity to promote neuronal survival can be recapitulated in cultured hippocampal neurons. Treating neurons with the GABA-A receptor antagonist bicuculline together with the potassium channel blocker 4-aminopyridine (Bic+4AP) activates glutamate neurotransmission and action potentials, turns on activity-dependent gene transcription - including the transcription of Kdm6b, and preconditions neuronal survival upon subsequent growth factor withdrawal (Zhang et al., 2007; Zhang et al., 2009). Thus this paradigm offers a useful model for discovery of the transcriptional mechanisms that underlie activity-induced neuroprotection.
We found that Kdm6b mRNA was significantly induced by Bic+4AP treatment of cultured hippocampal neurons, again similar to Bdnf (Fig. 5A). To test the function of Kdm6b induction in these neurons, we knocked down expression of Kdm6b with either of two independent lentiviral shRNAs. Compared with vector control infected neurons, the neurons expressing the Kdm6b shRNAs showed a significant reduction in Kdm6b mRNA both at baseline and after Bic+4AP (Fig. 5B). Expression of Kdm6b shRNA was also sufficient to lead to loss of Kdm6b protein (Fig 5C).
Figure 5.
Bic+4AP treatment induces Kdm6b expression in hippocampal neuron cultures. (A) Neuronal activity was in induced in hippocampal cultures (P0 + 11DIV) by the addition of the GABAA receptor antagonist bicuculline (1μM) along with the K channel blocker 4AP (2.5mM). Quantitative PCR was used to assess Kdm6b expression 0.5, 1, 3, and 6 hrs later, and Bdnf is shown as a positive control. All data were normalized to expression of the housekeeping gene Gapdh as a control for sample handling. Data are expressed as fold induction over no treatment (−). *p < 0.05 versus no treatment, error bars represent mean ± S.E.M., n=5–6 per group. (B) Knockdown of Kdm6b by two independent lentiviral shRNAs. Neurons infected with virus expressing either empty vector (vector control) or Kdm6b-specific shRNA (Kdm6b shRNA1 and Kdm6b shRNA2) were left untreated (−) or stimulated with Bic+4AP for 3hrs. Kdm6b mRNA was measured by quantitative RT-PCR and normalized to the expression of the housekeeping gene Gapdh. Data are expressed as fold induction compared with no treatment, vector control. *p < 0.05 versus no treatment, error bars represent ± S.E.M., n=3 per group. (C) Knockdown of Kdm6b protein expression by lentiviral shRNA plasmids. HEK 293T cells were cotransfected with either 1μg or 5μg of either the empty plasmid vector (control plasmid) or a vector expressing FLAG-tagged Kdm6b, along with either 10μg of the lentiviral vector expressing Kdm6b shRNA2 or the empty pLKO vector. GFP was cotransfected to confirm transfection efficiency. 3 days later, nuclear extracts were prepared for analysis of protein expression. The anti-FLAG antibody was used to detect Kdm6b expression. Actin is shown as a loading control. GFP is shown as a control for transfection efficiency.
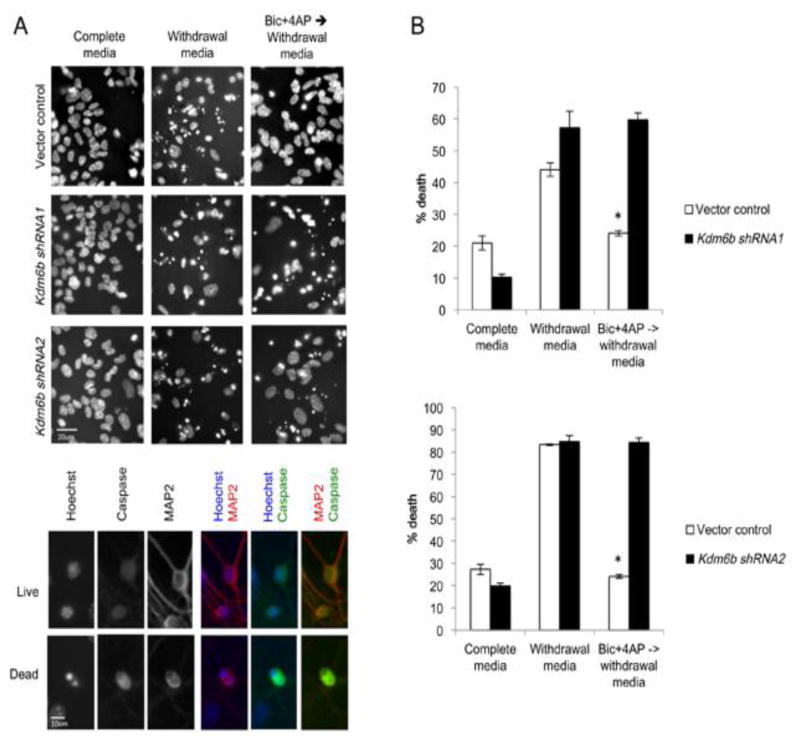
In vector control infected neurons, serum withdrawal led to a significant increase in neuronal death as indicated by the appearance of a condensed/fragmented nuclear morphology and confirmed by immunoreactivity for active caspase-3 (Fig. 6A–C). Pretreatment with Bic+4AP prevented death upon growth factor withdrawal in the control neurons, demonstrating the preconditioning effect of activity on neuronal survival (Fig. 6C). Knockdown with either of the two Kdm6b shRNAs did not increase neuronal death in full growth media, and similar to the control neurons, neurons expressing the Kdm6b shRNAs showed a significant increase in cell death upon growth factor withdrawal. However unlike control neurons, we found that Bic+4AP pretreatment failed to promote neuronal survival in neurons expressing either of the two Kdm6b shRNAs (Fig. 6C). These data demonstrate that expression of Kdm6b is required for activity-conditioned hippocampal neuronal survival.
Figure 6.
Kdm6b is required for neuronal-activity-dependent priming of neuronal survival. (A) Representative images of Hoechst-stained nuclei in cultured control (Vector control) and Kdm6b knockdown (shRNA1, shRNA2) hippocampal neurons under full medium and growth factor withdrawal conditions +/− pretreatment with Bic+4AP. Apoptotic cells were identified by their pyknotic nuclei seen with Hoeschst (blue) and confirmed by cleaved-caspase-3 staining (green). Neurons were identified by MAP2 staining (red). Scale bar 20μm top, 10μm bottom. (B) Quantification of growth factor withdrawal induced cell death with or without Bic+4AP pretreatment in vector control (white bars) and Kdm6b knockdown (black bars) neurons. *p<0.05 Bic+4AP pretreatment compared with vector control in growth factor withdrawal media. n=3 independent samples/treatment, 500–700 cells counted per sample.
Kdm6b is required for induction of a subset of activity-induced genes
Given that Kdm6b is a transcriptional regulator, and because activity-regulated gene transcription is thought to mediate the conditioning effects of neuronal activity on survival, we investigated whether Kdm6b is required for Bic+4AP-dependent induction of known pro-survival genes. Previous studies have identified a set of activity-inducible genes that are both transcriptionally induced by the activation of synaptic NMDA receptors and required for the promotion of neuronal survival upon growth factor withdrawal (Zhang et al., 2007; Zhang et al., 2009). Many of these pro-survival genes are targets of the Erk1/2-CREB transcriptional signaling pathway, which is strongly induced by both pilocarpine and Bic+4AP treatment of hippocampal neurons (Hardingham et al., 2002; Lee et al., 2009; Zhang et al., 2009). To determine whether Kdm6b is required for transcriptional regulation of these pro-survival genes, we used quantitative PCR to measure the Bic+4AP-dependent induction of Atf3, Bdnf, Btg2, Gadd45b, Gadd45g, Ifi202b, Inhba, Npas4, and Nr4a1 in both vector control and Kdm6b knockdown neurons. Surprisingly, despite the failure of Bic+4AP to promote neuronal survival in neurons lacking Kdm6b, we found that all of these pro-survival genes were induced to statistically indistinguishable levels in both control and Kdm6b knockdown neurons (Fig. S1). Importantly, these data establish that neurons lacking Kdm6b are capable of mounting a robust transcriptional response to Bic+4AP.
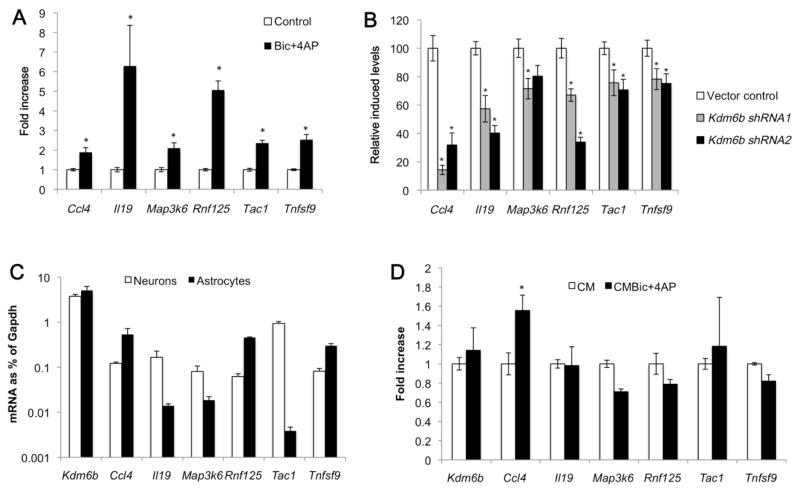
In addition to CREB, other transcription factors including NF-κB are induced by neuronal activity and are required for the preconditioning of neuronal survival in models of ischemia and epilepsy (Blondeau et al., 2001). Thus we considered the possibility that the induction of transcriptional targets other than those in the Erk1/2-CREB pathway might be affected by the loss of Kdm6b. To conduct an unbiased analysis of activity-induced gene transcription we performed RNA-Seq on control and Kdm6b knockdown neurons under unstimulated conditions or following Bic+4AP (Table S2). From all genes induced >1.5 fold in response to Bic+4AP (Table S3), we selected the top 100 that showed reduced Bic+4AP induction in Kdm6b knockdown neurons compared with vector infected control neurons (Table S4) and used the Database for Annotation, Visualization and Integrated Discovery (DAVID) to perform gene ontology (GO) classification and pathway analysis. This analysis identified two biological processes significantly enriched in this gene set: Response to Wounding and Inflammatory Response (Table 3). We first used quantitative PCR to validate the Bic+4AP induction of a subset of these and functionally related genes from the Kdm6b knockdown list. These genes included the chemokine Ccl4, the cytokine Il19, the JNK/p38 MAP kinase pathway upstream regulator Map3k6, the E3 ubiquitin ligase Rnf125, the proinflammatory neuropeptide Tac1, and the TNF ligand superfamily member Tnfsf9 (Fig. 7A). We then confirmed that each of these genes was significantly less Bic+4AP inducible in neuronal cultures lacking Kdm6b expression compared with vector control infected neuronal cultures (Fig. 7B).
Table 3. GO analysis of biological pathways dysregulated by knockdown of Kdm6b.
The top 100 genes showing reduced induction in neurons infected with Kdm6b shRNA compared to control were subjected to gene ontology (GO) analysis using DAVID.
| Enrichment score 4.57 | |||||
|---|---|---|---|---|---|
| Biological process | Gene count | P value | Benjamini | FDR | Genes |
| Response to wounding | 13 | 5.26E-07 | 0.0004 | 0.0008 | NOG, SPHK1, PPARG, TAC1, CCL4, CCL7, TNFRSF1B, PROCR, F3, HBEGF, KDM6B, CD14, FN1 |
| Inflammatory response | 9 | 3.93E-05 | 0.0167 | 0.0609 | TNFRSF1B, SPHK1, PPARG, TAC1, CCL4, KDM6B, CCL7, CD14, FN1 |
| Defense response | 10 | 9.39E-04 | 0.2347 | 1.4444 | TNFRSF1B, SPHK1, PPARG, TAC1, CCL4, KDM6B, CD74, CCL7, CD14, FN1 |
| Immune response | 10 | 0.0013 | 0.2479 | 2.0442 | RNF125, ICAM1, TNFRSF1B, PROCR, IL4RA, TNFSF9, CCL4, CD74, CCL7, CD14 |
The top functional annotation cluster is shown. Significant pathways are highlighted in gray.
Figure 7.
Impaired neuronal activity-inducible expression of a subset of activity-regulated genes in Kdm6b knockdown neurons. (A) Bic+4AP treatment induces expression of Ccl4, Il19, Map3k6, Rnf125, Tac1 and Tnfsf9 in cultured hippocampal neurons. Quantitative PCR was used to assess mRNA levels at baseline (control, white bars) or following 3hrs of Bic+4AP treatment (black bars) in control pLKO shRNA vector infected neurons. *p < 0.05 Bic+4AP compared with control, and n=6–9 per group. (B) Reduced Bic+4AP dependent induction of Ccl4, Il19, Map3k6, Rnf125, Tac1 and Tnfsf9 in Kdm6b knockdown neurons. Quantitative PCR was used to measure baseline and Bic+4AP induced expression of the indicated genes in neurons infected with the shRNA control vector (white bars) or either of two independent Kdm6b shRNAs (gray and black bars). Two fully independent experiments were used for each shRNA, and these samples were independent of the samples used for RNA sequencing. Only Bic+4AP induced levels of gene expression are shown and data are graphed as the percentage of Bic+4AP induced levels for each gene in the vector control infected cells. n=6–9 per group. *p < 0.05 Kdm6b knockdown compared with vector control, n=6–9 per group. (C) Gene expression in neuronal and astrocyte cultures. Expression of each gene is graphed as a percentage of the housekeeping gene Gapdh. n=3/cell type. (D) Bic+4AP regulates Ccl4 expression in astrocyte cultures. Astrocyte cultures were exposed for 3 hrs to conditioned medium from control neuronal cultures (CM) or conditioned medium from neuronal cultures that had been treated for 3 hrs with Bic+4AP (CMBic+4AP). Expression of each gene was normalized to control levels and fold change with Bic+4AP is graphed. n=6/treatment, *p<0.05 CMBic+4AP compared with CM.
Kdm6b is both induced by Bic+4AP and required for the Bic+4AP induction of specific genes. However these data do not indicate whether increasing the expression of Kdm6b alone is sufficient to drive gene expression. To address this question we overexpressed Kdm6b in HEK-293T cells. Of the genes shown in Fig. 7B, which required Kdm6b for their induction by Bic+4AP, four genes (MAP3K6, RNF125, TAC1, and TNFSF9) as well as KDM6B were found to be expressed in HEK-293T cells. However only TAC1 showed a small, though significant increase upon Kdm6b overexpression (Fig. S2). We interpret these data to imply that Kdm6b likely cooperates with other Bic+4AP regulated transcriptional mechanisms to modulate activity-dependent gene transcription.
Finally, in addition to being expressed in neurons, Kdm6b is also expressed in other brain cell types including astrocytes (Fig 7C), which comprise ~20% of the cells in our dissociated neuronal cultures (Fig. S3). Because our shRNA-expressing viruses infect both neurons and astrocytes in these cultures (data not shown) we sought to determine in which cell type(s) Kdm6b was acting in order to regulate Bic+4AP-dependent gene induction. To address this question, we prepared astrocyte cultures from postnatal mouse brain and compared gene expression and regulation in these cells to that in our neuronally enriched cultures. Of the Bic+4AP inducible genes shown in Fig. 7B, we found that all six were expressed in both neuronal and astrocyte cultures (Fig. 7C). Kdm6b was expressed at similar levels in both culture types. By contrast three of its candidate target genes (Il19, Map3k6, and Tac1) showed strong enrichment in neuronal cultures, whereas the other three (Ccl4, Rnf125, and Tnfsf9) were preferentially expressed in astrocyte cultures (Fig. 7C). We then asked Bic+4AP induced expression of these genes in astrocytes. Because the molecular targets of bicuculline (GABAA receptors) and 4-aminopyridine (KCNA family voltage-gated potassium channels) are primarily expressed in neurons, we first treated neuronal cultures with Bic+4AP and then transferred the neuronal conditioned medium onto astrocyte cultures. Interestingly, only Ccl4 showed significant Bic+4AP induction in astrocytes (Fig. 7D), whereas all six genes along with Kdm6b were induced by Bic+4AP treatment of neuronal cultures (Fig. 4B, 7A). These data strongly suggest that Kdm6b regulates Bic+4AP-dependent gene expression via its actions in neurons. However the possibility remains that Kdm6b contributes to brain function via its expression in astrocytes as well as neurons. In total, these data suggest that the activity- and Kdm6b-dependent regulation of a program of gene expression that is enriched for genes annotated for their role in inflammation may contribute to neuronal activity-dependent preconditioning of neuronal survival.
Discussion
Activity-dependent regulation of KDMs and KMTs in the adult hippocampus
In this study we have identified multiple KDMs and KMTs that show activity-dependent expression changes in the hippocampus of the adult mouse brain. Several of these gene products have previously been studied for their roles in nervous system development. For example, knockdown of the Zebrafish ortholog of the H3K9/K27 demethylase Jhdm1d results in early anatomical defects in the developing brain, mice lacking the JmjC-containing protein Jarid2 show neural tube defects, the H3K27 demethylase Kdm6b promotes neuronal differentiation, and the KMT Ezh2 regulates the process of cortical neurogenesis (Takeuchi et al., 1995; Jepsen et al., 2007; Burgold et al., 2008; Hirabayashi et al., 2009; Pereira et al., 2010; Tsukada et al., 2010). Our data now suggest that these regulatory enzymes play additional roles beyond development in post-mitotic, differentiated neurons of the adult brain. We identify one such function by demonstrating that neuronal activity-dependent induction of Kdm6b mRNA is required for activity-dependent survival of hippocampal neurons. Finally our analysis of activity-regulated gene transcription in hippocampal neurons lacking Kdm6b raises the possibility that Kdm6b-dependent, inflammation-related signaling pathways may contribute to the preconditioning of neuronal survival. Taken together these data comprise experimental support for the hypothesis that activity-dependent regulation of KDM/KMT expression contributes to transcription-dependent neuronal adaptations in the adult brain.
Five of the activity-regulated KDM/KMT genes we identified here have functional connections to histone H3K27 methylation. These gene products include the H3K27 demethylase Kdm6b; the Y chromosome paralog of Kdm6a called Uty, which has functions in cell differentiation but is thought to be enzymatically inactive (Shpargel et al., 2012); the mixed specificity H3K9/K27 demethylase Jhdm1d (Greer and Shi, 2012); the JmjC-domain containing protein Jarid2, which plays a non-enzymatic role in recruiting the polycomb PCR2 complex (Herz and Shilatifard, 2010); and Ezh2, which is the enzymatically active H3K27 methyltransferase in the PRC2 complex (Black et al., 2012). Dynamic changes in H3K27 methylation have been tightly correlated with the temporal regulation of gene expression programs during neuronal differentiation (Mikkelsen et al., 2007; Mohn et al., 2008). However, whether dynamic regulation of H3K27 methylation contributes to changes in gene transcription in mature neurons remains unknown. Only one study published to date has looked for activity-regulated changes in H3K27me3 genome-wide in neurons (Kim et al., 2010). This study failed to identify statistically significant changes in the global or local H3K27me3 patterns six hours following KCl-induced membrane depolarization of dissociated hippocampal neuron cultures. However we note that expression of the H3K27 KDMs/KMTs were not KCl-induced in the Kim et al. (2010) study. By contrast, at least two studies have identified stimulus-dependent changes in H3K27 methylation locally at promoters of the Bdnf gene in vivo, suggesting that there exist signaling cascades that can induce changes in this histone modification in neurons (Tsankova et al., 2006; Karpova et al., 2010). Although we found no global changes in H3K27me3 in hippocampal neurons following pilocarpine-induced seizures, whether this stimulus may lead to promoter- or enhancer-specific changes in histone H3 methylation remains to be addressed.
Functions of Kdm6b in the CNS
The robust activity-dependent induction of Kdm6b mRNA in the adult hippocampus led us to pursue an anatomical and functional characterization of this KDM. Previous studies in a number of different cell types have suggested that regulated expression of Kdm6b mRNA is an important means of transitioning through stages of cell fate determination. Kdm6b expression is transiently upregulated when ES cells are induced to differentiate into neural lineage precursors, and this upregulation of Kdm6b is necessary for the induction of neural lineage genes including Nestin (Burgold et al., 2008). Kdm6b is upregulated again when neural stem cells are induced to differentiate into immature neurons, and in this system the expression of Kdm6b has been shown to be sufficient to drive the expression of multiple neuronal genes (Jepsen et al., 2007). Outside of the nervous system, transiently induced expression of Kdm6b also heralds its function in differentiation of the macrophage lineage (Hu et al., 2006; De Santa et al., 2007; Chen et al., 2012). However although neurogenesis is ongoing and seizure-inducible in the subgranular zone (SGZ) of the dentate gyrus in the rodent hippocampus, instead of localizing to the SGZ, our in situ analysis of seizure-induced Kdm6b expression showed induction throughout the granule layer where mature neurons are found. These data do not rule out a role for Kdm6b in seizure-induced neurogenesis, however they strongly suggest Kdm6b has functions in mature, post-mitotic neurons.
In the macrophage lineage, in addition to its role during differentiation, Kdm6b expression shows rapid (time course of hours) induction in response to macrophage-activating stimuli, including bacterial lipopolysaccharides (LPS), cytokines, and TNFα (De Santa et al., 2007; De Santa et al., 2009; Ishii et al., 2009; Kruidenier et al., 2012). In these cells Kdm6b contributes to the regulation of several hundred LPS-induced genes (De Santa et al., 2009). Furthermore, Kdm6b knockout mice show impaired macrophage-mediated immunological responses to helminth infection, suggesting the biological relevance of this transcriptional dysregulation (Satoh et al., 2010).
In hippocampal neurons we find that activity-induced expression of Kdm6b is required for the preconditioning of neuronal survival upon subsequent growth factor withdrawal. It is well-established that low intensity exposure to certain environmental stressors can temporarily protect neurons against subsequent damage from otherwise harmful insults in a process called preconditioning (Kelly and McIntyre, 1994; Gidday, 2006). In the case of both ischemia and status epilepticus, which are pathologically-relevant insults in the adult brain, long-lasting insult tolerance requires the induction of new protein synthesis (Jimenez-Mateos et al., 2008). The preconditioning of neuronal survival is tightly linked with activation of synaptic NMDA-type glutamate receptors and MAP kinase signaling pathways, and inhibition of either Erk1/2 or p38 MAP kinases abrogates the protective effects of preconditioning seizures on status epilepticus induced hippocampal cell death in vivo (Jiang et al., 2005; Zhang et al., 2007). CREB is a key transcriptional transducer of the survival functions of the Erk1/2 pathway, via its ability to drive the transcription of pro-survival genes including Bdnf and Bcl2 (Bonni et al., 1999). In parallel, the transcription factor NF-κB is an important effector of the p38 MAP kinase pathway, and preconditioning with kainate or ischemia exhibits NF-κB dependent protection against neuronal death (Blondeau et al., 2001; Mattson and Meffert, 2006). However, knowledge of NF-κB gene targets in neurons is limited, with only a few genes, such as the mitochondrial antioxidant enzyme manganese superoxide dismutase (MnSOD) and pro-survival members of the Bcl2 family, having been established as NF-κB targets that protect against apoptosis (Mattson et al., 1997; Tamatani et al., 1999; Zong et al., 1999). Interestingly, the Kdm6b promoter contains binding sites for the NF-κB family of transcription factors, and in macrophages, the induction ofKdm6b by inflammatory stimuli occurs in an NF-κB-dependent manner (De Santa et al., 2007). NF-κB-dependent transcription is strongly activated in cultured hippocampal neurons following synaptic glutamate receptor activation (Meffert et al., 2003). This same stimulus is sufficient to induce expression of Kdm6b (Zhang et al., 2009). However we note that despite evidence that many of the genes induced by Bic+4AP treatment in hippocampal neurons require glutamate receptor activation, the generation of action potentials, and the elevation of nuclear calcium (Hardingham et al., 2001; Zhang et al., 2007), whether the induction of Kdm6b by either pilocarpine or Bic+4AP requires neuronal firing remains to be determined. If NF-κB mediates activity-dependent Kdm6b expression in hippocampal neurons, it is possible that Kdm6b may be required downstream of p38 MAP kinase and NF-κB to mediate preconditioning neuroprotection.
Transcriptional targets of Kdm6b in the preconditioning of neuronal survival
Hundreds of genes with a range of molecular functions show induced expression in parallel with the survival-promoting effects of Bic+4AP. A previous study focused on nine highly activity-induced genes and showed that each was sufficient to promote neuronal survival in the absence of activity (Zhang et al., 2009). However our data demonstrate that induction of this specific gene set is not sufficient to support activity-dependent neuronal survival when neurons are stimulated with Bic+4-AP in the absence of Kdm6b. These data highlight the importance of considering the full program of activity-induced gene regulation in context and suggest that a balance between different gene pathways may be required to promote neuronal survival. Interestingly, Kdm6b-dependent neuronal preconditioning of survival is associated with the induction of a set of genes (including Ccl4, Il19, Map3k6, Rnf125, Tac1, and Tnfsf9) that are annotated for their function in inflammation. Although inflammation can reflect a pathological response to tissue injury, many inflammatory factors are also synthesized and released in response to increased neuronal activity in the brain. For example, expression of Tac1 and Ccl4 is induced in the hippocampus after status epilepticus (Sperk et al., 1990; Kan et al., 2012), and in vivo activation of kainate receptors induces Map3k6 expression in the brain (Ryan et al., 2005). This process of “neurogenic neuroinflammation” is proposed to function in homeostasis and to promote an adaptive response to increased metabolic demands (Xanthos and Sandkuhler, 2014). Previous studies support the role of many inflammatory factors in cell survival. For example, Substance P, the gene product of the Tac1 gene, mediates neuroprotection through activation of the Erk and Akt signaling pathways (Amadoro et al., 2007) and outside of the CNS, Ccl4 promotes cell survival of immortalized lymphoblastoid cell lines (Tsai et al., 2013). Notably, inflammatory gene products including the complement factor C1q, several class I major histocompatibility receptors, and tumor necrosis factor α have all been found to mediate physiologically relevant aspects of activity-dependent synapse development and plasticity (Huh et al., 2000; Beattie et al., 2002; Schafer et al., 2012). Based on the data presented here, we propose that the activity-dependent induction of inflammatory gene products is an adaptive response that promotes neuronal survival, and that Kdm6b is an integral coordinator of this process.
Our data suggest that neurons are the major site of Kdm6b and Bic+4AP-regulated changes in gene expression, thus we think it is likely that Kdm6b is acting in neurons to promote activity-dependent preconditioning of neuronal survival. However Ccl4, Il19, Map3k6, Rnf125, Tac1, and Tnfsf9 are all components of intercellular signaling pathways (e.g. cytokine, interleukin, TNF, interferon, and neuropeptide), and as we have shown (Fig. 7C) these genes are expressed to different extents in both neurons and astrocytes. Thus we cannot rule out the possibility that Kdm6b-dependent interactions between neurons and non-neuronal cells could be required to promote neuronal survival. For example, a recent study showed that inhibition of Kdm6b expression in the substantia nigra in vivo changes microglial activation in a manner that results in enhanced neuronal death, though whether Kdm6b was acting in the neurons or the microglia of the substantia nigra was not determined (Tang et al., 2013). Future studies that address the cell-type specificity of Kdm6b expression and function will help resolve where and how this KDM is working to effect its functions in neuronal survival.
Finally, how does the knockdown of Kdm6b result in dysregulation of activity-induced gene expression? It is possible that the genes showing impaired inducibility in Kdm6b knockdown cells are direct targets of transcriptional activation by Kdm6b. Kdm6b has activity as a histone H3K27 demethylase, and trimethylation of H3K27 is correlated with transcriptional repression (Agger et al., 2007; Mikkelsen et al., 2007). However to what degree changes in H3K27 trimethylation are correlated with Kdm6b-dependent transcriptional activation remains unclear (De Santa et al., 2009), and in at least some cases Kdm6b has been shown to activate genes through mechanisms that are independent of its demethylase activity (Miller et al., 2010; Zhao et al., 2013). Future studies that quantitatively assess H3K27 trimethylation levels genome-wide in control and Kdm6b knockdown neurons will be required to determine whether this mechanism is important in the actions of Kdm6b described here. Regardless of the mechanisms by which it regulates transcription, Kdm6b is recruited to its binding sites in genomic DNA via protein-protein interactions with a variety of DNA binding proteins including T-box transcription factors, Smad proteins, and the estrogen receptor (Akizu et al., 2010; Miller et al., 2010; Svotelis et al., 2011). Overexpression of Kdm6b in heterologous cells was not sufficient to drive expression of most of the Kdm6b-dependent Bic+4AP-regulated genes we tested (Fig. S2), which raises the possibility that Kdm6b may interact with Bic+4AP regulated transcription factors in neurons. In this regard it is interesting that many cytokines, interleukins, and TNF family genes are targets of regulation by the activity-regulated transcription factor NF-κB (Mattson and Meffert, 2006). Given the potential of NF-κB to regulate Kdm6b expression as discussed above, we hypothesize that activity-induced expression of Kdm6b may function as a positive feedback loop to increase the gain of NF-κB signaling in the brain, driving a neuroinflammation-based homeostatic adaptive response to increased neuronal activity.
Supplementary Material
Oligonucleotide primers used in this study.
RNA-Seq data.
Bic+4AP induced genes. All expressed genes (FPKM > 0) were averaged for Vector control and Kdm6b shRNA1 knockdown conditions for either control or 3hr Bic+4AP treatment conditions.
Kdm6b-dependent Bic+4AP induced genes. All genes showing >1.5 fold induction by Bic-AP were analyzed for relative induction in Kdm6b shRNA compared with vector control conditions.
Neuronal activity-induced expression of a set of known pro-survival genes is unaffected by Kdm6b knockdown. Quantitative PCR was used to measure baseline and Bic+4AP induced expression of the indicated genes in neurons infected with the shRNA control vector (white bars) or either of two independent Kdm6b shRNAs (gray and black bars). Two fully independent experiments were used for each shRNA, and these samples were independent of the samples used for RNA sequencing. Only Bic+4AP induced levels of gene expression are shown and data are graphed as the percentage of Bic+4AP induced levels for each gene in the vector control infected cells. n=6–9 per group.
Overexpression of Kdm6b is not sufficient to drive strong expression of candidate target genes. Control or Kdm6b overexpressing 293T cells were analyzed by quantitative PCR for expression of the gene set from Figure 7B that shows Kdm6b dependence for Bic+4AP induction. Delta Ct analysis was used to calculate the expression of each gene as a percentage of the expression of the housekeeping gene GAPDH. n=6 samples/condition, *p<0.05 overexpressed compared with control for each gene.
Dissociated neuronal cultures contain both neurons and astrocytes. We immunostained dissociated mouse hippocampal cultures with markers of neurons (MAP2, green) and astrocytes (GFAP, red). Nuclei of all cells were identified by staining with the DNA binding dye Hoechst (blue). An investigator identified each Hoechst+ cell as a MAP2+ neuron or GFAP+ astrocyte in five independent fields on each of three independent coverslips. Counting 1000 cells showed 80.3% neurons and 19.7% astrocytes in these cultures. Scale bar 25μm.
Acknowledgments
This work was supported in part by grants from the Ruth K. and Shepherd Broad Biomedical Research Foundation (A.E.W.) and NIH grant 1R01DA033610 (A.E.W.). We thank Jalen Carter for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Akizu N, Estaras C, Guerrero L, Marti E, Martinez-Balbas MA. H3K27me3 regulates BMP activity in developing spinal cord. Development. 2010;137:2915–2925. doi: 10.1242/dev.049395. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Garcia-Verdugo JM. Neurogenesis in adult subventricular zone. J Neurosci. 2002;22:629–634. doi: 10.1523/JNEUROSCI.22-03-00629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadoro G, Pieri M, Ciotti MT, Carunchio I, Canu N, Calissano P, Zona C, Severini C. Substance P provides neuroprotection in cerebellar granule cells through Akt and MAPK/Erk activation: evidence for the involvement of the delayed rectifier potassium current. Neuropharmacology. 2007;52:1366–1377. doi: 10.1016/j.neuropharm.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- Beattie EC, Stellwagon D, Morishita W, Bresnahan JC, Ha BK, Von Zastrow M, Beattie MS, Malenka RC. Control of synaptic strength by TNFalpha. Science. 2002;295:2282–2285. doi: 10.1126/science.1067859. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau N, Widmann C, Lazdunski M, Heurteaux C. Activation of the nuclear factor-kappaB is a key event in brain tolerance. J Neurosci. 2001;21:4668–4677. doi: 10.1523/JNEUROSCI.21-13-04668.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castermans D, Vermeesch JR, Fryns JP, Steyaert JG, Van de Ven WJ, Creemers JW, Devriendt K. Identification and characterization of the TRIP8 and REEP3 genes on chromosome 10q21.3 as novel candidate genes for autism. Eur J Hum Genet. 2007;15:422–431. doi: 10.1038/sj.ejhg.5201785. [DOI] [PubMed] [Google Scholar]
- Castren E, da Penha Berzaghi M, Lindholm D, Thoenen H. Differential effects of MK-801 on brain-derived neurotrophic factor mRNA levels in different regions of the rat brain. Exp Neurol. 1993;122:244–252. doi: 10.1006/exnr.1993.1124. [DOI] [PubMed] [Google Scholar]
- Chawla S, Hardingham GE, Quinn DR, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Nature. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W, Lv R, Li X, Villen J, Gygi SP, Liu XS, Shi Y. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 2012;26:1364–1375. doi: 10.1101/gad.186056.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli A, Calza A, Santoru F, Grasso F, Concas A, Sassoe-Pognetto M, Giustetto M. Morphine withdrawal produces ERK-dependent and ERK-independent epigenetic marks in neurons of the nucleus accumbens and lateral septum. Neuropharmacology. 2013;70:168–179. doi: 10.1016/j.neuropharm.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Clifford DB, Olney JW, Maniotis A, Collins RC, Zorumski CF. The functional anatomy and pathology of lithium-pilocarpine and high-dose pilocarpine seizures. Neuroscience. 1987;23:953–968. doi: 10.1016/0306-4522(87)90171-0. [DOI] [PubMed] [Google Scholar]
- Curia G, Longo D, Biagini G, Jones RS, Avoli M. The pilocarpine model of temporal lobe epilepsy. J Neurosci Methods. 2008;172:143–157. doi: 10.1016/j.jneumeth.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S, Testa G, Sung WK, Wei CL, Natoli G. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidday JM. Cerebral preconditioning and ischaemic tolerance. Nat Rev Neurosci. 2006;7:437–448. doi: 10.1038/nrn1927. [DOI] [PubMed] [Google Scholar]
- Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklin AV, Deramus T, Wheelock M, Davis RL, McMahon LL, Lubin FD. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci. 2012;32:5440–5453. doi: 10.1523/JNEUROSCI.0147-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Arnold FJL, Bading H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat Neurosci. 2001;4:261–267. doi: 10.1038/85109. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- Herz HM, Shilatifard A. The JARID2-PRC2 duality. Genes Dev. 2010;24:857–861. doi: 10.1101/gad.1921610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hu LY, Tepper CG, Lo SH, Lin WC. An efficient strategy to identify early TPA-responsive genes during differentiation of HL-60 cells. Gene Expr. 2006;13:179–189. doi: 10.3727/000000006783991791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S-C, Chrivia J, Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. J Neurosci. 2002;22:8422–8428. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WFt, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114:3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Jiang W, Van Cleemput J, Sheerin AH, Ji SP, Zhang Y, Saucier DM, Corcoran ME, Zhang X. Involvement of extracellular regulated kinase and p38 kinase in hippocampal seizure tolerance. J Neurosci Res. 2005;81:581–588. doi: 10.1002/jnr.20566. [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Hatazaki S, Johnson MB, Bellver-Estelles C, Mouri G, Bonner C, Prehn JH, Meller R, Simon RP, Henshall DC. Hippocampal transcriptome after status epilepticus in mice rendered seizure damage-tolerant by epileptic preconditioning features suppressed calcium and neuronal excitability pathways. Neurobiol Dis. 2008;32:442–453. doi: 10.1016/j.nbd.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Kamikawa Y, Donohoe M. The localization of histone H3K27me3 demethylase Jmjd3 is dynamically regulated. Epigenetics. 2014;9 doi: 10.4161/epi.28524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan AA, van der Hel WS, Kolk SM, Bos IW, Verlinde SA, van Nieuwenhuizen O, de Graan PN. Prolonged increase in rat hippocampal chemokine signalling after status epilepticus. J Neuroimmunol. 2012;245:15–22. doi: 10.1016/j.jneuroim.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Karpova NN, Rantamaki T, Di Lieto A, Lindemann L, Hoener MC, Castren E. Darkness reduces BDNF expression in the visual cortex and induces repressive chromatin remodeling at the BDNF gene in both hippocampus and visual cortex. Cell Mol Neurobiol. 2010;30:1117–1123. doi: 10.1007/s10571-010-9544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly ME, McIntyre DC. Hippocampal kindling protects several structures from the neuronal damage resulting from kainic acid-induced status epilepticus. Brain Res. 1994;634:245–256. doi: 10.1016/0006-8993(94)91927-5. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruidenier L, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok RPS, Lundblad JR, Chrivia JC, Richards JP, Bächinger HP, Brennan RG, Roberts SGE, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;379:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lee B, Cao R, Choi YS, Cho HY, Rhee AD, Hah CK, Hoyt KR, Obrietan K. The CREB/CRE transcriptional pathway: protection against oxidative stress-mediated neuronal cell death. J Neurochem. 2009;108:1251–1265. doi: 10.1111/j.1471-4159.2008.05864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Liu H, Cao Y, Basbaum AI, Mazarati AM, Sankar R, Wasterlain CG. Resistance to excitotoxin-induced seizures and neuronal death in mice lacking the preprotachykinin A gene. Proc Natl Acad Sci U S A. 1999;96:12096–12101. doi: 10.1073/pnas.96.21.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog Neurobiol. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Meffert MK. Roles for NF-kappaB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13:852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Goodman Y, Luo H, Fu W, Furukawa K. Activation of NF-kappaB protects hippocampal neurons against oxidative stress-induced apoptosis: evidence for induction of manganese superoxide dismutase and suppression of peroxynitrite production and protein tyrosine nitration. J Neurosci Res. 1997;49:681–697. doi: 10.1002/(SICI)1097-4547(19970915)49:6<681::AID-JNR3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- Parent JM, Yu TW, Leibowitz RT, Geschwind DH, Sloviter RS, Lowenstein DH. Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus. J Neurosci. 1997;17:3727–3738. doi: 10.1523/JNEUROSCI.17-10-03727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa E, Ye K, Nolan KA, Morrell L, Okun JM, Persky AD, Saito T, Lachman HM. Positive association of schizophrenia to JARID2 gene. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:45–51. doi: 10.1002/ajmg.b.30386. [DOI] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploski JE, Newton SS, Duman RS. Electroconvulsive seizure-induced gene expression profile of the hippocampus dentate gyrus granule cell layer. J Neurochem. 2006;99:1122–1132. doi: 10.1111/j.1471-4159.2006.04156.x. [DOI] [PubMed] [Google Scholar]
- Ryan JC, Morey JS, Ramsdell JS, Van Dolah FM. Acute phase gene expression in mice exposed to the marine neurotoxin domoic acid. Neuroscience. 2005;136:1121–1132. doi: 10.1016/j.neuroscience.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Kastner R, Humpel C, Wetmore C, Olson L. Cellular hybridization for BDNF, trkB, and NGF mRNAs and BDNF-immunoreactivity in rat forebrain after pilocarpine-induced status epilepticus. Exp Brain Res. 1996;107:331–347. doi: 10.1007/BF00230416. [DOI] [PubMed] [Google Scholar]
- Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperk G, Marksteiner J, Saria A, Humpel C. Differential changes in tachykinins after kainic acid-induced seizures in the rat. Neuroscience. 1990;34:219–224. doi: 10.1016/0306-4522(90)90315-u. [DOI] [PubMed] [Google Scholar]
- Svotelis A, Bianco S, Madore J, Huppe G, Nordell-Markovits A, Mes-Masson AM, Gevry N. H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ERalpha ligand dependency. EMBO J. 2011;30:3947–3961. doi: 10.1038/emboj.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Tamatani M, Che YH, Matsuzaki H, Ogawa S, Okado H, Miyake S, Mizuno T, Tohyama M. Tumor necrosis factor induces Bcl-2 and Bcl-x expression through NFkappaB activation in primary hippocampal neurons. J Biol Chem. 1999;274:8531–8538. doi: 10.1074/jbc.274.13.8531. [DOI] [PubMed] [Google Scholar]
- Tang Y, Li T, Li J, Yang J, Liu H, Zhang’ J, Le W. Jmjd3 is essential for the epigenetic modulation of microglia phenotypes in the immune pathogenesis of Parkinson’s disease. Cell Death Diff. 2013 doi: 10.1038/cdd.2013.159. ePub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SC, Lin SJ, Lin CJ, Chou YC, Lin JH, Yeh TH, Chen MR, Huang LM, Lu MY, Huang YC, Chen HY, Tsai CH. Autocrine CCL3 and CCL4 induced by the oncoprotein LMP1 promote Epstein-Barr virus-triggered B cell proliferation. J Virol. 2013;87:9041–9052. doi: 10.1128/JVI.00541-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–437. doi: 10.1101/gad.1864410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE. Activity-Dependent Regulation of Brain-derived neurotrophic factor Transcription. In: Dukek S, editor. Transcriptional Regulation by Neuronal Activity. New York, NY: Springer; 2008. pp. 155–174. [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005744. pii: a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LL, Chao A, Bilbo SD. Environmental enrichment alters glial antigen expression and neuroimmune function in the adult rat hippocampus. Brain Behav Immun. 2012;26:500–510. doi: 10.1016/j.bbi.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthos DN, Sandkuhler J. Neurogenic neuroinflammation: inflammatory CNS reactions in response to neuronal activity. Nat Rev Neurosci. 2014;15:43–53. doi: 10.1038/nrn3617. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Steijaert MN, Lau D, Schutz G, Delucinge-Vivier C, Descombes P, Bading H. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron. 2007;53:549–562. doi: 10.1016/j.neuron.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5:e1000604. doi: 10.1371/journal.pgen.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q, Chen Y, Wang HY, Wang RF. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell. 2013;152:1037–1050. doi: 10.1016/j.cell.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 1999;13:382–387. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers used in this study.
RNA-Seq data.
Bic+4AP induced genes. All expressed genes (FPKM > 0) were averaged for Vector control and Kdm6b shRNA1 knockdown conditions for either control or 3hr Bic+4AP treatment conditions.
Kdm6b-dependent Bic+4AP induced genes. All genes showing >1.5 fold induction by Bic-AP were analyzed for relative induction in Kdm6b shRNA compared with vector control conditions.
Neuronal activity-induced expression of a set of known pro-survival genes is unaffected by Kdm6b knockdown. Quantitative PCR was used to measure baseline and Bic+4AP induced expression of the indicated genes in neurons infected with the shRNA control vector (white bars) or either of two independent Kdm6b shRNAs (gray and black bars). Two fully independent experiments were used for each shRNA, and these samples were independent of the samples used for RNA sequencing. Only Bic+4AP induced levels of gene expression are shown and data are graphed as the percentage of Bic+4AP induced levels for each gene in the vector control infected cells. n=6–9 per group.
Overexpression of Kdm6b is not sufficient to drive strong expression of candidate target genes. Control or Kdm6b overexpressing 293T cells were analyzed by quantitative PCR for expression of the gene set from Figure 7B that shows Kdm6b dependence for Bic+4AP induction. Delta Ct analysis was used to calculate the expression of each gene as a percentage of the expression of the housekeeping gene GAPDH. n=6 samples/condition, *p<0.05 overexpressed compared with control for each gene.
Dissociated neuronal cultures contain both neurons and astrocytes. We immunostained dissociated mouse hippocampal cultures with markers of neurons (MAP2, green) and astrocytes (GFAP, red). Nuclei of all cells were identified by staining with the DNA binding dye Hoechst (blue). An investigator identified each Hoechst+ cell as a MAP2+ neuron or GFAP+ astrocyte in five independent fields on each of three independent coverslips. Counting 1000 cells showed 80.3% neurons and 19.7% astrocytes in these cultures. Scale bar 25μm.