Abstract
Background:
Pseudomonas aeruginosa is a leading cause of nosocomial infections worldwide. Resistance of P. aeruginosa strains to the broad-spectrum cephalosporins may be caused by extended-spectrum β-lactamases (ESBLs).
Objectives:
The aim of this study was to determine the antimicrobial resistance patterns and prevalence of PER-1 and VEB-1 type genes among ESBL producing strains of P. aeruginosa.
Material and Methods:
A total of 106 P. aeruginosa isolates were collected from two university hospitals in Hamadan, Iran, during a7-month study (2009). The antimicrobial susceptibility of isolates was determined by disc diffusion method and interpreted according to the clinical and laboratory standards institute (CLSI) recommendations. Production of ESBL was determined by combined disk test and presence of PER-1 and VEB-1 type ESBL genes was identified by PCR.
Results:
The resistance against broad-spectrum cephalosporins and monobactames were: cefepime (97%), cefotaxime (92.5%) ceftazidime (51%), and aztreonam (27%). Ciprofloxacin (91.5%), imipenem (84.9%) and meropenem (82.1%) were the most effective anti-pseudomonas agents in this study. The results revealed that 88.7% of the isolates were multidrug resistant, 58.25% of those were ESBL positive. Sixteen (26.6%), 9 (15%) and 3 (5%) strains among ESBL-producing strains contained blaPER-1, blaVEB and blaPER-1-blaVEB, respectively.
Conclusions:
This study highlighted the need to establish antimicrobial resistance surveillance networks for P. aeruginosa to determine the appropriate empirical treatment regimens. The high prevalence of multidrug resistance and production of ESBLs in P. aeruginosa isolates confirms the necessity of protocols considering these issues in the hospitals.
Keywords: Pseudomonas aeruginosa, Antimicrobial Drug Resistance, Beta-lactamase
1. Background
Pseudomonas aeruginosa is an important cause of nosocomial infections, including pneumonia, burn infection, urinary tract infections, meningitis and bacteremia. The infections can be particularly develop to a severe form in immune deficient patients (1). Antibiotics have been used successfully for several decades, but resistance genes have emerged and disseminated particularly in the last few years (2).
Extended-spectrum β-lactamases (ESBLs) mediate resistance to various broad-spectrum cephalosporins, including cefotaxime, ceftriaxone, ceftazidime, and aztreonam (3). These enzymes originally collected from Klebsiella pneumoniae and Escherichia coli and recently from P. aeruginosa (4-7). Most of the methods for detection of ESBLs are used in bacterial species such as Klebsiella and E. coli lacking chromosomal β-lactamase activity (8, 9), However, the detection of ESBL production in P. aeruginosa has some difficulties, because this bacterium not only has an inducible AmpC enzyme but also has an efflux-mediated resistance and a higher degree of impermeability than Enterobacteriaceae (10, 11). The PER-1 and VEB-1 type ESBLs belong to class A of β- lactamases and is associated with high level of resistance to cephems, monobactams and ceftazidime (12, 13).
2. Objectives
The aim of this study was to determine the antibacterial resistance patterns and prevalence of PER-1 and VEB type ESBLs among P. aeruginosa isolated from patients in west of Iran.
3. Materials and Methods
3.1. Bacterial Isolates
A total of 106 isolates of P. aeruginosa recovered from various clinical specimens (Table 1) in Beasat Teaching Hospital at the Hamadan University of Medical Sciences during a 7-month study in 2009. The identification were carried out by colonial morphology, positive oxidase test, pigment formation; growth test at 42ºC on nutrient agar, Gram staining and motility test.
Table 1. Distribution of P. seudomonas aeruginosa by Site of Isolation.
| Source | Isolates, No. (%) |
|---|---|
| Burn wounds | 55 (51.9) |
| Trashes | 20 (18.9) |
| Urine | 12 (11.3) |
| Blood | 7 (6.6) |
| Feces | 6 (5.7) |
| Sputum | 5 (4.7) |
| CSFa | 1 (0.9) |
| Total | 106 (100) |
a Abbreviation: CFS, Cerebral spinal fluid
3.2. Antibiotic Susceptibility Test
Antibiotic susceptibility of the isolates was determined by standard disk diffusion method (14). The following antibiotics were used: gentamicin (30 µg), aztreonam (30 µg), meropenem (10 µg), imipenem (10 µg), amikacin (30 µg), tobramycin (30 µg), piperacillin (30 µg), ceftazidime (30 µg), ciprofloxacin (5 µg), ofloxacin (5 µg), cefepime (30 µg) and cefotaxime (30 µg) (Himedia, India). P. aeruginosa ATCC 27853 was used as control. The results were interpreted according to the clinical and laboratory standards institute (CLSI) (15).
3.3. Phenotypic Detection of Beta-Lactamase
The isolates were tested for the ESBLs production by using combine disk test (CDT) as CLSI recommendations. CDT were performed on ceftazidime, cefotaxime, cefepime and aztreonam resistant strains by placing disks of ceftazidime, and cefotaxime (30 µg each) at a 20 mm distance from a disk containing ceftazidime-clavulanic acid (30/10μg), cefotaxime-clavulanic acid (30/10µg) and cefepime-clavulanic acid (30/10 µg) (16). ESBL production was inferred when the cephalosporin inhibitory zones were expanded by the clavulanate.
3.4. PCR Amplification
PCR amplifications were done using specific primers for the β-lactamases PER-1, and VEB-1 genes, as described previously (17). PCR was performed for all ESBL-producers which their resistance to cephalosporins and phenotypic was identified by confirmatory tests. DNA was extracted by the boiling method as previously described (17). The DNA amplification program consisted of an initial denaturation (94 °C, 5 minutes) followed by 35 cycles of denaturation (94 °C, 60 seconds), annealing (50 °C for PER-1 and 55 °C for VEB-1, 60 seconds), extension (72 °C, 45 seconds) and a single final extension for 5 minutes at 72 °C. Reaction mixtures for PCR contained 1.5mM MgCl2, 0.5 mM of each primer, 0.2 mM of dNTPs, 1 U of Taq polymerase, 1X PCR buffer and 2 µL of DNA. Primers PER-F (5-AATTTGGGCTTAGGGCAGAA-3') and PER-R (5'-ATGAATGTCATTATAAAAGC-3') were used for blaPER-1; and VEB-F (5'-CGACTTCCATTTCCCGATGC-3') and VEB-R (5'-GGACTCTGCAACAAAT AC GC-3') were used for blaVEB-1 amplification.
4. Results
4.1. Antimicrobial Susceptibility Test
Table 2 shows the antimicrobial susceptibility pattern of P. aeruginosa strains. Ciprofloxacin (91.5%), imipenem.
Table 2. Antimicrobial Susceptibility Pattern of P. aeruginosa Strains.
| Antimicrobials | Resistance, No. (%) | Sensitive, No. (%) | Intermediate, No. (%) |
|---|---|---|---|
| Amikacin | 32 (30.2) | 70 (66) | 4 (3.8) |
| Ceftazidime | 54 (51) | 40 (37.7) | 12(11.3) |
| Aztreonam | 29 (27.4) | 70 (66) | 7 (6.6) |
| Cefepime | 103 (97.2) | 1 (0.9) | 2 (1.9) |
| cefotaxime | 53 (50) | 10 (9.4) | 43 (40.6) |
| Ciprofloxacin | 5 (4.7) | 97 (91.5) | 4 (3.8) |
| Gentamicin | 39 (36.8) | 55 (51.9) | 12 (11.3) |
| Imipenem | 8 (7.5) | 90 (84.9) | 8 (7.5) |
| Meropenem | 14 (13.2) | 87 (82.1) | 5 (4.7) |
| Ofloxacin | 31 (29.2) | 72 (67.9) | 3 (2.8) |
| Piperacillin | 99 (93.4) | 7 (6.6) | 0 |
| Tobramycin | 39 (36.8) | 67 (63.2) | 0 |
(84.9%) and meropenem (82.1%) were the most active antimicrobial agents followed by ofloxacin (67.9% susceptibility). Aztreonam, a monobactam, and amikacin showed antibiotic activity against 66% of the strains. Susceptibility to the cephalosporins was reduced to 37.7% for ceftazidime, followed by cefotaxime (9.4%) and Cefepime (0.9%).
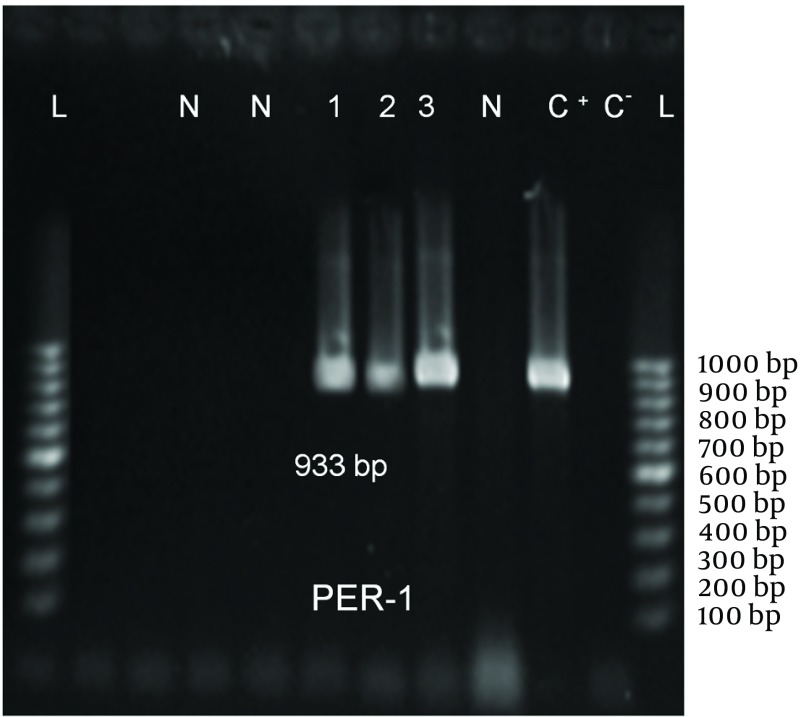
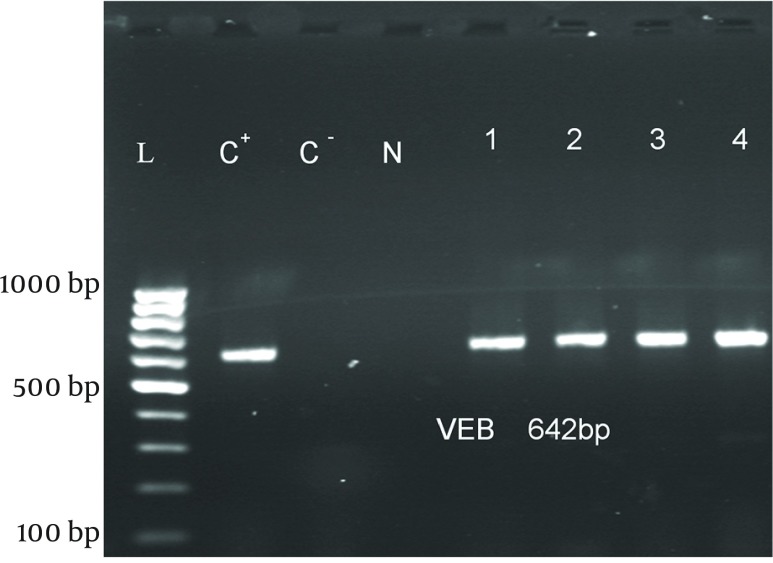
ESBL Production: Among 106 isolates, 94 (88.7%) were multidrug resistant and 60(58.3%) were putative ESBL producers using phenotypic confirmatory tests (Table 3). Sixteen (26.6%), 9 (15%) and 3 (5%) strains among 60 ESBL-producing strains had blaPER-1 (Figure 1), blaVEB-1 (Figure 2), and blaPER-1- blaVEB related genes, respectively (Table 4).
Table 3. Susceptibility Pattern to Antimicrobial Agents in ESBLs and non ESBLs-producing P. aeruginosa Strains.
| ESBL Positive (n=60) | ESBL Negative (n=46) | |||||
|---|---|---|---|---|---|---|
| Antimicrobials | Resistant, No. (%) | Susceptible, No. (%) | Intermediate, No. (%) | Resistant, No. (%) | Susceptible, No. (%) | Intermediate, No. (%) |
| Amikacin | 29 (48.3) | 30 (50) | 1 (1.7) | 4 (9.3) | 37 (86) | 2 (4.7) |
| Ceftazidime | 40 (66.7) | 18 (30) | 2 (3.3) | 13 (28.3) | 24 (55.2) | 6 (13) |
| Aztreonam | 25 (41.7) | 32 (53.3) | 3 (5) | 3 (6.97) | 39 (90.69) | 1 (2.32) |
| Cefepime | 58 (96.7) | 0 | 2 (3.3) | 42 (97.67) | 1 (2.32) | 0 |
| cefotaxime | 41 (68.3) | 6 (10) | 13 (21.7) | 12 (27.90) | 3 (6.97) | 28 (65.11) |
| Ciprofloxacin | 5 (8.3) | 50 (83.3) | 5 (8.3) | 0 | 42 (97.67) | 1 (2.32) |
| Gentamicin | 32 (53.3) | 28 (46.7) | 0 | 7 (16.27) | 36 (83.72) | 0 |
| Imipenem | 7 (11.7) | 45 (75) | 8 (13.3) | 1 (2.32) | 41 (95.34) | 1 (2.32) |
| Meropenem | 13 (21.7) | 44 (73.3) | 3 (5) | 2 (4.65) | 39 (90.69) | 2 (4.65) |
| Ofloxacin | 30 (50) | 29 (48.3) | 1 (1.7) | 4 (9.30) | 37 (86.04) | 2 (4.65) |
| Piperacillin | 54 (90) | 6 (10) | 0 | 41 (95.34) | 2 (4.65) | 0 |
| Tobramycin | 31 (51.6) | 28 (46.7) | 1 (1.7) | 4 (9.30) | 39 (90.69) | 0 |
Figure 1. PCR Amplification of blaPER-1 Gene.
L: 100 bp DNA ladder, C-: negative control, C+: positive control, N: PER-1 negative strains, 1–3: PCR products from PER-1positive strains.
Figure 2. PCR Amplification of blaVEB-1 Gene.
L: 100 bp DNA ladder, C-: negative control, C+: positive control, N: negative strain, 1–4: PCR products fromVEB-1 positive strains.
Table 4. Association of Antimicrobial Resistance Pattern and ESBL Genotypes.
| PER-1:16 (26.6%) | VEB: 9 (15%) | |||||
|---|---|---|---|---|---|---|
| Antimicrobials | Resistant, No. (%) | Susceptible, No. (%) | Intermediate, No. (%) | Resistant, No. (%) | Susceptible, No. (%) | Intermediate, No. (%) |
| Amikacin | 10 (62.5) | 5 (31.25) | 1(6.25) | 5 (55.5) | 2 (22.2) | 2 (22.2) |
| Ceftazidime | 12 (75) | 4 (25) | 0 | 9 (100) | 0 | 0 |
| Aztreonam | 10 (62.5) | 4 (25) | 2 (12.5) | 9 (100) | 0 | 0 |
| Cefepime | 16 (100) | 0 | 0 | 9 (100) | 0 | 0 |
| cefotaxime | 87.5 (14) | 0 | 2 (12.5) | 9 (100) | 0 | 0 |
| Ciprofloxacin | 2 (12.5) | 13 ( 81.25) | 1 (6.25) | 2 (22.2) | 6 (66.6) | 1 (11.11) |
| Gentamicin | 10(62.5) | 6 ( 37.5 ) | 0 | 6 (66.6) | 3 (33.3) | 0 |
| Imipenem | 5 (31.25) | 5 (31.25) | 6 (37.5) | 1 (11.11) | 6 (66.6) | 2 (22.2) |
| Meropenem | 7 ( 43.75) | 8 (50) | 1 (6.25) | 6 (66.6) | 2 (22.2) | 1 (11.11) |
| Ofloxacin | 11 (68.8) | 5 (31.25) | 0 | 8 (88.8) | 1 (11.11) | 0 |
| Piperacillin | 16 (100) | 0 | 0 | 9 (100) | 0 | 0 |
| Tobramycin | 10 (62.5) | 6 ( 37.5 ) | 0 | 6 (66.6) | 3 (33.3) | 0 |
5. Discussion
P. aeruginosa has a high resistance to antibiotics and is a common cause of morbidity and mortality in hospitalized and immunocompromised patients (18). Treatment of P. aeruginosa infections is complicated by the inhered and acquired resistance to the most of commonly used antimicrobial agents (19). The results from this study showed the high resistance of P. aeruginosa to most of used antimicrobial agents. It was also demonstrated that the prevalence of antibiotic resistance of the isolates was very high in comparison to other studies and most of P. aeruginosa isolates (88.7%) were multi-drug resistant (resistant to ≥ 3 different antibiotic classes) (20-25).
The prevalence of ESBL- producing P. aeruginosa isolates in this study was also higher than other investigations (5, 20, 26, 27). Among 60 ESBL-positive strains, 16 (26.6%), 9 (15%) and 3 (5%) contained PER-1, VEB-1 and PER-1-VEB-1 genes, respectively. These results indicated that the prevalence of VEB-1 gene in our area, is higher than Turkey and Korea, but the prevalence of VEB-1 and PER-1 genes, is lower than in Thailand (94.44% blaVEB-1) and Italy (34.61% blaPER-1) (5, 13, 26). Data on the prevalence of ESBL- producing P. aeruginosa strains in our area is limited. In the study performed by Shahcheraghi and colleges on P. aeruginosa isolates in Tehran, the rate of blaVEB and blaPER ESBLs were reported 24% and 17%, respectively, that was similar to our results (28). The high prevalence of PER-1 and VEB-1 indicated the high resistance to penicillins, ceftazidime and cefotaxime, as reported by other studies (26, 28).
This is the first report about the presence of these enzymes in P. aeruginosa isolates from west of Iran. It has shown that ESBL production in strains of P. aeruginosa can greatly complicate the clinical management of infection if advanced care is not taken. However, further studies are needed to determine other ESBL- types in P.aeruginosa strains in this area and their role in resistance to other antibiotic classes. The results of this study emphasizes on the need for a surveillance network to monitor the trends and emerge of new resistance mechanism in P. aeruginosa from different geographic regions. Therefore, the improvement in antibiotic prescription policies and infection control programs are of high necessity to prevent the spread of such resistant infectious agents.
Acknowledgments
No Acknowledgments.
Footnotes
Implication for health policy/practice/research/medical education:This study highlights the need for an antimicrobial resistance surveillance network for P. aeruginosa to monitor the trends and new types of resistance mechanism emerging.
Authors’ Contribution:All authors contributed in the research equally.
Financial disclosure:There is not any financial interest.
Funding/Support:This work was financially supported by Hamadan and Tabriz University of Medical Sciences.
References
- 1.Pagani L, Mantengoli E, Migliavacca R, Nucleo E, Pollini S, Spalla M, et al. Multifocal detection of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum beta-lactamase in Northern Italy. J Clin Microbiol. 2004;42(6):2523–9. doi: 10.1128/JCM.42.6.2523-2529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isturiz RE, Carbon C. Antibiotic use in developing countries. Infect Control Hosp Epidemiol. 2000;21(6):394–7. doi: 10.1086/501780. [DOI] [PubMed] [Google Scholar]
- 3.Livermore DM, Brown DF. Detection of beta-lactamase-mediated resistance. J Antimicrob Chemother. 2001;48 Suppl 1:59–64. doi: 10.1093/jac/48.suppl_1.59. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Mahrez Kamel, Rejiba Samia, Belhadj Cherifa, Belhadj Omrane. β-lactamase-mediated resistance to extended spectrum cephalosporins among clinical isolates of Pseudomonas aeruginosa. Res Microbiol. 1999;150(6):403–406. doi: 10.1016/s0923-2508(99)80075-8. [DOI] [PubMed] [Google Scholar]
- 5.Lee S, Park YJ, Kim M, Lee HK, Han K, Kang CS, et al. Prevalence of Ambler class A and D beta-lactamases among clinical isolates of Pseudomonas aeruginosa in Korea. J Antimicrob Chemother. 2005;56(1):122–7. doi: 10.1093/jac/dki160. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Lebessi E, Castro M, Fevre C, Foustoukou M, Nordmann P. Nosocomial outbreak of extended-spectrum beta-lactamase SHV-5-producing isolates of Pseudomonas aeruginosa in Athens, Greece. Antimicrob Agents Chemother. 2004;48(6):2277–9. doi: 10.1128/AAC.48.6.2277-2279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weldhagen GF. Rapid detection and sequence-specific differentiation of extended-spectrum beta-lactamase GES-2 from Pseudomonas aeruginosa by use of a real-time PCR assay. Antimicrob Agents Chemother. 2004;48(10):4059–62. doi: 10.1128/AAC.48.10.4059-4062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essack SY. Laboratory detection of extended-spectrum β-lactamases (ESBLs)—The need for a reliable, reproducible method. Diag Microbiol Infect Dis. 2000;37(4):293–295. doi: 10.1016/s0732-8893(00)00152-8. [DOI] [PubMed] [Google Scholar]
- 9.Jacoby GA, Han P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34(4):908–11. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel L, Menuteau O, Agoli N, Cattoen C, Nordmann P. Outbreak of extended-spectrum beta-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J Clin Microbiol. 2003;41(8):3542–7. doi: 10.1128/JCM.41.8.3542-3547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolska MK, Bukowski K, Jakubczak A. [Occurrence of beta-lactamase type ESBL and IBL in Pseudomonas aeruginosa rods]. Med Dosw Mikrobiol. 2001;53(1):45–51. [PubMed] [Google Scholar]
- 12.Aubert D, Poirel L, Chevalier J, Leotard S, Pages JM, Nordmann P. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001;45(6):1615–20. doi: 10.1128/AAC.45.6.1615-1620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endimiani A, Luzzaro F, Pini B, Amicosante G, Rossolini GM, Toniolo AQ. Pseudomonas aeruginosa bloodstream infections: risk factors and treatment outcome related to expression of the PER-1 extended-spectrum beta-lactamase. BMC Infect Dis. 2006;6:52. doi: 10.1186/1471-2334-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–6. [PubMed] [Google Scholar]
- 15.Wayne PA. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement NCCLS document M100-S9. National Committee for Clinical Laboratory Standards. 2008 [Google Scholar]
- 16.Yousefi S, Farajnia S, Nahaei MR, Akhi MT, Ghotaslou R, Soroush MH, et al. Detection of metallo-beta-lactamase-encoding genes among clinical isolates of Pseudomonas aeruginosa in northwest of Iran. Diagn Microbiol Infect Dis. 2010;68(3):322–5. doi: 10.1016/j.diagmicrobio.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Sohrabi N, Farajnia S, Akhi MT, Nahaei MR, Naghili B, Peymani A, et al. Prevalence of OXA-type beta-lactamases among Acinetobacter baumannii isolates from Northwest of Iran. Microb Drug Resist. 2012;18(4):385–9. doi: 10.1089/mdr.2011.0077. [DOI] [PubMed] [Google Scholar]
- 18.Todar Kenneth. Todar's online textbook of bacteriology. University of Wisconsin-Madison Department of Bacteriology; 2006. [Google Scholar]
- 19.Hauser AR, Sriram P. Severe Pseudomonas aeruginosa infections. Tackling the conundrum of drug resistance. Postgrad Med. 2005;117(1):41–8. doi: 10.3810/pgm.2005.01.1571. [DOI] [PubMed] [Google Scholar]
- 20.Chayakulkeeree M, Junsriwong P, Keerasuntonpong A, Tribuddharat C, Thamlikitkul V. Epidemiology of extended-spectrum beta-lactamase producing gram-negative bacilli at Siriraj Hospital, Thailand, 2003. Southeast Asian J Trop Med Public Health. 2005;36(6):1503–9. [PubMed] [Google Scholar]
- 21.Kaushik Robin, Kumar Sandeep, Sharma Rajeev, Lal Pawanindra. Bacteriology of burn wounds — the first three years in a new burn unit at the Medical College Chandigarh. Burns. 2001;27(6):595–597. doi: 10.1016/s0305-4179(01)00023-7. [DOI] [PubMed] [Google Scholar]
- 22.Mirsalehian A, Feizabadi M, Nakhjavani FA, Jabalameli F, Goli H, Kalantari N. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2010;36(1):70–4. doi: 10.1016/j.burns.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Nikbin VS, Abdi-Ali A, Feizabadi MM, Gharavi S. Pulsed field gel electrophoresis & plasmid profile of Pseudomonas aeruginosa at two hospitals in Tehran, Iran. Indian J Med Res. 2007;126(2):146–51. [PubMed] [Google Scholar]
- 24.Ozumba UC, Jiburum BC. Bacteriology of burn wounds in Enugu, Nigeria. Burns. 2000;26(2):178–180. doi: 10.1016/s0305-4179(99)00075-3. [DOI] [PubMed] [Google Scholar]
- 25.Celenza G, Pellegrini C, Caccamo M, Segatore B, Amicosante G, Perilli M. Spread of bla(CTX-M-type) and bla(PER-2) beta-lactamase genes in clinical isolates from Bolivian hospitals. J Antimicrob Chemother. 2006;57(5):975–8. doi: 10.1093/jac/dkl055. [DOI] [PubMed] [Google Scholar]
- 26.Chanawong A, M'Zali FH, Heritage J, Lulitanond A, Hawkey PM. SHV-12, SHV-5, SHV-2a and VEB-1 extended-spectrum beta-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J Antimicrob Chemother. 2001;48(6):839–52. doi: 10.1093/jac/48.6.839. [DOI] [PubMed] [Google Scholar]
- 27.Shahcheraghi F, Nikbin VS, Feizabadi MM. Prevalence of ESBLs genes among multidrug-resistant isolates of Pseudomonas aeruginosa isolated from patients in Tehran. Microb Drug Resist. 2009;15(1):37–9. doi: 10.1089/mdr.2009.0880. [DOI] [PubMed] [Google Scholar]
- 28.Tsakris A, Vatopoulos AC, Tzouvelekis LS, Legakis NJ. Diversity of resistance phenotypes and plasmid analysis in multi-resistant 0:12 Pseudomonas aeruginosa. Eur J Epidemiol. 1992;8(6):865–70. doi: 10.1007/BF00145334. [DOI] [PubMed] [Google Scholar]