Abstract
The advent of the next-generation sequencing data has made it possible to cost-effectively detect and characterize genomic variation in human genomes. Structural variation, including deletion, duplication, insertion, inversion and translocation, is of great importance to human genetics due to its association with many genetic diseases. BreakDancer is a bioinformatics tool that relates paired-end read alignments from a test genome to the reference genome for the purpose of comprehensively and accurately detecting various types of structural variation.
Keywords: Genomics, Next Generation Sequencing, BreakDancer, Structural Variation, Discordant Read Pair
INTRODUCTION
Structural variation (SV), defined as aberrations that change the size, copy number, location, orientation, and sequence content of genomic DNA, is an important topic in genetic studies. Similar to single nucleotide variants (SNVs), many SVs are normal polymorphisms that have little to no biological significance. However, other SVs are found to be related to inheritable diseases such as neurological disorders and cancer (Feuk, Carson, & Scherer, 2006; Lupski & Ph, 2007; Sharp, Cheng, & Eichler, 2006). BreakDancer (Chen et al., 2009) takes the paired-end alignment of next-generation sequencing (NGS) data as the input, detects SVs with respect to the reference genome and returns predicted breakpoints, SV type, and confidence scores. It has been widely used in various projects, including the 1000 Genomes Project and The Cancer Genome Atlas (TCGA) (Ding et al., 2010; Mills et al., 2011; Welch et al., 2012).
Since NGS reads are sequenced from random fragments of DNA, they need to be aligned to a reference genome in order to identify their originating genomic locations. Such alignment processes generate a map file containing the alignment results of each read or read-pair, including the chromosome, chromosomal position, orientation, insert size, and mapping quality. For BreakDancer, the input map files are expected to be in BAM format. Detailed information regarding read mapping can be found in (Langmead, Trapnell, Pop, & Salzberg, 2009; Langmead, 2010; Li & Durbin, 2009). A paired-end read derived from short insert libraries, if concordantly mapped, has a specific pattern of orientation: the first read aligns in the positive orientation, while the second read aligns in the negative orientation. Furthermore, since the fragment size (a.k.a. the insert size), usually follows a uni-modal distribution, concordantly aligned paired-end reads should conform to this distribution. On the other hand, discordant read-pairs (DRPs)—defined as reads with abnormal orientations or insert sizes (either too small or too large)—are indicative of structural variations or alignment errors. From the input map files, BreakDancer estimates the parameters of the insert size distribution, goes through reads to identify DRPs, and clusters them to identify SV breakpoints. The output of BreakDancer contains a list of putative SV breakpoints, each of which contains a pair of genomic positions, supporting DRP counts, a predicted SV type, a confidence score, and some extra features as addressed in the following sections.
BASIC PROTOCOL 1: Using BreakDancer to Identify SVs in a Single Genome
There are two sequential steps to run BreakDancer. First, the “bam2cfg.pl” script analyzes the first few thousand reads in each input BAM file and generates a configuration file containing statistics for each read group (RG; see Unit 11.10). Second, the “breakdancer-max” program uses these statistics to identify and cluster DRPs, ultimately yielding a list of putative SVs. The C++ version (1.0 and up) of breakdancer-max directly utilizes the samtools C library to achieve a significant performance increase over the original Perl implementation. However, it only supports properly formatted BAM files and has been tested using BAM files produced by BWA(Li & Durbin, 2009), Bowtie(Langmead et al., 2009), Novoalign, and Mapsplice(Wang et al., 2010). To obtain correct results, it is important to have the read group (@RG) tag in both the header and each alignment in the BAM files. It is also critical to clearly indicate the biological and technical origins of every read using the read group (RG) and library (LB) tags, particularly when providing multiple BAM files as input.
Necessary Resources
Hardware
A 64-bit Linux cluster is preferred. The memory requirement depends on the size of the BAM file and the extent of SVs. For a cancer genome sequenced at a coverage of 30X, usually less than 4 GB memory is needed for a whole genome analysis. BreakDancer can also be run on a desktop or laptop computer that has a Unix-like operating system such as OS X.
Software
The BreakDancer package. Users should download and install the package, as described in Support Protocol 1.
CPAN’s Statistics::Descriptive and GD::Graph modules, which are discussed in Support Protocol 2.
-
1
Run bam2cfg. In the downloaded BreakDancer directory, go to the “perl” subfolder. Run the following command:
perl bam2cfg.pl -g -h BAM_files > config_file
The BAM_files are the full paths of a set of BAM files to be processed, separated by spaces. The config_file is a tab-delimited output file containing the relevant statistics inferred from the BAM files.
It is strongly recommended that you perform a quality control assessment after running pam2cfg.pl and before breakdancer-max. This will catch some common BAM formatting problems and flag issues involving library construction artifacts. Support Protocol 3 describes the recommended quality control assessment.
-
2
Run breakdancer-max:
breakdancer-max [options] config_file > BreakDancer_output
The output of this step is a tab-delimited text file containing the list of putative SV breakpoints with detailed information. See Guidelines for Understanding Results for an explanation of how to interpret the filtered file.
BASIC PROTOCOL 2: Using BreakDancer to Identify Somatic SVs in Matched Tumor and Normal Genomes
BreakDancer can be used to detect somatic SVs when matched tumor and normal samples are sequenced. Usually, one starts with two BAM files, one from the tumor sample and the other from the matched normal sample. The procedure begins with BASIC PROTOCOL 1, but further analysis is required to identify alleles that are altered in the tumor samples. The following example is a simple one. Users can customize the protocol according to the data and application at hand.
Protocol steps
Run BASIC PROTOCOL 1 on a matched pair of tumor and normal samples.
-
Remove calls that are supported by at least one read from the normal sample. Suppose the breakdancer-max output file name is a.sv, and the name of the normal library is called n1. The following Linux command can be used to filter out germline calls:
grep –v n1 a.sv > filtered_file
The output file has the same format as a.sv, and it contains a subset of the calls calls supported by reads from only the tumor sample.
BASIC PROTOCOL 3: Using BreakDancer to Identify Segregating SVs in a Population of Samples
For population sequencing projects such as the 1000 Genomes Project and the Cancer Cell Line Encyclopedia (Barretina et al., 2012), it is useful to group together all samples from a continental or a disease population to achieve more sensitive detection. A straightforward way to do this is to inform bam2cfg.pl of the locations of all the BAM files from the population sequencing project in order to create a configuration file that contains a global set of valid read groups. When inter-chromosomal translocations are not expected, it is efficient to run breakdancer-max one chromosome at a time. The current implementation has allowed simultaneous analysis of several hundred BAM files in the 1000 Genomes phase 1–3 projects. After the results are obtained, it is useful to perform further filtering by testing the sample-specific copy number values estimated by breakdancer-max. A true copy number variant (CNV, such as deletion and duplication) is expected to have variable copy numbers across the samples and at least one sample should have an estimated copy number diverging substantially from the neutral value 2.0. The following is a simple example to filter out false CNV calls that have no evidence in any of the samples given a user-specified threshold t.
-
Run BASIC PROTOCOL 1 on all samples in the same population. Make certain to utilize the “-a” option in BASIC PROTOCOL 1, Step 2 as follows.
breakdancer-max -a config_file > BreakDancer_output
To detect true copy number variants, the estimated copy numbers starting from column #12 are compared with the neutral value 2.0. Suppose the breakdancer-max output file is a.sv, and the threshold for a copy number variant is t. The following Perl command will filter out putative CNVs that have no evidence in any of the samples.
perl –ane ‘foreach $a ( @F[11 .. $#F] ) { if( $a ne ‘NA’ && abs( $a – 2 ) > t ) { print $_; last } }’ a.sv > filtered_file
The selection of t determines how stringent the filter is. The larger t is, the more stringent the filter. Besides copy number analysis, filtering calls by origin (source library) or supporting read count may also be useful.
SUPPORT PROTOCOL 1: Download and Install BreakDancer
The C/C++ library dependencies required to install BreakDancer are bundled with the software in the latest version, but there are still a few tools needed in order to checkout and compile the software. These tools (listed below) should be available in most package managers on recent operating systems (e.g., yum for RedHat, apt for Debian/Ubuntu, homebrew for OS X).
Necessary Resources
Hardware
A computer running Linux or OS X with at least 4GB of RAM.
Software
Git
A C/C++ compiler such as GCC or Clang.
cmake v2.8 or above (http://www.cmake.org)
-
Clone a copy of the BreakDancer repository with git.
git clone --recursive https://github.com/genome/breakdancer.git
This creates a ‘breakdancer’ directory in the current working directory. We refer to this directory as $BD_ROOT from now on.
Note: failure to provide the --recursive option to git will cause errors when cmake is run.
-
Create and enter a temporary build directory in the newly created ‘breakdancer’ directory:
mkdir $BD_ROOT/build cd $BD_ROOT/build
-
Run cmake and make to build the software
cmake .. make
This creates the $BD_ROOT/bin/ directory containing the executables:
breakdancer-max samtools
Add this bin directory to your path. For Bourne shell derivatives:
export PATH=$BD_ROOT/build/bin:$PATH
For csh/tcsh shell derivatives:
set path = ($BD_ROOT/build/bin $path)
SUPPORT PROTOCOL 2: Download and Install Perl Modules from CPAN
The “bam2cfg.pl” script depends on the following Perl modules, available from CPAN:
Statistics::Descriptive
-
GD::Graph
The procedure of downloading CPAN modules is described on the CPAN website (http://www.cpan.org/modules/INSTALL.html).
SUPPORT PROTOCOL 3: Quality Control Checks
We recommend that you perform a few quality control checks after bam2cfg is finished and before running breakdancer-max.
Below are some guidelines to aid in this process.
-
1
Check if the configuration file generated by bam2cfg.pl contains one line for each read group or library in every input BAM file. For each BAM file, run the following command,
samtools view -H BAM_file| grep -c @RG grep -c BAM_file config_file
Compare the two outputs from the above two commands, which indicate the (RG, LB) pair numbers from the BAM file and the configuration file, respectively. If the first number is smaller than the second one, then this input BAM file may be missing RG or LB information in the header.
-
2
Check if the configuration file contains “NA” as read group.
grep -cw readgroup:NA config_file
If output is greater than 0, then the RG or LB information is missing in at least one of the BAM files.
-
3
Check the coefficient of variation (standard deviation divided by mean) of the insert size for each read group.
perl -ane ‘ ($mean)=($_=~/mean:(\S+)/);($std)=($_=~/std:(\S+)/);print $std/$mean .”\n” ‘ config_file
The above command prints the coefficient of variation for each read group. Normally, they should be < 0.2 or 0.3.
-
4
Check the percentage of inter-chromosomal read pairs.
perl -ane ‘ ($CTX)=($_=~/32\((\S+?)\)/);print $CTX.”\n” ‘ config_file
The above command prints the percentage of the reads with tag 32 for each read group, which corresponds to inter-chromosomal read pairs. This percentage should normally be smaller than 3%. Higher values may indicate problems with library construction or sequencing. This step requires that the option “-g” in running bam2cfg.pl to generate the distribution over numeric read flags in the configuration file.
-
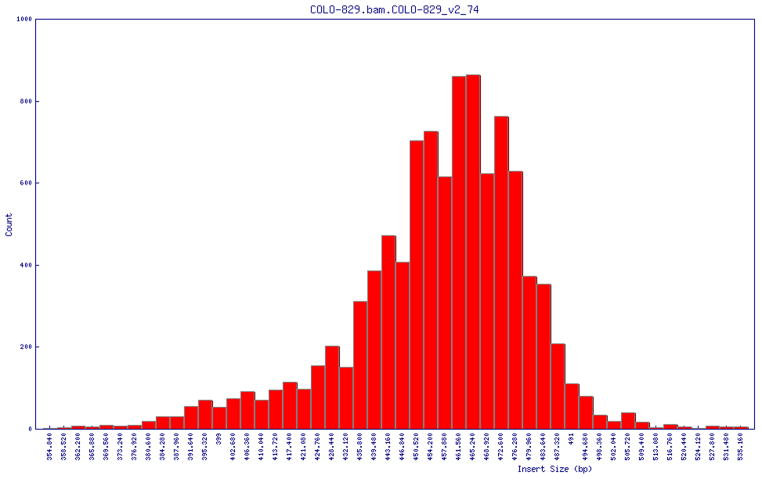
5
(optional). Manually check the histograms of the insert size distribution for each group or library. A normal distribution is expected, as seen in Figure 1, whereas a bimodal distribution is undesirable. This step requires option “-h” when running bam2cfg.pl.
Figure 1.
histogram of the insert size distribution showing a normal distribution.
It is possible to use a manually created configuration file for “breakdancer-max” and thus skip the bam2cfg.pl step. Formats and contents regarding the configuration file are described in Guidelines for understanding results section.
GUIDELINES FOR UNDERSTANDING RESULTS
Bam2cfg.pl output
An example tab-delimited configuration file produced by bam2cfg.pl looks like the following:
read-group:2825107881 platform:illumina map:tumor.bam readlen:75.00 lib:H_KA-189941-0921313gsc-lib4 ⍰num:10001 lower:86 upper:443 mean:315.09 std:43.92 exe:samtools view ⍰
read-group:2843249908 platform:illumina map:tumor.bam readlen:75.00 lib:H_KA-189941-0921313gsc-lib4 ⍰num:10001 lower:86 upper:443 mean:315.09 std:43.92 exe:samtools view ⍰
Each row contains six or more key:value pairs (separated by colon) that specify:
. map:the location of the map file
. mean:the mean insert size
. std:the standard deviation insert size
. readlen:the average read length
. lib:the library name
. exe:a command line that can run by Perl system-calls to produce alignment views (only required by the deprecated Perl version of BreakDancer, i.e., version 1.0)
In addition to the above 6 keys, users are allowed to explicitly specify the insert size cutoffs using the two keys: upper and lower, as shown in the above two examples. This will instruct breakdancer-max to detect deletions using read pairs that are at least 443 bp apart (outer distance) and detect insertions using read pairs that are at most 86 bp apart.
Breakdancer-max output
A standard BreakDancer output file consists of the following columns:
Chromosome 1
Position 1
Orientation 1
Chromosome 2
Position 2
Orientation 2
Type of a SV
Size of a SV
Confidence Score
Total number of supporting read pairs
Total number of supporting read pairs from each map file
(and above). The copy number estimated for each BAM
Columns 1–3 and 4–6 are used to specify the coordinates of the two breakends. The orientation is a string that records the number of reads mapped to the plus (+) or the minus (−) strand in the anchoring regions.
Column 7 is the type of SV detected: DEL (deletion), INS (insertion), INV (inversion), ITX (intra-chromosomal translocation), CTX (inter-chromosomal translocation), and UN (unknown).
Column 8 is the size of the SV in bp. It is meaningless for inter-chromosomal translocations.
Column 9 is the confidence score associated with the prediction (Chen et al., 2009).
Column 11 describes the origins of the supporting read pairs, which is useful in pooled analysis. For example, one may want to give SV breakpoints that are supported by more than one library higher confidence than those detected in only one library. It can also be used to distinguish somatic events from germline events, i.e., those detected in only the tumor libraries versus those detected in both the tumor and the normal libraries. An example of this can be found in Basic Protocol 2.
Columns 12 and above are copy number estimates for this SV region, one column per input (BAM) file, in an order specified by the header-line (starting with #). The copy number in the region is estimated as twice the observed number of normally mapped reads between the two breakends divided by the expected number in the same region of a diploid genome. The estimation here is relatively straightforward without any further corrections. For more accurate estimation, please refer to the section “Suggestions for Further Analysis”.
The followings are the examples and explanations of the output of “breakdancer-max”.
Example 1:
1 10000 10+0- 2 20000 7+10- CTX -296 99 10 tB|10 1.02
An inter-chromosomal translocation that starts from chr1:10000 and goes into chr2:20000 with 10 supporting read pairs from the library tB, a confidence score of 99, and an estimated copy number of 1.02.
Example 2:
1 59257 5+1- 1 60164 0+5- DEL 862 99 5 nA|2:tB|1 1.03 2.05
A deletion between chr1:59257 and chr1:60164 connected by 5 read pairs, among which 3 support the deletion hypothesis including 2 in library nA and 1 in library tB . Copy numbers of 1.03 and 2.05 were estimated for the two samples.
Note that real SV breakpoints are expected to reside within the predicted boundaries with 95% confidence intervals equal to twice standard deviation insert size.
Critical Parameters and Troubleshooting
Bam2cfg parameters
-c FLOAT
This specifies how far the “upper” and the “lower” insert size cutoffs lie from the mean in units of standard deviation (std). For example, with the default value of 4, the upper and lower cutoffs are estimated as mean + 4 ×std and mean – 4 ×std, respectively. It is often useful to adjust these values to more refined estimates based on inspection of the insert size histograms (produced by bam2cfg.pl -h or other analysis) because insert size distributions from real DNA libraries are often asymmetric and can substantially differ from normal distribution. For example, we found that better results can be achieved by setting the upper and lower cutoffs to 999 permilles (99.9%) and 1 permille (0.1%), respectively, for some low coverage samples in the 1000 Genomes Project.
-v FLOAT
Libraries with insert size distributions that differ significantly from a normal distribution (e.g., bimodal, severely skewed) are not generally suitable for BreakDancer analysis. By default, libraries where the coefficient of variation (std/mean) exceeds 1.0 are excluded from the configuration files and will not be further analyzed. To disable this filtering (e.g., when analyzing RNA-seq libraries), users can specify a cutoff much larger than the default of 1.0 (e.g., -v 10000).
-f STRING
For a single BAM file containing multiple libraries, BreakDancer depends on the correct specification of read group (RG) and library (LB) information in the BAM files to perform correct analysis. If you have a single BAM file that contains multiple libraries, make sure the BAM header contains complete RG and LB specification and each read is assigned to a RG. Otherwise bam2cfg.pl may fail to produce a configuration file that correctly specifies the biological and technical origin of the reads, resulting in incorrect analysis. Note that Reads that do not carry RG tags are assumed to come from a “NA” library.
On another hand, listing multiple map files in a single configuration file would automatically enable pooled analysis: reads from all the map files are jointly analyzed to find unified SV hypotheses across all inputs. In this case, it is even more important to correctly assign RG and LB information such that reads can be correctly pooled in analysis. Although alternative configurations are possible, breakdancer-max only reports statistics at library (LB) and map file levels. Therefore, the most convenient configuration is to use one map file for one DNA sample and use LBs and RGs to track DNA libraries and sequencing lanes, respectively.
If the alignment records carry the RG but the BAM header does not, the “-f” option can be used to inform bam2cfg.pl of the missing RG and LB specification by a two-column tab-delimited text file that specifies (RG, LB) pairs created by user, so that bam2cfg can work with BAM files having incomplete headers.
In detail, a valid BAM header should describe the relationships between read RG and LB tags, e.g.,
@RG ID:20FUK.1 PL:illumina PU:20FUKAAXX100202.1 LB:Solexa-18483 DT:2010-02-02T00:00:00-0500 SM:NA12878 CN:BI @RG ID:20FUK.2 PL:illumina PU:20FUKAAXX100202.2 LB:Solexa-18484 DT:2010-02-02T00:00:00-0500 SM:NA12878 CN:BI
When they are missing, bam2cfg fails to associate RG: 20FUK.1 with LB:Solexa-18483 and RG:20FUK.2 with LB:Solexa-18484 in the above examples. To remedy such an issue without having to regenerate the entire BAM file, users can create a tab-delimited text file (e.g., rglib.txt) that contains 2 columns, e.g.,
20FUK.1 Solexa-18483 20FUK.2 Solexa-18484
and supplement it through the -f option in the command line, e.g.,
bam2cfg.pl -f rglib.txt my.bam > my.cfg
For more information on adding read groups to BAM files, please follow instructions on the samtools website (http://samtools.sourceforge.net).
-h
When the Perl GD::Graph module is properly installed, this option will output a png file for each library (LB), which contains a plot of insert size histogram estimated from the first n (specified by -n) properly aligned read pairs (Figure 1).
Breakdancer-max parameters
-o
In multi-core or distributed computing environments, it is highly recommended to divide the work to be done by chromosome and run multiple jobs in parallel. The “-o chromosome-name” option to “breakdancer-max” facilitates this. For example, the command of running breakdancer-max on chromosome 1 of “sample1.bam” with configuration file “sample1.cfg” is:
breakdancer-max -o chr1 sample1.cfg > output_file
Note that “chr1” could also be “1” or “Chr1” etc., depending on the format of chromosome name in the BAM file. The best way to check this is to read the third column (1-based) of the first few lines of the BAM file using the command “samtools view sample1.bam | cut -f3,3 | less”.
-t
While running in parallel by chromosome greatly reduces the analysis time, only intra-chromosomal SV breakpoints can be detected. The command-line option, “-t”, is provided to skip intra-chromosomally mapped reads and only detect inter-chromosomal SV breakpoints, i.e., fusions of genetic materials from different chromosomes. This option runs faster and requires less memory than a complete analysis because it only examines read pairs mapped onto different chromosomal pairs. Note that a union of the SV breakpoints from running breakdancer-max on options “-t” and “-o”, respectively renders the final output file.
-c INT
This option is similar to the -c option of bam2cfg.pl. However, it is only functional when the upper and the lower cutoffs are not specified in the configuration file.
-a
Specifies whether to report detailed library-specific copy numbers and supporting reads. By default, copy numbers and supporting reads are reported at map (BAM) file level.
Suggestions for Further Analysis
For those users interested not only in SV breakpoints and types, but also in the fraction of variant alleles in heterogeneous DNA samples, it is highly recommended that users download BreakDown (Fan et al., n.d.) from SourceForge (https://sourceforge.net/projects/breakdown/) for further analysis. BreakDown is a Perl module that utilizes multi-modal alignment information to accurately estimate the genotype of an SV and the fraction of the variant allele in each sample. Users can feed BreakDown with BreakDancer’s output and utilize it as a filter to reduce the number of false positives and increase specificity. In cancer genome studies, users can further infer variant allele fractions to identify sub-clonal SVs and estimate clonal diversity.
Acknowledgments
This work was supported in part by the National Cancer Institute (NCI) grant R01-CA172652-01 to K.C., National Human Genome Research Institute (NHGRI) grants U41-HG007497-01 and U01-HG006517, and the National Cancer Institute Cancer Center Support Grant P30-CA016672.
Footnotes
INTERNET RESOURCES
http://sourceforge.net/projects/samtools/files/samtools/0.1.6/ A link to download samtools 0.1.6, which is being supported by BreakDancer1.1.2.
https://github.com/genome/breakdancer.git
http://sourceforge.net/projects/breakdancer/files/
Links to download BreakDancer package.
http://sourceforge.net/apps/mediawiki/samtools/?title=SAM_protocol#Install_software
LITERATURE CITED
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modeling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Getz G. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30(5):413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, Mardis ER. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature methods. 2009;6(9):677–81. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Fulton LL. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Nakhleh L, Chen K. BreakDown: integrative estimation of allele fraction from structural variation breakpoints n.d [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nature reviews Genetics. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Current protocols in bioinformatics. 2010 Dec;Chapter 11(Unit 11.7) doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR., PhD clinical implications of basic research Structural Variation in the Human Genome. 2007:1169–1171. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Cheetham RK. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annual review of genomics and human genetics. 2006;7:407–42. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, Liu J. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic acids research. 2010;38(18):e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Xia J. The origin and evolution of mutations in Acute Myeloid Leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023.The. [DOI] [PMC free article] [PubMed] [Google Scholar]