Abstract
Bacteriophage lambda uses site-specific recombination to move its DNA into and out of the Escherichia coli genome. The recombination event is mediated by the recombinase integrase (Int) together with several accessory proteins through short specific DNA sequences known as attachment sites. A gel mobility shift assay has been used to show that, in the absence of accessory proteins, Int can align and hold together two DNA molecules, each with an attachment site, to form stable non-covalent 'bimolecular complexes'. Each attachment site must have both core and arm binding sites for Int to participate in a bimolecular complex. These stable structures can be formed between pairs of attL and attP attachment sites, but cannot include attB or attR sites; they are inhibited by integration host factor (IHF) protein. The bimolecular complexes are shown to represent a synaptic intermediate in the reaction in which Int protein promotes the IHF-independent recombination of two attL sites. These complexes should enable a detailed analysis of synapsis for this pathway.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberti S., Oehler S., von Wilcken-Bergmann B., Müller-Hill B. Genetic analysis of the leucine heptad repeats of Lac repressor: evidence for a 4-helical bundle. EMBO J. 1993 Aug;12(8):3227–3236. doi: 10.1002/j.1460-2075.1993.tb05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin A. A., Beatty L. G., Sadowski P. D. Synaptic intermediates promoted by the FLP recombinase. J Mol Biol. 1990 Jul 5;214(1):55–72. doi: 10.1016/0022-2836(90)90146-D. [DOI] [PubMed] [Google Scholar]
- Amin A., Roca H., Luetke K., Sadowski P. D. Synapsis, strand scission, and strand exchange induced by the FLP recombinase: analysis with half-FRT sites. Mol Cell Biol. 1991 Sep;11(9):4497–4508. doi: 10.1128/mcb.11.9.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. E., Gardner J. F., Gumport R. I. Extent of sequence homology required for bacteriophage lambda site-specific recombination. J Mol Biol. 1985 Jan 20;181(2):187–197. doi: 10.1016/0022-2836(85)90084-1. [DOI] [PubMed] [Google Scholar]
- Bear S. E., Clemens J. B., Enquist L. W., Zagursky R. J. Mutational analysis of the lambda int gene: DNA sequence of dominant mutations. J Bacteriol. 1987 Dec;169(12):5880–5883. doi: 10.1128/jb.169.12.5880-5883.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Better M., Lu C., Williams R. C., Echols H. Site-specific DNA condensation and pairing mediated by the int protein of bacteriophage lambda. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5837–5841. doi: 10.1073/pnas.79.19.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Better M., Wickner S., Auerbach J., Echols H. Role of the Xis protein of bacteriophage lambda in a specific reactive complex at the attR prophage attachment site. Cell. 1983 Jan;32(1):161–168. doi: 10.1016/0092-8674(83)90506-8. [DOI] [PubMed] [Google Scholar]
- Burgin A. B., Jr, Nash H. A. Symmetry in the mechanism of bacteriophage lambda integrative recombination. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9642–9646. doi: 10.1073/pnas.89.20.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. W., Lee J., Jayaram M. DNA cleavage in trans by the active site tyrosine during Flp recombination: switching protein partners before exchanging strands. Cell. 1992 May 15;69(4):647–658. doi: 10.1016/0092-8674(92)90228-5. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Echols H. Integrative and excisive recombination by bacteriophage lambda: evidence for an excision-specific recombination protein. J Mol Biol. 1970 Feb 14;47(3):575–583. doi: 10.1016/0022-2836(70)90324-4. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. F., Nash H. A. Role of Escherichia coli IHF protein in lambda site-specific recombination. A mutational analysis of binding sites. J Mol Biol. 1986 Sep 20;191(2):181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Nash H. Communication between segments of DNA during site-specific recombination. 1987 Jan 29-Feb 4Nature. 325(6103):401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Han Y. W., Gumport R. I., Gardner J. F. Complementation of bacteriophage lambda integrase mutants: evidence for an intersubunit active site. EMBO J. 1993 Dec;12(12):4577–4584. doi: 10.1002/j.1460-2075.1993.tb06146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A., Flamm E., Weisberg R. A. An Escherichia coli mutant unable to support site-specific recombination of bacteriophage lambda. J Mol Biol. 1985 May 25;183(2):129–140. doi: 10.1016/0022-2836(85)90207-4. [DOI] [PubMed] [Google Scholar]
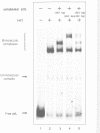
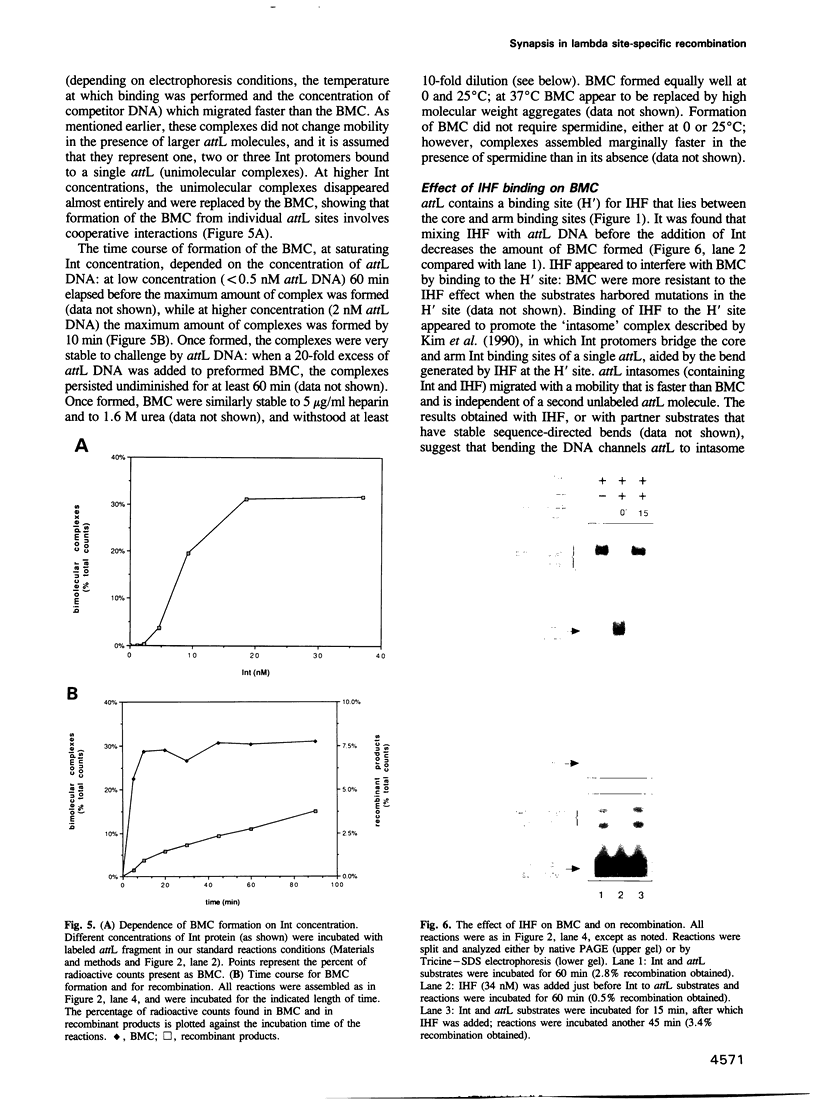
- Kim S., Landy A. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science. 1992 Apr 10;256(5054):198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Moitoso de Vargas L., Nunes-Düby S. E., Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in lambda attL. Cell. 1990 Nov 16;63(4):773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. An intermediate in the phage lambda site-specific recombination reaction is revealed by phosphorothioate substitution in DNA. Nucleic Acids Res. 1988 Jul 25;16(14B):6839–6856. doi: 10.1093/nar/16.14.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J Mol Biol. 1988 Nov 5;204(1):95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
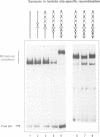
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller H. I., Kikuchi A., Nash H. A., Weisberg R. A., Friedman D. I. Site-specific recombination of bacteriophage lambda: the role of host gene products. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1121–1126. doi: 10.1101/sqb.1979.043.01.125. [DOI] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Pargellis C. A., Hasan N. M., Bushman E. W., Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988 Sep 23;54(7):923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Heteroduplex substrates for bacteriophage lambda site-specific recombination: cleavage and strand transfer products. EMBO J. 1989 Nov;8(11):3523–3533. doi: 10.1002/j.1460-2075.1989.tb08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numrych T. E., Gumport R. I., Gardner J. F. A comparison of the effects of single-base and triple-base changes in the integrase arm-type binding sites on the site-specific recombination of bacteriophage lambda. Nucleic Acids Res. 1990 Jul 11;18(13):3953–3959. doi: 10.1093/nar/18.13.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numrych T. E., Gumport R. I., Gardner J. F. Characterization of the bacteriophage lambda excisionase (Xis) protein: the C-terminus is required for Xis-integrase cooperativity but not for DNA binding. EMBO J. 1992 Oct;11(10):3797–3806. doi: 10.1002/j.1460-2075.1992.tb05465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Half-att site substrates reveal the homology independence and minimal protein requirements for productive synapsis in lambda excisive recombination. Cell. 1989 Oct 6;59(1):197–206. doi: 10.1016/0092-8674(89)90881-7. [DOI] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Qian X. H., Inman R. B., Cox M. M. Protein-based asymmetry and protein-protein interactions in FLP recombinase-mediated site-specific recombination. J Biol Chem. 1990 Dec 15;265(35):21779–21788. [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988 Jan 15;52(1):9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell. 1986 Sep 26;46(7):1011–1021. doi: 10.1016/0092-8674(86)90700-2. [DOI] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Ross W., Landy A. Bacteriophage lambda int protein recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983 May;33(1):261–272. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Shulman M., Landy A. Biochemical analysis of att-defective mutants of the phage lambda site-specific recombination system. J Mol Biol. 1982 Apr 15;156(3):505–522. doi: 10.1016/0022-2836(82)90263-7. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Kuwabara M. D., Chen C. H., Bruice T. W. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 1991;208:414–433. doi: 10.1016/0076-6879(91)08022-a. [DOI] [PubMed] [Google Scholar]
- Stark W. M., Sherratt D. J., Boocock M. R. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989 Aug 25;58(4):779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Weisberg R. A., Enquist L. W., Foeller C., Landy A. Role for DNA homology in site-specific recombination. The isolation and characterization of a site affinity mutant of coliphage lambda. J Mol Biol. 1983 Oct 25;170(2):319–342. doi: 10.1016/s0022-2836(83)80151-x. [DOI] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]