Abstract
AIM: To investigate whether the presence of human leukocyte antigen (HLA) marker could add new information to discriminated atypical diabetic type 2 patients.
METHODS: We analyzed 199 patients initially diagnosed as type 2 diabetes who are treated in special care diabetes clinics (3rd level). This population was classified in “atypical” (sample A) and “classic” (sample B) according to HLA typing. We consider “classic patient” when has absence of type 1 diabetes associated HLA alleles and no difficulties in their diagnosis and treatments. By the other hand, we considered “atypical patient” when show type 1 diabetes associated HLA alleles and difficulties in their diagnosis and treatments. The standard protocol Asociacion Latinoamericana de Diabetes 2006 was used for patients follow up. To analyze differences between both populations in paraclinical parameters we used unpaired t tests and contingence tables. Bivariate and multivariate analyses were carried out using the SPSS software program. In all studies we assume differences statistically significant, with a P-value < 0.05 corrected and 95%CI.
RESULTS: The typing HLA in the “atypical” populations show that 92.47% patients presented at list one type 1 diabetes associated HLA alleles (DQB1*0201-0302 and DR 3-4) and 7.53% had two of its. The results showed for categorical variables (family history, presence or absence of hypertension and/or dyslipidemia, reason for initial consultation) the only difference found was at dyslipidemia (OR = 0.45, 0.243 < OD < 0.822 (P < 0.001). In relation to continuous variables we found significant differences between atypical vs classic only in cholesterol (5.07 ± 1.1 vs 5.56 ± 1.5, P < 0.05), high density lipoproteins (1.23 ± 0.3 vs 1.33 ± 0.3, P < 0.05) and low density lipoproteins (2.86 ± 0.9 vs 3.38 ± 1.7, P < 0.01). None of the variables had discriminating power when logistic regression was done.
CONCLUSION: We propose an algorithm including HLA genotyping as a tool to discriminate atypical patients, complementing international treatment guidelines for complex patients.
Keywords: Atypical diabetes, Clinical algorithm, Immunity molecular marker
Core tip: There are evidences that exists a lot of patients who were diagnosed as type 2 diabetics but present difficult management, don’t have good responses to treatment and don’t achieve the metabolic goals. We include the study of human leukocyte antigen markers typically associated whit type 1 diabetes to characterize these patients. This paper provides information about the possibility of incorporate a standardized molecular diagnosis in the clinical practice to identify complex or atypical type 2 diabetic patient.
INTRODUCTION
Diabetes mellitus is a chronic disease that requires ongoing medical care to prevent acute complications and reduce the risk of long-term complications. While recognizing two major groups of diabetic patients, type 1 and 2, the clinical presentation and disease progression vary considerably in both types of diabetes. However, ADA Position Statement establishes that there are patients who cannot be classified as type 1 or type 2[1]. The true diagnosis may be more obvious only over time. There is growing evidence that emphasize the existence of a significant overlap between diabetes type 1 and 2[2-12].
Despite the increasing incidence of the disease and the efforts made to establish diagnostic guidelines some patients do not qualify strictly into the given definitions.
Such patients which can be simultaneously classified in more than one group significantly complicate the medical treatment. They generally require the assistance of a multidisciplinary team in second or third level centers. It is in these patients considered “atypical”, where it is necessary to deepen the diagnosis with other complementary examinations with additional technologies. In these cases the classical diagnostic markers and risk factors analysis for various chronic complications, are not sufficient by themselves for a clinical differentiation. In a previous paper we found a high proportion of type 2 diabetes patients who presented HLA susceptibility alleles for type 1 diabetes[13]. Therefore, we propose to add the usage of a molecular marker (HLA) to the international standard criteria
According to the ADA type 1 diabetes is strongly associated with specific HLA groups while in type 2 diabetes does not exist this association[14]. Of all of the type 1 diabetes associated genes and regions revealed by different studies, the HLA association remains the strongest by far, with reported ORs ranging from 0.02 to .11 for specific DR-DQ haplotypes[15,16].
The presence of these genetic variants in patients diagnosed as type 2 let us assign them the “atypical” label. We propose this clinical, biochemical and molecular study to keep deepening in the characterization of HLA as a tool for their differentiation.
In this paper we pretend to provide the Clinicians with a tool to identify those patients at atypical presentation in whom the algorithms have not been useful. We present the basis for a possible new algorithm that can contribute to the early identification of these problematic patients.
MATERIALS AND METHODS
Population design
We analyzed a population of 199 patients seen in 3rd level Clinics for Diabetes from two centers: public (Pasteur Hospital) and private initially diagnosed with type 2 diabetes[14]. For the preparation of this study were considered only those patients receiving comprehensive care of their diabetes, following a nutritional plan and presenting a good adherence to physical activity according to their functional ability within the recommendations of Asociacion Latinoamericana de Diabetes (ALAD)/ADA and medicated with one or more oral antidiabetic drugs. In turn, this population was classified based on the presence or absence of type 1 diabetes HLA susceptibility alleles described in the Uruguayan population[13].
Sample A: 93 “atypical” patients that met the following inclusion criteria: (1) Patients who had good adherence to the treatment; (2) They fulfilled the objectives of education and nutrition plans according to international guidelines; (3) Present doubts on classification of diabetic type and/or no good therapeutic response (two consecutive measurements of glicated hemoglobin within three months not reduced in 1.5%[17]) to ADA, ALAD algorithms; and (4) Patients with susceptibility HLA alleles for autoimmune disease. We considered DQB1*0201-0302 and DR 3-4 as susceptible ones in the Uruguayan population[18].
Sample B: 106 “classic” patients fulfilling the same requirements a, b of sample A but which do not have diagnostic doubts, responded to treatment and do not present HLA alleles associated with autoimmune disease.
Patient of both samples who had other endocrine disorders or tumors were excluded.
All subjects were interviewed by medical doctors following ALAD guidelines on diagnosis treatment and control of type 2 diabetes with evidence-based medicine[19].
All patients were assessed for the following items: (1) Family history of diabetes; (2) Personal history: chronological age, age at diagnosis, time of evolution; (3) Motive of initial consultation: patients were categorized into five groups: incidental finding by fasting glucose, oral glucose tolerance test, presence of typical symptoms, acute debut with ketoacidosis without precipitating cause, and patients referred by other specialists for the presence of complications; (4) Presence or absence of classical risk factors associated with type 2 diabetes (hypertension and/or dyslipidemia); (5) Body mass index (BMI) was calculated and categorized according to the World Health Organization[20]: overweight (25-29.9 kg/m2) and obesity (≥ 30 kg/m2); and (6) Clinical evaluation and metabolic parameters: glicated hemoglobin, cholesterol, low density lipoproteins (LDL), high density lipoproteins (HDL), triglycerides (TG), TG/HDL ratio as insulin-resistance index (> 3)[21,22]. To analyze levels of dyslipidemia, both samples were stratified according to the 2° Dyslipidemia Consensus in Uruguay (Table 1)[23]. We analyzed the phenotypic classification of dyslipidemia respect to Table 2[24].
Table 1.
Reference values (mmol/L) of parameters stratified
| Dyslipidemia parameters | Desirable | Limit | Abnormal |
| Total cholesterol | < 5.2 | 5.2-6.19 | > 6.2 |
| HDL | > 1.2 | 1.2-0.9 | < 0.9 |
| LDL | < 3.4 | 3.4-4.0 | > 4.1 |
| Triglycerides | 2.3 | 2.3-2.99 | > 3.0 |
HDL: High density lipoproteins; LDL: Low density lipoproteins.
Table 2.
Phenotypic classification of dyslipidemia
| Total Cholesterol | LDL | Triglycerides | HDL | |
| Hypercholesterolemia | ≥ 6.2 | ≥ 4.1 | < 2.3 | |
| Combined hyperlipidemia | ≥ 4.1 | ≥ 2.3 | ||
| Hipo alfa lipoproteinemia | > 4 | < 2.3 | < 1 |
Reference values (mmol/L). HDL: High density lipoproteins; LDL: Low density lipoproteins.
Molecular analysis
DNA was obtained from peripheral blood using standard (phenol/chloroform) technique. The HLA typing was performed by reverse ASO technique (Innogenetics Ltd, Belgium, UE).
All patients gave written informed consent and the study protocol was approved by the Ethical Committee of Ministry of Public Health and the corresponding Ethical Committee of each participant Institution.
Statistical analysis
Continuous variables were expressed as the means and standard deviations. Differences between groups were determined by unpaired t tests after checking the normal distribution or converted to normalize of the data. Categorical variables were described using proportion and 2 × 2 contingence table. Bivariate and multivariate analyses were based on dependent variables (two categories sample A, sample B). Logistic regression with all variables was done. All tests were carried out using the SPSS software program. In all studies we assume differences statistically significant, with a P-value < 0.05 corrected and 95%CI.
RESULTS
Population characterizes
The total population consisted of 94 women (47.24%) and 105 men (52.76%). The gender distribution was similar in samples A and B. In the statistical analysis of categorical variables (family history, presence or absence of hypertension and/or dyslipidemia, reason for initial consultation) the only difference found was at dyslipidemia (ODDs 0.45, CI: 0.243-0.822 (P < 0.001)). In relation to values of cholesterol, HDL, LDL and TG, only the last parameter not showed statistical differences (Table 3). Subsequently each of these variables was analyzed, separating into classes in accordance to the Uruguayan Dyslipidemia Consensus. Sample A showed a higher proportion of normal values for cholesterol and LDL (55.9% vs 37.7%, 70.7% vs 54.8%, respectively). In relation to dyslipidemia phenotypic classification, hypercholesterolemia was the only parameters statistically significant: 12.3% atypical patients vs 2.2%, classic patients with ODDs 0.07 (CI: 0.009-0.54).
Table 3.
Clinical characteristics expressed by media and standard deviation
| Sample A n = 93 | Sample B n = 106 | P value | |
| Age (yr) | 62.01 ± 11.65 | 66.02 ± 9.55 | 0.060 |
| Age onset (yr) | 47.18 ± 12.61 | 49.54 ± 10.13 | 0.131 |
| Years of evolution | 16.41 ± 9.72 | 15.45 ± 9.22 | 0.528 |
| BMI (kg/mts2) | 32.07 ± 5.26 | 31.45 ± 5.95 | 0.430 |
| HbA1c (%)1 | 8.31 ± 1.87 | 8.16 ± 1.65 | 0.545 |
| Total cholesterol (nmol/L) | 5.07 ± 1.1 | 5.56± 1.5 | 0.010a |
| HDL (nmol/L) | 1.23 ± 0.3 | 1.33 ± 0.3 | 0.010a |
| LDL (nmol/L) | 2.86 ± 0.9 | 3.38 ± 1.7 | 0.009b |
| Triglycerides (nmol/L) | 2.29 ± 1.4 | 2.81 ± 0.9 | 0.864 |
| TG/HDL | 2.10 ± 1.5 | 1.96 ± 2.0 | 0.572 |
At beginning of the study. Sample A vs Sample B:
P < 0.05,
P < 0.01 for all parameters. BMI: Body mass index; LDL: Low-density lipoprotein-cholesterol; HDL: High-density lipoprotein-cholesterol; TG/HDL: Insulin resistance index; HbA1c (%): Glycated hemoglobin percentage.
Furthermore, we found that only part of the patients from the sample A (atypical) presented classical risk factors associated with type 2 diabetes (hypertension and/or dyslipidemia).
Analyzing the qualitative variables (Table 4) the only difference found was also in the lipid profile. In relation to BMI no difference between both samples were observed. It is important to point out those only 4 individuals in sample A had a normal weight in spite of having HLA alleles associated with type 1 diabetes.
Table 4.
χ2 analysis
| OR | 95%CI | P value | |
| Dyslipidemia | 0.45 | 0.24-0.82 | < 0.01 |
| Total cholesterol | 0.48 | 0.27-0.84 | < 0.01 |
| HDL | 1.84 | 1.04-3.23 | < 0.05 |
| LDL | 0.50 | 0.28-0.91 | < 0.05 |
LDL: Low-density lipoprotein-cholesterol; HDL: High-density lipoprotein-cholesterol.
None of the variables had discriminating power when logistic regression was done. The P value of the χ2 test was > 0.05.
HLA marker
The typing HLA in the “atypical” populations show that 92.47% patients presented at list one type 1 diabetes associated HLA alleles (DQB1*0201-0302 and DR 3-4) and 7.53% had two of its.
DISCUSSION
The usual elements that are taken into consideration in the diagnosis and treatment of atypical diabetic patients are not sufficient for identify individuals considered “atypical” for presentation, evolution and/or poor therapeutic response according to international guidelines. For this reason, we investigate whether the inclusion of an immunity molecular marker would provide conclusive information that helps the Clinician with an appropriate individualized therapeutic classification in this group of patients.
According to the consensus this marker differentiates two major types of diabetes. Type 1 diabetes is strongly associated with HLA while type 2 diabetes is not[14]. Our study demonstrated that the clinical, biochemical and molecular-genetic characterization of atypical patient population and their comparison with classic type 2 diabetes patients showed that although a few differences were found to be statistically significant, they are not individually sufficient to clarify the situation of each patient. We propose here to add the usage of HLA typing to the international standard criteria.
Despite the enormous efforts that have been made to identify gene variants associated with type 2 diabetes, until present no one has fulfill the expectations to prevent or improve the treatment of diabetes. The addition of the genotypic variants risk score to clinical prediction models, only moderately (minimally) improve the statistical results[25,26]. In a previous paper analyzing the genotype-phenotype relation, observed the existence of a high proportion of patients that despite being classified as type 2 diabetes according to the diagnostic guidelines, they presented HLA alleles strongly associated with type 1 diabetes[27].
The observed statically differences in the lipid profiles of atypical patients are insufficient to define changes in classification, treatment and/or monitoring. In these complex patients usual clinical markers used for diagnosis and for the risk factors analysis for various complications were not sufficient by themselves to differentiate classic type 2 diabetics.
BMI is usually considered as an important marker to differentiate between types of diabetes but, no differences were observed between classical and atypical patients. As in these patients a fast increment of the obesity rate has been observed, the presence of this factor has been considered as an important factor in reducing the described differences between type 1 and 2 diabetes[12]. The presences of overweight or obesity would induce the Clinician not to look for the presence of HLA susceptibility to autoimmune disease. In fact, in the sample of atypical patients only 4 of them had normal weight despite having HLA alleles associated with type 1 diabetes. This finding is not consistent with international classifications where, although there may be exceptions, defines the patient with type 2 diabetes as overweight or with abdominal fat distribution without autoimmunity, while rarely type 1 diabetics are obese[1].
Based on these data, we believe that this molecular marker analysis provide valuable data to clarify these patients. It is also clear that the mere presence of molecular marker is not indicative of the evolution of each patient’s disease or how pancreatic reserve presents in each individual.
From the results, we consider that the study should be complemented with the search for other clinical or evolution markers to enable an accurate differentiation. Dosage of peptide C could be a very good parameter to evaluate the stage of beta cell. This factor was not included in this study because it is not standardized in Uruguay.
At present, we have not enough evidence to answer a crucial question on these atypical patients, at what point the genetic study should be done? (1) to debut; (2) after adopt changes in lifestyle and no achieve control objectives were observed; (3) after 6 mo of no response to treatment plan indicated by international guidelines; and (4) at any time of evolution. We think that is important know the genotype of the patient when, after adjusting nutritional plan and changes in lifestyle, no clinical improvements were observed. This question should be answered with new evidence that address the issues raised in this work.
Here, we simply propose a new tool for the Clinician. We are aware that the genetic typing of HLA is a costly analysis but, the information presented here justifies its implementation in a very specific group of patients. From our point of view, the addition of such study to the actually used algorithm would clearly help to Clinicians in making a different evaluation of atypical patients (Figure 1).
Figure 1.
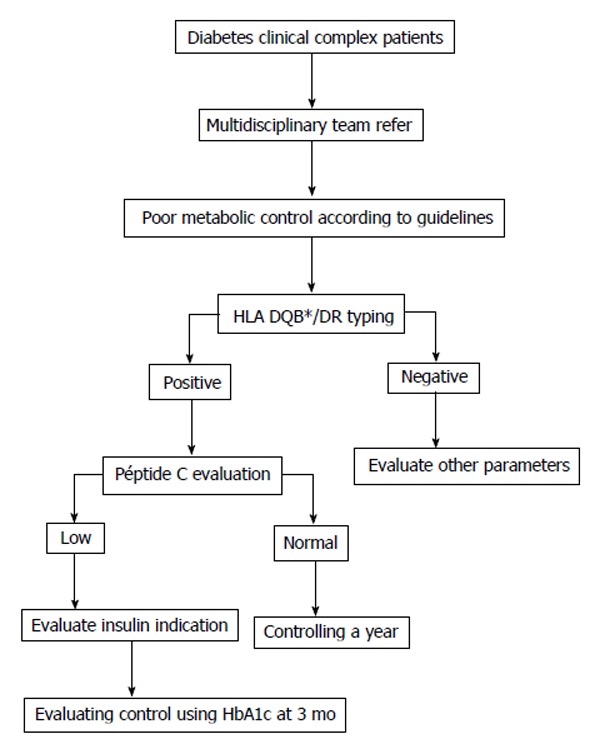
Algorithm for complex diabetes patients with difficulties in diagnosis, evolution, poor therapeutic response where international algorithms have been fulfilled. HbA1c: Glycated hemoglobin.
All authors are requested to disclose any actual or potential conflict of interest including any financial, personal or other relationships with other people or organizations within three years of beginning the submitted work that could inappropriately influence, or be perceived to influence, their work.
COMMENTS
Background
The type 2 diabetes is a pathology that represents serious challengers for the clinicians. Now a day more and more patient cannot be classified in neither of the type 1 or type 2 diabetes. Their clinical presentation, evolution and difficulty in achieving therapeutic goals make them atypical patients. According to previous data most atypical patients, classically diagnosed as type 2 diabetes, show type 1 diabetes associated human leukocyte antigen (HLA) alleles. These genetics variants can appear like markers for atypical type 2 diabetic patients.
Research frontiers
The HLA marker was classically associated whit type 1 diabetes. Their presence in diabetic type 2 patient is reported in about 10%. This study show that type 2 diabetic patients with the same adherence to indications and treatments will have a different develop of their disease when have type 1 diabetes associated HLA alleles. The authors try to demonstrate that this marker is a potentially way to differentiate patient who will be out of guise of treatments, in response to drugs and in achieve metabolic goals.
Innovations and breakthroughs
Recent reports have highlighted the importance of improve the knowledge of type 2 diabetes, their etiology, diagnosis and treatments. The global grow tendency of this pathology and the difficulties observed in this treatments makes experts check over algorithm for a good follow up of this patients. This is the first study to report a standardized marker to include in the algorithm in order to identify uncharacteristically type 2 diabetics.
Applications
By understanding how the development of the type 2 diabetes in atypical patients is the authors have to recognize them early. This study may represent a future strategy for discriminates them and use the guides of treatments in an individualized way.
Terminology
HLA marker implicates genetics variants which had being studding since a lot of year for their associated to autoimmunity showed for type 1 diabetes. There are susceptibility and protectant alleles. Non-surprisingly, these variants were reported in type 2 diabetes but are unknown there influence in the development of the pathology.
Peer review
Overall an interesting manuscript, which helps to shed some discriminatory light on a growing sub-population of diabetic patients who cannot be readily classified as type 1 or type 2 based upon their medical history and metabolic profile.
Footnotes
P- Reviewer: Romani A, Velasco I, Zhao JB S- Editor: Wen LL L- Editor: A E- Editor: Liu SQ
References
- 1.American Diabetes Association. Standards of medical care in diabetes--2012. Diabetes Care. 2012;35 Suppl 1:S11–S63. doi: 10.2337/dc12-s011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naik RG, Brooks-Worrell BM, Palmer JP. Latent autoimmune diabetes in adults. J Clin Endocrinol Metab. 2009;94:4635–4644. doi: 10.1210/jc.2009-1120. [DOI] [PubMed] [Google Scholar]
- 3.Libman IM, Becker DJ. Coexistence of type 1 and type 2 diabetes mellitus: “double” diabetes? Pediatr Diabetes. 2003;4:110–113. doi: 10.1034/j.1399-5448.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 4.Kolb H, Mandrup-Poulsen T. An immune origin of type 2 diabetes? Diabetologia. 2005;48:1038–1050. doi: 10.1007/s00125-005-1764-9. [DOI] [PubMed] [Google Scholar]
- 5.Tuomi T. Type 1 and type 2 diabetes: what do they have in common? Diabetes. 2005;54 Suppl 2:S40–S45. doi: 10.2337/diabetes.54.suppl_2.s40. [DOI] [PubMed] [Google Scholar]
- 6.Pozzilli P, Buzzetti R. A new expression of diabetes: double diabetes. Trends Endocrinol Metab. 2007;18:52–57. doi: 10.1016/j.tem.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Pozzilli P, Guglielmi C, Pronina E, Petraikina E. Double or hybrid diabetes associated with an increase in type 1 and type 2 diabetes in children and youths. Pediatr Diabetes. 2007;8 Suppl 9:88–95. doi: 10.1111/j.1399-5448.2007.00338.x. [DOI] [PubMed] [Google Scholar]
- 8.Mimbacas A, Javiel G. Diabetes type 1 and type 2: What is behind a classification? In: Chih-Pin Liu eds. Type 1 Diabetes - Complications, Pathogenesis, and Alternative Treatments; 2011. pp. 287–304. [Google Scholar]
- 9.Mimbacas A, Vitarella G, Souto J, Reyes AL, Farias J, Fernandez M, Fabregat M, Javiel G. The phenotype masks the genotype: a possible new Diabetes expression. J Pediatr Genet. 2012;2:131–134. doi: 10.3233/PGE-2012-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone MA, Camosso-Stefinovic J, Wilkinson J, de Lusignan S, Hattersley AT, Khunti K. Incorrect and incomplete coding and classification of diabetes: a systematic review. Diabet Med. 2010;27:491–497. doi: 10.1111/j.1464-5491.2009.02920.x. [DOI] [PubMed] [Google Scholar]
- 11.Inzucchi SE, Bergenstal RM, Buse JB, Diamant M, Ferrannini E, Nauck M, Peters AL, Tsapas A, Wender R, Matthews DR. Management of hyperglycemia in type 2 diabetes: a patient-centered approach: position statement of the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) Diabetes Care. 2012;35:1364–1379. doi: 10.2337/dc12-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inzucchi SE NHS. Royal College of General Practitioners. Available from: http: //www.webicina.com/
- 13.Mimbacas A, García L, Zorrilla P, Acosta M, Airaudo C, Ferrero R, Pena A, Simonelli B, Soto E, Vitarella G, et al. Genotype and phenotype correlations in diabetic patients in Uruguay. Genet Mol Res. 2009;8:1352–1358. doi: 10.4238/vol8-4gmr667. [DOI] [PubMed] [Google Scholar]
- 14.American Diabetes Association. Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 2011;34:s62–S69. doi: 10.2337/dc11-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2:a007732. doi: 10.1101/cshperspect.a007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, Mychaleckyj JC, Todd JA, Bonella P, Fear AL, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathan DM, Buse JB, Davidson MB, Ferrannini E, Holman RR, Sherwin R, Zinman B. Medical management of hyperglycemia in type 2 diabetes: a consensus algorithm for the initiation and adjustment of therapy: a consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care. 2009;32:193–203. doi: 10.2337/dc08-9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mimbacas A, Pérez-Bravo F, Hidalgo PC, Javiel G, Pisciottano C, Grignola R, Jorge AM, Gallino JP, Gasagoite J, Cardoso H. Association between diabetes type 1 and DQB1 alleles in a case-control study conducted in Montevideo, Uruguay. Genet Mol Res. 2003;2:29–35. [PubMed] [Google Scholar]
- 19.Asociacion Latinoamericana de Diabetes. 2012 Guias ALAD de diagnóstico, control y tratamiento de la diabetes tipo 2. Available from: http: //issuu.com/alad-diabetes/docs/guias_alad_2012.
- 20.World Health Organization. Global Database on Body Mass Index an interactive surveillance tool for monitoring nutrition transition. 2011. Available from: http: //apps.who.int/bmi/index.jsp. [Google Scholar]
- 21.McLaughlin T, Abbasi F, Cheal K, Chu J, Lamendola C, Reaven G. Use of metabolic markers to identify overweight individuals who are insulin resistant. Ann Intern Med. 2003;139:802–809. doi: 10.7326/0003-4819-139-10-200311180-00007. [DOI] [PubMed] [Google Scholar]
- 22.Marotta T, Russo BF, Ferrara LA. Triglyceride-to-HDL-cholesterol ratio and metabolic syndrome as contributors to cardiovascular risk in overweight patients. Obesity (Silver Spring) 2010;18:1608–1613. doi: 10.1038/oby.2009.446. [DOI] [PubMed] [Google Scholar]
- 23.2o Dislipidemia Consensus in Uruguay SUDEAT (Sociedad Uruguaya de Artereosclerrosis). 2o Consenso uruguayo sobre dislipidemias. El Taller de Willy, Eds. Montevideo, Uruguay. El Taller de Willy, Eds. Montevideo, Uruguay: 1998. [Google Scholar]
- 24.Lismann S. Tratamiento actual de las dislipemia. Available from: http: //www.intramed.net /UserFiles/Files/dislip.pdf.
- 25.Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, Berglund G, Altshuler D, Nilsson P, Groop L. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 26.Vassy JL, Meigs JB. Is genetic testing useful to predict type 2 diabetes? Best Pract Res Clin Endocrinol Metab. 2012;26:189–201. doi: 10.1016/j.beem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;27 Suppl 1:S5–S10. doi: 10.2337/diacare.27.2007.s5. [DOI] [PubMed] [Google Scholar]


