Abstract
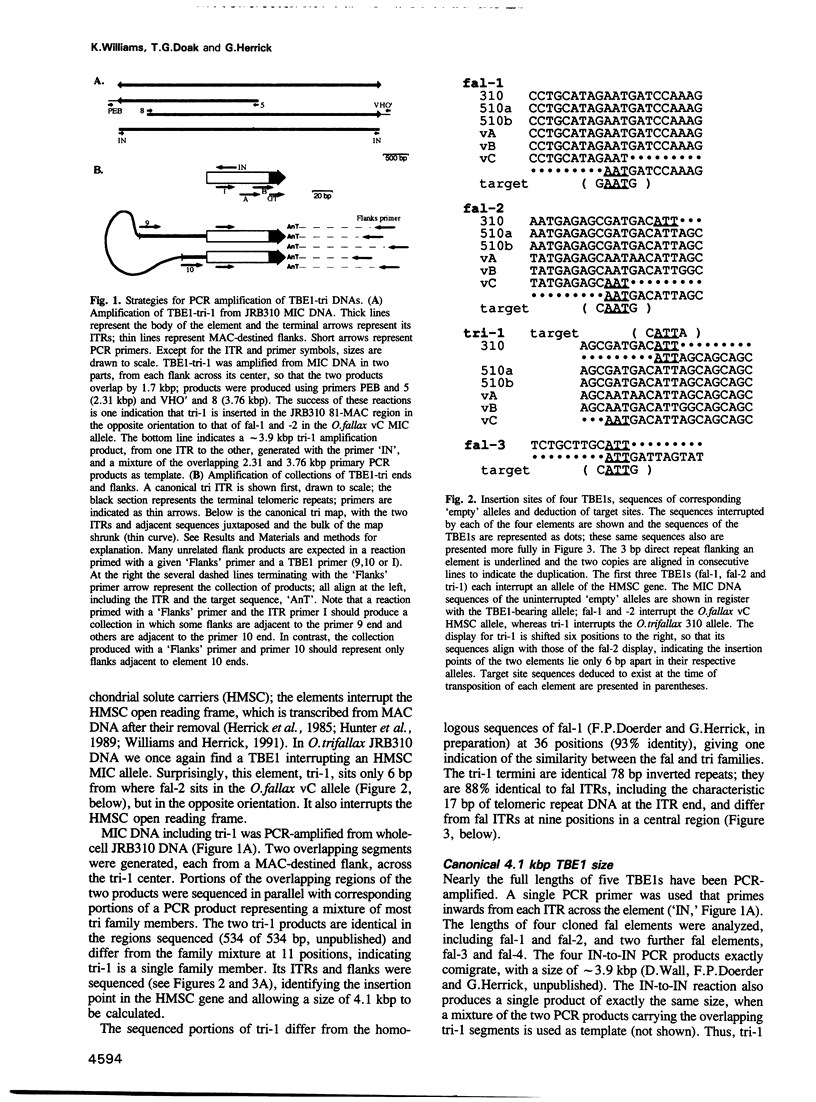
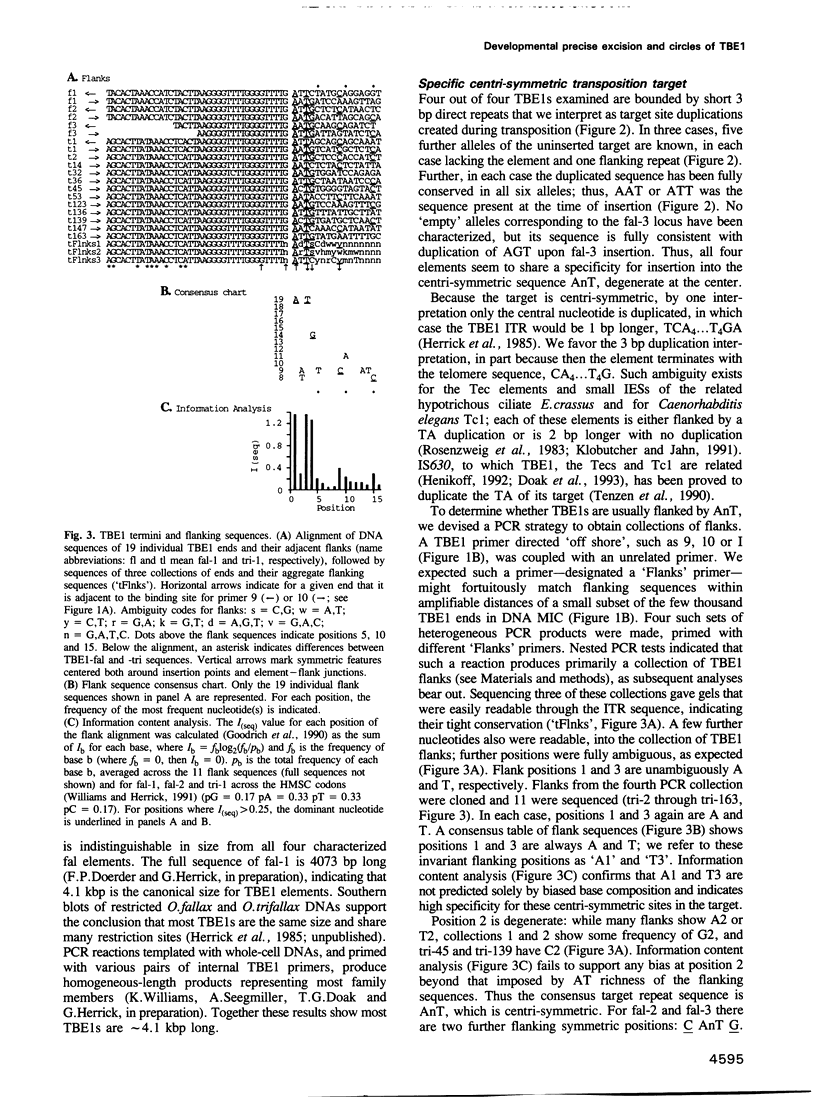
The 4.1 kbp TBE1 elements of Oxytricha fallax and Oxytricha trifallax are deduced to transpose into a centrisymmetric target, CAnTG, and to duplicate the central AnT. Despite conserved C(A4C4)2 telomeric repeats at their tips, free TBE1s found during macronuclear development are not linear but 4.1 kbp circles closed on one copy of the AnT target duplication. The macronucleus-destined flanks are rejoined to regenerate the target, effecting efficient and precise somatic reversion of the germline transpositional mutation. A model is presented in which transposase catalyzes concerted precise rejoining of the flanks and cyclization of the excised element.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender J., Kleckner N. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):7996–8000. doi: 10.1073/pnas.89.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartinhour S. W., Herrick G. A. Three different macronuclear DNAs in Oxytricha fallax share a common sequence block. Mol Cell Biol. 1984 May;4(5):931–938. doi: 10.1128/mcb.4.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson D., Herrick G. Micronuclear DNA sequences of Oxytricha fallax homologous to the macronuclear inverted terminal repeat. Nucleic Acids Res. 1982 May 11;10(9):2911–2924. doi: 10.1093/nar/10.9.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels W. R., Johnson-Schlitz D. M., Eggleston W. B., Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990 Aug 10;62(3):515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- Godiska R., Yao M. C. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell. 1990 Jun 29;61(7):1237–1246. doi: 10.1016/0092-8674(90)90688-b. [DOI] [PubMed] [Google Scholar]
- Goff S. P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann A. T., Craig N. L. Tn7 transposition creates a hotspot for homologous recombination at the transposon donor site. Genetics. 1993 Jan;133(1):9–16. doi: 10.1093/genetics/133.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Detection of Caenorhabditis transposon homologs in diverse organisms. New Biol. 1992 Apr;4(4):382–388. [PubMed] [Google Scholar]
- Herrick G., Cartinhour S. W., Williams K. R., Kotter K. P. Multiple sequence versions of the Oxytricha fallax 81-MAC alternate processing family. J Protozool. 1987 Nov;34(4):429–434. doi: 10.1111/j.1550-7408.1987.tb03207.x. [DOI] [PubMed] [Google Scholar]
- Herrick G., Cartinhour S., Dawson D., Ang D., Sheets R., Lee A., Williams K. Mobile elements bounded by C4A4 telomeric repeats in Oxytricha fallax. Cell. 1985 Dec;43(3 Pt 2):759–768. doi: 10.1016/0092-8674(85)90249-1. [DOI] [PubMed] [Google Scholar]
- Hunter D. J., Williams K., Cartinhour S., Herrick G. Precise excision of telomere-bearing transposons during Oxytricha fallax macronuclear development. Genes Dev. 1989 Dec;3(12B):2101–2112. doi: 10.1101/gad.3.12b.2101. [DOI] [PubMed] [Google Scholar]
- Jaraczewski J. W., Jahn C. L. Elimination of Tec elements involves a novel excision process. Genes Dev. 1993 Jan;7(1):95–105. doi: 10.1101/gad.7.1.95. [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Jahn C. L. Developmentally controlled genomic rearrangements in ciliated protozoa. Curr Opin Genet Dev. 1991 Oct;1(3):397–403. doi: 10.1016/s0959-437x(05)80306-5. [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Jahn C. L., Prescott D. M. Internal sequences are eliminated from genes during macronuclear development in the ciliated protozoan Oxytricha nova. Cell. 1984 Apr;36(4):1045–1055. doi: 10.1016/0092-8674(84)90054-0. [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Turner L. R., LaPlante J. Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev. 1993 Jan;7(1):84–94. doi: 10.1101/gad.7.1.84. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Moerman D. G., Kiff J. E., Waterston R. H. Germline excision of the transposable element Tc1 in C. elegans. Nucleic Acids Res. 1991 Oct 25;19(20):5669–5672. doi: 10.1093/nar/19.20.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H. The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 1991 Jul;10(7):1919–1925. doi: 10.1002/j.1460-2075.1991.tb07718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P., Prère M. F., Fayet O., Chandler M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992 Dec;11(13):5079–5090. doi: 10.1002/j.1460-2075.1992.tb05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott D. M. The unusual organization and processing of genomic DNA in hypotrichous ciliates. Trends Genet. 1992 Dec;8(12):439–445. doi: 10.1016/0168-9525(92)90328-2. [DOI] [PubMed] [Google Scholar]
- Rae P. M., Spear B. B. Macronuclear DNA of the hypotrichous ciliate Oxytricha fallax. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4992–4996. doi: 10.1073/pnas.75.10.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig B., Liao L. W., Hirsh D. Target sequences for the C. elegans transposable element Tc1. Nucleic Acids Res. 1983 Oct 25;11(20):7137–7140. doi: 10.1093/nar/11.20.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausta S. L., Turner L. R., Buckley L. K., Klobutcher L. A. High fidelity developmental excision of Tec1 transposons and internal eliminated sequences in Euplotes crassus. Nucleic Acids Res. 1991 Jun 25;19(12):3229–3236. doi: 10.1093/nar/19.12.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T., Matsutani S., Ohtsubo E. Site-specific transposition of insertion sequence IS630. J Bacteriol. 1990 Jul;172(7):3830–3836. doi: 10.1128/jb.172.7.3830-3836.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., Herrick G. Expression of the gene encoded by a family of macronuclear chromosomes generated by alternative DNA processing in Oxytricha fallax. Nucleic Acids Res. 1991 Sep 11;19(17):4717–4724. doi: 10.1093/nar/19.17.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]