Abstract
The aim of this study was to isolate and identify Actinobacteria from Malaysia mangrove forest and screen them for production of antimicrobial secondary metabolites. Eighty-seven isolates were isolated from soil samples collected at 4 different sites. This is the first report to describe the isolation of Streptomyces, Mycobacterium, Leifsonia, Microbacterium, Sinomonas, Nocardia, Terrabacter, Streptacidiphilus, Micromonospora, Gordonia, and Nocardioides from mangrove in east coast of Malaysia. Of 87 isolates, at least 5 isolates are considered as putative novel taxa. Nine Streptomyces sp. isolates were producing potent antimicrobial secondary metabolites, indicating that Streptomyces isolates are providing high quality metabolites for drug discovery purposes. The discovery of a novel species, Streptomyces pluripotens sp. nov. MUSC 135T that produced potent secondary metabolites inhibiting the growth of MRSA, had provided promising metabolites for drug discovery research. The biosynthetic potential of 87 isolates was investigated by the detection of polyketide synthetase (PKS) and nonribosomal polyketide synthetase (NRPS) genes, the hallmarks of secondary metabolites production. Results showed that many isolates were positive for PKS-I (19.5%), PKS-II (42.5%), and NRPS (5.7%) genes, indicating that mangrove Actinobacteria have significant biosynthetic potential. Our results highlighted that mangrove environment represented a rich reservoir for isolation of Actinobacteria, which are potential sources for discovery of antimicrobial secondary metabolites.
1. Introduction
Actinobacteria represent a significant component of the microbial population in most soils including the mangrove region [1]. This phylum of bacteria has been extremely useful to the pharmaceutical industry due to their seemingly unlimited capacity to produce secondary metabolites with diverse biological activities and chemical structures [1–4]. Approximately 50% of Actinobacteria are from the genus Streptomyces, and approximately 75% of commercially useful antibiotics are derived from this genus [5].
In recent years, the chances of discovering novel biologically active molecules from various known soil bacteria (including Actinobacteria) have reduced, implying that a saturation effect could be occurring. The isolation of known Actinobacteria such as Streptomyces from various environments was found to be producing similar compounds [6]. Furthermore, the emergence of multidrug resistant pathogenic bacteria such as MRSA and fungus has resulted in critical demand for new natural products and chemical compounds in pharmacology, which in turn has made the exploration of poorly exploited areas such as the mangrove environments essential to discover novel Actinobacteria and novel metabolites [1, 7, 8].
The mangrove forests are highly productive ecosystems that provide vital protections to the coastline located in the tidal zones in the tropical and subtropical areas. These ecosystems are habitat to diverse flora and fauna of marine, freshwater, and terrestrial species [9]. In contrast to the well-documented species diversity of larger animals and plants in these mangrove ecosystems, the diversity of microbial community in the mangrove environments needs to be improved [1, 10, 11].
The constant changes in the environmental factors such as tidal gradient and salinity in the mangrove environments are understood to be the driving force for metabolic pathway adaptations that could lead to the production of unusual metabolites. Therefore, there had been increasing exploitation of the mangrove microorganism resources [1, 12]. Many researchers discovered that the poorly explored mangrove environments contain high populations of novel Actinobacteria, as demonstrated by the isolation of Asanoa iriomotensis [13], Nonomuraea maheshkhaliensis [14], and Streptomyces xiamenensis [10]. Furthermore, many strains are also prolific producers of useful antibiotics [1, 11, 15]. Many Actinobacteria isolates from marine environments contain polyketide synthetase (PKS) and nonribosomal polyketide synthetase (NRPS) pathways, the characteristics of secondary metabolites production [16].
The Tanjung Lumpur mangrove forest located on the east coast of Peninsular Malaysia is mostly unexplored; therefore, this location is anticipated to be able to provide a rich source of Actinobacteria, the prolific producers of antimicrobial secondary metabolites. To our knowledge, no studies have reported the diversity and antimicrobial activities of Actinobacteria from Tanjung Lumpur mangrove environment. Therefore, there is a high possibility to identify novel Actinobacteria and discover valuable antimicrobial secondary metabolites. The aim of this study was to isolate and identify the Actinobacteria and screen them to discover potential sources for antimicrobial secondary metabolites.
2. Materials and Methods
2.1. Environmental Sampling
Soil sediments were collected from Tanjung Lumpur mangrove forest located in the city of Kuantan, State of Pahang, in December of the year 2012. Four different mangrove sediments were collected at site MUSC-TLS1 (3°48′3.2′′N 103°20′11.0′′E) until MUSC-TLS4 (3°48′21.3′′N 103°20′3.3′′E). At each site, five-sediment core samples were collected at a depth of 0–30 cm within a 50 m2 area. The sediments from each site were bulked and homogenized to prepare the composite samples. Sediments were placed into sterile plastic bags using an aseptic metal trowel and kept in the dark for transport to the laboratory. The physicochemical parameters such as temperature and pH of the sampling area were determined using soil temperature profile sensor ST01 and ph meter. There was little fluctuation in temperature and pH of the sampling locations. The temperature of soil samples ranged between 23 and 25°C and slightly acidic pH were observed in all the samples (6.1–6.4).
2.2. Selective Isolation of Actinobacteria
Air-dried soil sediment (~7 days) was ground with mortar and pestle. Selective pretreatment of the samples was performed using a phenol solution (1.5%, 30 min at 30°C) [17] or wet heat in sterilized water (15 min at 50°C) [18]. The pretreated samples were diluted 1 : 10 v/v with sterile 25% Ringer's solution and serial dilution to 10−4. One hundred μL of the 10−1, 10−2, 10−3, and 10−4 suspensions was spread in triplicate onto isolation media.
Dilutions of soil suspensions were spread onto 6 different types of isolation media: ISP 2 (yeast malt agar), ISP 7 (tyrosine agar) [19], starch casein agar (SCA) [20], Streptomyces agar (SA) [21], Actinomycetes isolation agar (AIA) [21], and nutrient agar [22]. All media were supplemented with cycloheximide (50 mg/L), nystatin (50 mg/L), and nalidixic acid (20 mg/L) [23] and incubated at 28°C for 1–4 weeks. Purified cultures were maintained on ISP medium 2 [19] slants at room temperature for short-term storage and as glycerol suspensions (20%, v/v) at −80°C for long-term storage.
2.3. Morphological, Physiological, and Biochemical Characterizations of 87 Isolates of Actinobacteria
The cultural, morphological, biochemical, and physiological characterizations of the Actinobacteria isolates were performed as described by Shirling and Gottlieb [19]. Light microscopy (80i, Nikon) and scanning electron microscopy (JEOL-JSM 6400) were used to observe the morphologies of selected isolates after incubation on ISP 2 medium at 28°C for 7 days. Using the light microscope, the formation of aerial and substrate mycelium and spore arrangement was observed. Cultural characteristics of isolates, which include growth, colony color on different isolation media, the presence of aerial and substrate mycelium, distinctive reverse colony color, and diffusible pigment, were determined using six different isolation media including ISP 2, ISP 7, SCA, SA, AIA, and nutrient agar with procedures as described by International Streptomyces Project (ISP). The production of melanoid pigments was examined using tyrosine agar (ISP 7). The ISCC-NBS colour charts were used to determine the names and designations of the colony colours [24].
Biochemical characterizations such as Gram staining and blood hemolysis were performed. Gram staining was performed by standard Gram reaction and was confirmed by using KOH lysis [25]. Hemolytic activity was performed in blood agar medium containing 5% (w/v) peptone, 3% (w/v) yeast extract, 5% (w/v) NaCl, and 5% (v/v) human blood [26]. Plates were examined for hemolysis after incubation at 32°C for 5 days. The presence of clear zone around colonies signifies the potential of isolates for surfactant production. Physiological characterization such as growth temperature was performed. The growth temperature was tested at 12–52°C at intervals of 4°C on ISP 2. The Biolog GenIII MicroPlates were used according to manufacturer's instructions to determine a total of 71 carbon-source utilization assays and 23 chemical sensitivity assays for selected isolates.
2.4. Preliminary Screening of Actinobacteria Isolates for Antimicrobial Activity
Eighty-seven isolates were preceded to preliminary screening for antimicrobial activity by using the cross streak method [27]. The isolates were cross streaked on ISP 2 medium and incubated at room temperature for 5–7 days. After observing a good growth of Actinobacterial cultures, overnight cultures of 12 different pathogens were used for the screening; namely, Bacillus subtilis ATCC 31098T, Bacillus cereus NBRC 13494T, Enterococcus faecalis NBRC 12965T, methicillin-resistant Staphylococcus aureus (MRSA) ATCC BAA-44T, Staphylococcus epidermidis ATCC 12228T, Aeromonas hydrophila ATCC 7966T, Acinetobacter calcoaceticus NBRC 13006T, Klebsiella oxytoca NBRC 12582T, Klebsiella pneumonia NBRC 14440T, Pseudomonas aeruginosa NRBC 12582T, Salmonella typhi ATCC 19430T, and Yersinia pseudotuberculosis NBRC 105692T were streaked at the right angle of Actinobacterial cultures. Plates were incubated at 28°C for 48 hrs and the zone of inhibition was recorded. ISP 2 plates without Actinobacteria isolates but streaked with the same stock of pathogens were used as control.
2.5. Crude Extracts Preparation and Screening for Secondary Antimicrobial Metabolites
Eighty-seven Actinobacteria isolates were subjected to subsequent investigation by antimicrobial screening of their secondary metabolites. The method of Thakur et al. [31] was modified for screening bioactive antimicrobial metabolites. The same pathogens used during preliminary screening were used for this screening. These pathogens were cultured overnight at 37°C at 200 rpm in nutrient broth. The cultures were diluted with their respective media to 0.8–1.2 × 106 CFU/mL, and 100 μL aliquots of this inoculum were transferred to nutrient agar media.
The fermentation medium used was FM3 [32]; the medium was autoclaved at 121°C for 20 min. Each of the 87 purified isolates was transferred to a test tube (30 mm × 200 mm) containing 20 mL of the relevant fermentation medium and cultured at 200 rpm, at an angle of 45°, for 7–10 days at 28°C. The resulting fermentation media obtained from each of the isolates were separated from the mycelium by centrifugation at 10,000 rpm at 4°C for 15 min. The supernatants were filtered (Whatman number 1 filter paper) and tested for extracellular antimicrobial activity against different target pathogens using agar well diffusion method [33]. A sterile cork borer was used to puncture well in appropriate agar medium plates previously seeded with one of the pathogen strains. One hundred μL from the fermentation supernatant of the isolates was added to each of the wells. The inoculated plates were kept at 4°C for at least 2 hrs to allow the diffusion of produced antimicrobial metabolites. The diameters of inhibition are determined after 24 hrs of incubation at 37°C and verified active substance extraction. Each experiment was repeated three times and average value of inhibitory zones was reported. Blank wells without the fermentation medium were taken as control.
2.6. Molecular Identification of Actinobacteria Isolates
2.6.1. Genomic DNA Extraction and PCR Amplification of 16S rDNA
Genomic DNA extractions for 87 isolates of Actinobacteria were performed as described by Hong et al. [1]. The primer pair 27F-1492R [1, 28] was used for PCR amplification using the Kyratec PCR Supercycler (Kyratec, Australia). The PCR reaction mixture that consisted of 20–200 ng bacteria genomic DNA, 10.0 μL of 2X Prime Taq Premix (Genet Bio, Korea), 10 pmoles primer 27F and primer 1492R, and sterile ultrapure water was added to final volume of 20 μL. The cycling parameters were as described by Hong et al. [1].
2.6.2. Phylogenetic Analysis of 16S rRNA Gene Sequences
The PCR products were purified using the GeneAll Expin Gel SV purification kit (GeneAll, South Korea); and then molecular cloning was performed using the InsTAclone PCR cloning kit (Thermo Scientific, USA) according to the manufacturer's protocols. The insertions were verified using colony-PCR and colonies with transformations were preceded to plasmid DNA extraction using the Eppendorf FastPlasmid Minikit. Purified plasmid DNA served as templates for PCR to confirm the insertion of the gene of interest. Plasmid DNAs were sequenced using an ABI PRISM 3100 DNA sequencer (Applied Biosystems, USA).
The cloned 16S rRNA gene sequences were aligned manually using sequences from the closest related genera retrieved from the GenBank/EMBL/DDBJ databases using CLUSTAL-X [34]. The alignment was manually verified and adjusted prior to the construction of a phylogenetic tree. The calculation for the level of sequence similarity was performed using the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/) [35]. The phylogenetic tree was inferred using the neighbor joining algorithms [36] via molecular evolutionary genetic analysis (MEGA version 5.2) [37]. The stabilities of the resultant tree topologies were evaluated using bootstrap analysis [38]. The pairwise distances between sequences were generated using Kimura's 2-parameter model [39].
2.6.3. DNA-DNA Hybridization for Putative Novel Isolates
Biomass for chemotaxonomic studies was obtained after growing in tryptic soy broth (TSB) at 28°C for 7 days on a rotary shaker. The extraction of genomic DNA for DNA-DNA hybridization of putative novel isolates (MUSC 115T and MUSC 135T) and their closely related type strains was carried out by the identification service of the DSMZ, Braunschweig, Germany. Genomic DNA extractions from the isolates were performed as described by Cashion et al. [40]. DNA-DNA hybridization was carried out as described by de Ley et al. [41] under consideration of the modifications described by Huss et al. [42] using a model Cary 100 Bio UV/VIS-spectrophotometer equipped with a Peltier-thermostatted 6 × 6 multicell changer and a temperature controller with in situ temperature probe (Varian).
2.6.4. PCR Detection of PKS-I, PKS-II, and NRPS Sequences
Three sets of degenerate primers were used for the amplification of genes encoding polyketide synthases I and II (PKS-I and PKS-II) and nonribosomal peptide synthetases (NRPS) (Table 1). The PCR reaction mixture that consisted of 20–200 ng bacteria genomic DNA, 10.0 μL of 2X Prime Taq Premix (Genet Bio, Korea), 10 pmoles of different primer sets (Table 1), and sterile ultrapure water was added to final volume of 20 μL. The PCR was performed using the Kyratec PCR Supercycler (Kyratec, Australia) with the following cycling conditions: (i) 94°C for 5 min; (ii) 30 cycles of 94°C for 1 min, 57°C (for K1F-M6R and KSα-KSβ) or 62°C (for A3F-A7R) for 1 min, and 72°C for 2 min; and (iii) 72°C for 5 min. The PCR amplification products were resolved using electrophoresis in 1.5% agarose gel (Promega, USA) and stained with ethidium bromide (0.5 μg mL−1) and viewed using Molecular Imager Chemidoc XRS System (Biorad, USA).
Table 1.
PCR primers used in this study.
| Primer name | Sequence (5′-3′) | Target gene | Product size (bp) | Reference |
|---|---|---|---|---|
| 27F | GTTTGATCCTGGCTCAG | 16S rRNA | 1400–1500 | [1, 28] |
| 1492R | TACGGCTACCTTGTTACGACTT | |||
| K1F | TSAAGTCSAACATCGGBCA | PKS-I | 1200–1400 | [29] |
| M6R | CGCAGGTTSCSGTACCAGTA | |||
| KSα | TSGCSTGCTTGGAYGCSATC | PKS-II | 600 | [30] |
| KSβ | TGGAANCCGCCGAABCCTCT | |||
| A3F | GCSTACSYSATSTACACSTCSGG | NRPS | 700–800 | [29] |
| A7R | SASGTCVCCSGTSCGGTAS |
3. Results and Discussion
3.1. Selective Isolation of Actinobacteria
Based on the distinct morphology of Actinobacteria isolates on the media plate, a total of 87 isolates of Actinobacteria were successfully isolated from four composite mangrove sediments collected from Tanjung Lumpur of the State of Pahang, Malaysia. Sediments samples were named as TLS1, TLS2, TLS3, and TLS4 that were contributing to 47, 12, 15, and 13 isolates, respectively, of the total number of isolates. Eighty-seven isolates were isolated from 6 types of isolation media supplemented with cycloheximide, nystatin, and nalidixic acid; these media are, namely, SCA (n = 35), ISP 2 (n = 18), ISP 7 (n = 18), AIA (n = 11), NA (n = 3), and SA (n = 2). These results indicated that starch casein agar (SCA) was the most suitable medium for isolating Actinobacteria in this study, and this result is in agreement with others [43].
3.2. Diversity of Actinobacteria Isolates
Eighty-seven Actinobacteria isolates were identified to the genera level based on the molecular and morphological characteristics. The 16S rRNA gene sequences of these isolates were compared with 16S rRNA sequences of type strains retrieved from DDBJ/EMBL/GenBank. Results showed that Actinobacteria isolated in this study exhibited high level of diversity, as the 87 isolates were distributed among 5 suborders: Streptomycineae (n = 53), Micrococcineae (n = 16), Corynebacterineae (n = 16), Micromonosporineae (n = 1), and Propionibacterineae (n = 1) within the phylum Actinobacteria. A total of 11 genera were identified and each genus was distinguished by its 16S rRNA gene sequences. Of the 87 isolates, 59.8% (n = 52) of the isolates were assigned as genus Streptomyces, 14.9% (n = 13) as the genus Mycobacterium, and the remaining isolates as Leifsonia (n = 6), Microbacterium (n = 4), Sinomonas (n = 4), Nocardia (n = 2), Terrabacter (n = 2), Gordonia (n = 1), Micromonospora (n = 1), Nocardioides (n = 1), and Streptacidiphilus (n = 1). In this study, it is evident that some genera not commonly found in the mangrove environment were discovered, such as Streptacidiphilus, Sinomonas, Terrabacter, and Leifsonia. So far, the different species of Streptacidiphilus were isolated from area such as Pinus soils [44], acidic rhizosphere soil [45], and rice field soil [46], while Sinomonas was mostly discovered in forest soils [47], polluted forest soil [48], and volcanic soil [49].
The isolation of Actinobacteria from eleven genera showed a wide distribution of Actinobacteria in mangrove environment, especially in soil and sediments [1]. The presence of predominant number of Streptomyces isolates (59.8%) in this study is in agreement with results reported by Hong et al. [1] which isolated substantial Streptomyces isolates from mangrove soils in China. The identification of substantial number of Streptomyces isolates was extremely important for the antimicrobial bioactivities screening in this study, as this genus is proven to be the prolific producers of novel antibiotics [50, 51], with approximately 75% of commercially useful antibiotics being derived from Streptomyces [5].
The analysis of 16S rRNA sequences is used to determine higher taxonomic relationships of Actinobacteria [52, 53]. In this study, the 16S rRNA gene sequences were used to build a phylogenetic tree for 35 isolates of non-Streptomyces isolates together with their closely related type strains (Figure 1), whereas another phylogenetic tree was built for 52 isolates assigned to genus Streptomyces and their closely related type strains (Figure 2). The percentages of the 16S rRNA gene sequence similarities (95.1 to 100%) of these isolates to the closest type strains are shown in Table 2. The taxonomic studies showed that some isolates were highly potential to be assigned as novel genus or species based on the phylogenetic and pairwise comparison of 16S rRNA gene sequences of the novel isolates with the type strains. Four isolates from three genera (Sinomonas, Microbacterium, and Streptomyces) showed high possibilities of novel species discovery and one isolate (MUSC 201T) was identified as a novel genus in the family Nocardioidaceae.
Figure 1.
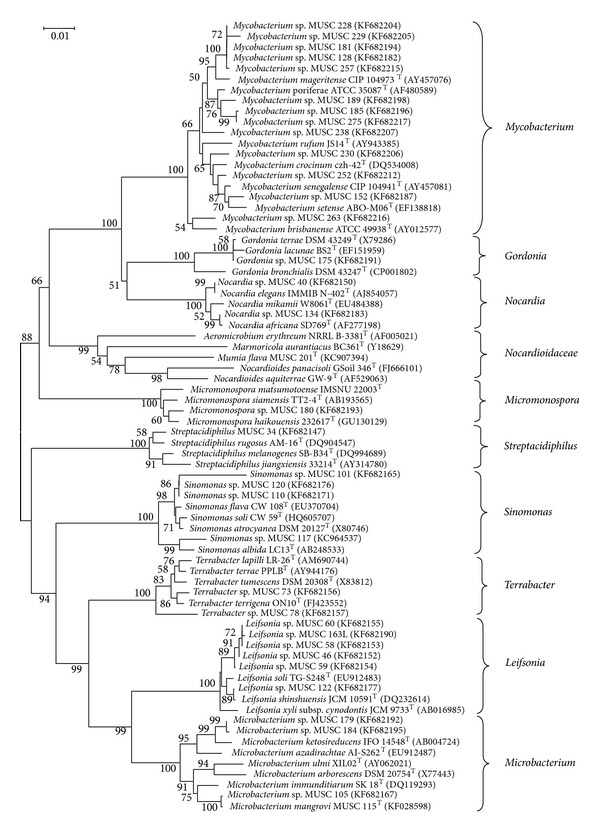
Phylogenetic tree based on 16S rRNA sequences using neighbour-joining method for 35 isolates of non-Streptomyces Actinobacteria and their closely related type strains. Bootstrap values (>40%) based on 1000 resampled datasets are shown at branch nodes. Bar, 1 substitution per 100 nucleotide positions.
Figure 2.
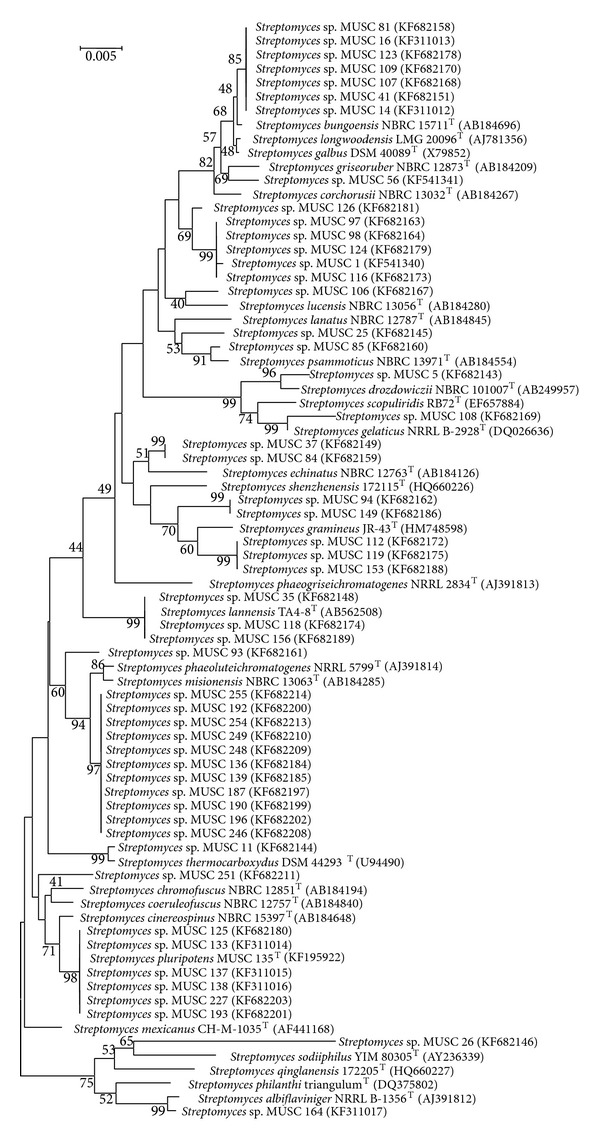
Phylogenetic tree based on 16S rRNA sequences using neighbour-joining method for 52 isolates of Streptomyces sp. and their closely related type strains. Bootstrap values (>40%) based on 1000 resampled datasets are shown at branch nodes. Bar, 5 substitutions per 1000 nucleotide positions.
Table 2.
Identification and antimicrobial activities and presence of PKS and NRPS genes in Actinobacteria isolates from Tanjung Lumpur, Malaysia.
| Isolate number (Genbank accession number) | Closest relative | Sequence identity (%) | Activitya (mm) against pathogens | Presence of gene | ||
|---|---|---|---|---|---|---|
| PKS-I | PKS-II | NRPS | ||||
| MUSC 1 (KF541340) | Streptomyces corchorusii | 98.8 | BS (9) | + | − | − |
| MUSC 5 (KF682143) | Streptomyces drozdowiczii | 99.4 | − | − | − | − |
| MUSC 11 (KF682144) | Streptomyces thermocarboxydus | 99.9 | − | − | + | − |
| MUSC 14 (KF311012) | Streptomyces bungoensis | 99.4 | MRSA (2), SE (2) | + | − | − |
| MUSC 16 (KF311013) | Streptomyces bungoensis | 99.5 | MRSA (3), SE (3) | + | − | − |
| MUSC 25 (KF682145) | Streptomyces philanthi | 96.3 | − | − | − | − |
| MUSC 26 (KF682146) | Streptomyces qinglanensis | 96.2 | − | − | − | − |
| MUSC 34 (KF682147) | Streptacidiphilus rugosus | 99.4 | − | − | − | − |
| MUSC 35 (KF682148) | Streptomyces lannensis | 100.0 | − | − | + | − |
| MUSC 37 (KF682149) | Streptomyces phaeogriseichromatogenes | 97.9 | − | − | − | − |
| MUSC 40 (KF682150) | Nocardia elegans | 99.9 | − | − | + | − |
| MUSC 41 (KF682151) | Streptomyces bungoensis | 99.6 | − | + | − | − |
| MUSC 46 (KF682152) | Leifsonia shinshuensis | 99.4 | − | − | − | − |
| MUSC 56 (KF541341) | Streptomyces antibioticus | 100.0 | BS (4.5), BC (4), EF (3) | − | + | − |
| MUSC 58 (KF682153) | Leifsonia shinshuensis | 99.3 | − | − | − | − |
| MUSC 59 (KF682154) | Leifsonia shinshuensis | 99.3 | − | − | − | − |
| MUSC 60 (KF682155) | Leifsonia shinshuensis | 99.4 | − | − | − | − |
| MUSC 73 (KF682156) | Terrabacter tumescens | 99.0 | − | − | − | − |
| MUSC 78 (KF682157) | Terrabacter lapilli | 98.3 | − | − | − | − |
| MUSC 81 (KF682158) | Streptomyces bungoensis | 97.8 | − | + | − | + |
| MUSC 84 (KF682159) | Streptomyces echinatus | 98.7 | − | − | − | − |
| MUSC 85 (KF682160) | Streptomyces psammoticus | 99.5 | − | − | + | − |
| MUSC 93 (KF682161) | Streptomyces phaeogriseichromatogenes | 97.0 | − | − | + | − |
| MUSC 94 (KF682162) | Streptomyces gramineus | 98.6 | − | − | − | − |
| MUSC 97 (KF682163) | Streptomyces phaeogriseichromatogenes | 97.7 | − | + | − | − |
| MUSC 98 (KF682164) | Streptomyces corchorusii | 98.9 | − | − | − | − |
| MUSC 101 (KF682165) | Sinomonas soli | 96.8 | − | − | − | − |
| MUSC 105 (KF682166) | Microbacterium immunditiarum | 98.0 | − | − | − | − |
| MUSC 106 (KF682167) | Streptomyces lucensis | 99.2 | − | − | + | − |
| MUSC 107 (KF682168) | Streptomyces bungoensis | 98.8 | − | + | − | − |
| MUSC 108 (KF682169) | Streptomyces scopuliridis | 97.1 | − | − | + | − |
| MUSC 109 (KF682170) | Streptomyces bungoensis | 99.6 | − | + | − | − |
| MUSC 110 (KF682171) | Sinomonas atrocyanea | 99.4 | − | − | − | − |
| MUSC 112 (KF682172) | Streptomyces gramineus | 98.4 | − | − | + | − |
| MUSC 115 (KF028598) | Microbacterium immunditiarum | 98.1 | − | − | − | − |
| MUSC 116 (KF682173) | Streptomyces phaeogriseichromatogenes | 97.3 | − | − | − | − |
| MUSC 117 (KC964537) | Sinomonas atrocyanea | 98.0 | − | − | − | − |
| MUSC 118 (KF682174) | Streptomyces lannensis | 97.4 | − | − | − | − |
| MUSC 119 (KF682175) | Streptomyces gramineus | 97.8 | − | + | + | − |
| MUSC 120 (KF682176) | Sinomonas soli | 99.1 | − | − | − | − |
| MUSC 122 (KF682177) | Leifsonia soli | 99.5 | − | − | − | − |
| MUSC 123 (KF682178) | Streptomyces bungoensis | 97.1 | − | + | − | − |
| MUSC 124 (KF682179) | Streptomyces phaeogriseichromatogenes | 97.9 | − | − | − | − |
| MUSC 125 (KF682180) | Streptomyces cinereospinus | 99.3 | − | − | + | − |
| MUSC 126 (KF682181) | Streptomyces phaeogriseichromatogenes | 98.3 | − | + | − | − |
| MUSC 128 (KF682182) | Mycobacterium rufum | 96.7 | − | − | − | − |
| MUSC 133 (KF311014) | Streptomyces cinereospinus | 99.3 | MRSA (6) | − | + | − |
| MUSC 134 (KF682183) | Nocardia africana | 99.8 | − | + | + | + |
| MUSC 135 (KF195922) | Streptomyces cinereospinus | 99.2 | BC (4), MRSA (12), AH (4), ST (4) | − | + | − |
| MUSC 136 (KF682184) | Streptomyces phaeoluteichromatogenes | 98.5 | − | − | + | − |
| MUSC 137 (KF311015) | Streptomyces cinereospinus | 99.2 | MRSA (4) | − | + | − |
| MUSC 138 (KF311016) | Streptomyces cinereospinus | 99.2 | MRSA (11) | − | + | − |
| MUSC 139 (KF682185) | Streptomyces phaeogriseichromatogenes | 97.5 | − | − | + | − |
| MUSC 149 (KF682186) | Streptomyces gramineus | 97.9 | − | − | + | − |
| MUSC 152 (KF682187) | Mycobacterium setense | 97.9 | − | + | − | + |
| MUSC 153 (KF682188) | Streptomyces gramineus | 98.1 | − | − | + | − |
| MUSC 156 (KF682189) | Streptomyces lannensis | 97.3 | − | − | + | − |
| MUSC 163L (KF682190) | Leifsonia shinshuensis | 99.2 | − | − | − | − |
| MUSC 164 (KF311017) | Streptomyces albiflaviniger | 98.4 | BC (2), EF (1), MRSA (4) | + | − | − |
| MUSC 175 (KF682191) | Gordonia terrae | 99.9 | − | − | − | − |
| MUSC 179 (KF682192) | Microbacterium ketosireducens | 97.7 | − | − | − | − |
| MUSC 180 (KF682193) | Micromonospora siamensis | 97.9 | − | − | + | − |
| MUSC 181 (KF682194) | Mycobacterium rufum | 96.6 | − | − | − | − |
| MUSC 184 (KF682195) | Microbacterium azadirachtae | 97.7 | − | − | − | − |
| MUSC 185 (KF682196) | Mycobacterium crocinum | 97.9 | − | − | + | − |
| MUSC 187 (KF682197) | Streptomyces phaeogriseichromatogenes | 97.9 | − | − | + | − |
| MUSC 189 (KF682198) | Mycobacterium poriferae | 98.7 | − | − | − | − |
| MUSC 190 (KF682199) | Streptomyces phaeogriseichromatogenes | 97.9 | − | − | − | − |
| MUSC 192 (KF682200) | Streptomyces phaeoluteichromatogenes | 98.7 | − | + | + | − |
| MUSC 193 (KF682201) | Streptomyces mexicanus | 98.0 | − | − | + | − |
| MUSC 196 (KF682202) | Streptomyces phaeogriseichromatogenes | 97.8 | − | − | + | − |
| MUSC 201 (KC907394) | Nocardioides panacisoli | 95.1 | − | − | + | − |
| MUSC 227 (KF682203) | Streptomyces cinereospinus | 97.0 | − | − | + | − |
| MUSC 228 (KF682204) | Mycobacterium mageritense | 98.8 | − | − | − | − |
| MUSC 229 (KF682205) | Mycobacterium mageritense | 97.5 | − | − | + | − |
| MUSC 230 (KF682206) | Mycobacterium crocinum | 98.3 | − | − | + | − |
| MUSC 238 (KF682207) | Mycobacterium crocinum | 97.6 | − | − | − | + |
| MUSC 246 (KF682208) | Streptomyces phaeogriseichromatogenes | 97.5 | − | − | + | − |
| MUSC 248 (KF682209) | Streptomyces phaeogriseichromatogenes | 97.9 | − | − | + | − |
| MUSC 249 (KF682210) | Streptomyces misionensis | 98.3 | − | + | + | − |
| MUSC 251 (KF682211) | Streptomyces coeruleofuscus | 98.7 | − | − | + | − |
| MUSC 252 (KF682212) | Mycobacterium senegalense | 99.4 | − | − | − | − |
| MUSC 254 (KF682213) | Streptomyces phaeogriseichromatogenes | 97.5 | − | + | − | − |
| MUSC 255 (KF682214) | Streptomyces phaeogriseichromatogenes | 97.6 | − | − | + | − |
| MUSC 257 (KF682215) | Mycobacterium mageritense | 98.7 | − | − | − | + |
| MUSC 263 (KF682216) | Mycobacterium brisbanense | 98.2 | − | − | − | − |
| MUSC 275 (KF682217) | Mycobacterium poriferae | 99.0 | − | − | − | − |
aActivity estimated by measuring the diameter of the clear zone of the growth inhibition. −: no activity.
BS: Bacillus subtilis; BC: Bacillus cereus; EF: Enterococcus faecalis; MRSA: methicillin-resistant Staphylococcus aureus; SE: Staphylococcus epidermidis; AH: Aeromonas hydrophila; ST: Salmonella typhi.
Isolate MUSC 201T was obtained from soil sample MUSC-TLS4 (3°48′21.3′′N 103°20′3.3′′E) pretreated with wet heat method [18]; the mixture was spread onto ISP 2 supplemented with cycloheximide and nystatin and incubated at 28°C for 7 days. The 16S rRNA gene sequences were determined for isolate MUSC 201T (1486 bp) [GenBank: KC907394]. The comparison of isolate MUSC 201T to the closely related phylogenetic neighbors indicated that it was closely related to the type strains of different genera within the family Nocardioidaceae: Nocardioides (95.1% to 91.9% similarity), Aeromicrobium (94.6 to 92.7% similarity), Marmoricola (93.1% to 92.5% similarity), and Kribbella (92.4% to 91.5% similarity). Isolate MUSC 201T formed a distinct monophyletic clade within the family Nocardioidaceae (Figure 1), most closely related to type strain Nocardioides panacisoli GSoil 346T, and Actinobacteria were isolated from soil of ginseng field [54] at high bootstrap value of 78%. Isolate MUSC 201T had low 16S rRNA gene sequence similarities with members of the closest related genus Nocardioides (94.1–95.1%) and was separated from them by a long evolutionary distance in the phylogenetic tree (Figure 1). On the basis of phylogenetic, genotypic, and chemotaxonomic profiles, isolate MUSC 201T is truly different from any existing genera in the family Nocardioidaceae; hence, isolate MUSC 201T represented a novel species in a new genus of the family Nocardioidaceae, for which the name “Mumia flava gen. nov., sp. nov.” is proposed and published [55].
The Microbacterium sp. isolates (MUSC 179, MUSC 184, MUSC 105, and MUSC 115T) formed distinct phylogenetic clade in the neighbour-joining tree but were separated from other members of the genus Microbacterium (Figure 1). Isolates MUSC 179 and MUSC 184 were closely related to Microbacterium ketosireducens IFO 14548T (97.7% similarity) and Microbacterium azadirachtae AI-S262T (97.7% similarity) (Table 2). Another 2 isolates (MUSC 105 and MUSC 115) were assigned to Microbacterium immunditiarum SK 18T, with 98.0 and 98.1% similarity (Table 2). Isolate MUSC 115T was selected for further analysis to determine its potential as novel species.
Isolate MUSC 115T was obtained from soil sample MUSC-TLS1 (3°48′3.2′′N 103°20′11.0′′E) pretreated with wet heat method [18]; the mixture was spread onto starch casein agar. The 16S rRNA gene sequence was established for strain MUSC 115T (1484 bp) [GenBank: KF028598]. Phylogenetic analysis of isolate MUSC 115T indicated that it formed a subclade with Microbacterium immunditiarum SK 18T at high bootstrap supported value of 75% (Figure 1). Pairwise comparison of the 16S rRNA gene sequences from isolate MUSC 115T showed similarities to Microbacterium immunditiarum SK 18T, Microbacterium ulmi XIL02T, and Microbacterium arborescens NBRC 3750T at 98.1, 97.8, and 97.5%, respectively. The DNA-DNA relatedness values between strain MUSC 115T and Microbacterium immunditiarum SK 18T (23.6 ± 0.5%), Microbacterium ulmi XIL02T (26.2 ± 2.7%), and Microbacterium arborescens DSM 20754T (16.3 ± 1.1%) were significantly lower than 70%, the threshold value for the delineation of genomic species [56]. The DDH results suggested that strain MUSC 115T does not belong to any of these species. The comparison of phenotypic characterization between isolate MUSC 115T with its closely related type strains showed substantial differences between them [57]. Hence, isolate MUSC 115T represented a novel species within the genus Microbacterium with lineages distinct from other members of the genus Microbacterium (Figure 1), proposed as “Microbacterium mangrovi sp. nov.” [57].
The Sinomonas sp. isolates (MUSC 101, MUSC 110, MUSC 117, and MUSC 120) formed a distinct phylogenetic clade that was distinct from other members of the phylum Actinobacteria with high bootstrap value (100%) (Figure 1). Two isolates (MUSC 101 and MUSC 120) were assigned to Sinomonas soli, with 96.8 and 99.1% similarity (Table 2). Isolate MUSC 101 was separated from the rest of the isolates by a long evolutionary distance in the phylogenetic tree; this association is supported by bootstrap value of 86% (Figure 1). The 16S rRNA gene sequence similarity value was lower than 97%, which is considered to be the cutoff value for species identity [58]. Therefore, isolate MUSC 101 could serve as a highly potential candidate as a novel species of the genus Sinomonas. Phylogenetic analysis of isolates Sinomonas sp. MUSC 117 indicated that it formed a distinct clade with type strains from genus Sinomonas at high bootstrap value of 100% (Figure 1). Isolate Sinomonas sp. MUSC 117 formed subclade with Sinomonas albida, an association supported by robust bootstrap value of 99% (Figure 1). The pairwise comparison of 16S rRNA gene sequence analysis showed that isolate MUSC 117 is assigned to type strains Sinomonas atrocyanea, Sinomonas albida, and Sinomonas soli with 98, 97.9, and 97.8% similarity, respectively. These 16S rRNA gene similarity values are less than the similarity values between closely related Sinomonas species, such as S. atrocyanea and its closest related species, S. soli (99.5% similarity). Therefore, isolate MUSC 117 serves as a good candidate for novel species in the genus Sinomonas.
3.3. Diversity and Phylogeny of Bioactive Actinobacteria Isolates
Of 87 isolates, 9 isolates which belonged to genus Streptomyces were producing bioactive metabolites (Table 2). The relationships between taxonomic and metabolic diversity are being highlighted for these bioactive isolates. As more novel Actinobacteria isolated from various environments are able to offer precious sources of new bioactive metabolites and compounds [59–61], the analysis of the 16S rRNA gene sequences of the bioactive isolates showed that isolates Streptomyces sp. MUSC 14 and Streptomyces sp. MUSC 16 shared the highest similarities to Streptomyces bungoensis NBRC 15711T with 99.5 to 99.4% identities (Table 2). They formed distinct subclade with Streptomyces bungoensis NBRC 15711T at 48% bootstrap value (Figure 2). Isolate MUSC 164 showed 98.4% identities to Streptomyces albiflaviniger NRRL B-1356T (Table 2) and they formed a monophyletic clade at 99% bootstrap value (Figure 2).
Isolates Streptomyces sp. MUSC 133, MUSC 135T, MUSC 137, and MUSC 138 exhibited the highest similarities to sequences of type strain Streptomyces cinereospinus with 99.2 to 99.3% identities (Table 2). The phylogenetic analysis showed that isolates MUSC 133, MUSC 135T, MUSC 137, and MUSC 138 formed a distinct clade with type strains Streptomyces cinereospinus NBRC 15397T at bootstrap value of 71% (Figure 2). Furthermore, these Streptomyces isolates (MUSC 133, MUSC 135T, MUSC 137, and MUSC 138) formed a distinct monophyletic subclade supported by robust bootstrap values of 98% (Figure 2). The 16S rRNA gene sequence similarities between isolate MUSC 135T and isolates MUSC 133, MUSC 137, and MUSC 138 were 100, 100, and 99.9%, respectively. Therefore, isolates MUSC 133, MUSC 135T, MUSC 137, and MUSC 138 should be considered as the same species. Isolate MUSC 135T was chosen as the representative for subsequent characterization as this isolate could produce diverse and potent antimicrobial secondary metabolites. Isolate MUSC 135T showed similarities to type strains Streptomyces cinereospinus NBRC 15397T, Streptomyces mexicanus CH-M-1035T, and Streptomyces coeruleofuscus NBRC 12757T with 99.18, 99.17, and 98.97% similarity, respectively (Table 2). These similarities values are lower as compared to the similarity value of type strain Streptomyces cinereospinus NBRC 15397T to its closest related species Streptomyces coeruleofuscus NBRC 12757T (99.4% similarity). Furthermore, the DNA-DNA relatedness values between strain MUSC 135T and Streptomyces cinereospinus NBRC 15397T (26.3 ± 2.1%), coeruleofuscus NBRC 12757T (49.5 ± 2.9%), and Streptomyces mexicanus NBRC 100915T (49.6 ± 2.5%) were notably below 70%, the threshold value for the delineation of genomic species [56]. Polyphasic characterizations results supported the observation that isolate Streptomyces sp. MUSC 135T represented a novel species, proposed as “Streptomyces pluripotens sp. nov.,” [62] that exhibited merits in producing antimicrobial secondary metabolites.
3.4. Antibacterial Potential of Isolates
The 87 isolates were preceded to preliminary screening for antimicrobial activity against 12 pathogens by using the cross streak method [31]. Of 87 isolates, 48 isolates (55.2%) exhibited inhibitory against at least 1 pathogen used in this study. Of 48 isolates, 20.8% (n = 10) exhibited inhibitory activity against Gram negative bacteria and 2.1% (n = 1) inhibited Gram positive bacteria. The remaining 77.1% (n = 37) showed excellent inhibition towards both Gram negative and Gram positive bacteria.
3.5. Antimicrobial Activity of Culture Extracts from Mangrove Actinobacteria Isolates
The 87 Actinobacteria isolates were subjected to subsequent investigation by antimicrobial screening of their secondary metabolites. The secondary metabolites of these isolates were tested for antimicrobial activities against the same group of 12 pathogenic bacteria used during preliminary screening. Of 87 isolates, 9 isolates (10.3%) which belonged to the genus Streptomyces were exhibiting activity against at least one of the pathogenic bacteria tested. Of the 9 isolates, 55.6% (n = 5) were active against more than 1 pathogen. Two isolates (MUSC 14 and MUSC 16) were inhibiting 2 pathogens. Three isolates (MUSC 56, MUSC 135, and MUSC 164) exhibited broad spectrum of antibacterial activity, with MUSC 135 showing the strongest inhibition effects against MRSA (inhibition zone of 12 mm), Bacillus cereus (4 mm), Acinetobacter calcoaceticus (4 mm), and Salmonella typhi (4 mm) in this study (Table 2). The strong inhibitory activities of these Actinobacteria isolates towards an array of pathogens are a good indicator that these isolates could be the potential candidates for production of highly valuable bioactive secondary metabolites. Some of the bioactive metabolites are currently being chemically analyzed to identify the novelty of the active compounds.
The identification of Streptomyces as the most bioactive genus in this study is in line with other researchers [1] as Streptomyces has the ability to catabolize a wide range of compounds and produce secondary metabolites with diverse biological activities and chemical structure [4]. Streptomyces is the largest genus of the Actinobacteria and over two-thirds of all natural antibiotics are derived from this group of bacteria [5]. Streptomyces has a huge biosynthetic potential that remains unchallenged among other microbial groups. This is proven as some Streptomyces species, whose biosynthetic repertoire was considered to be only three to five secondary metabolites, in fact possess more than 20 genomic regions encoding known or predicted biosynthetic pathways [63, 64].
Total of 10% (9/87) of Actinobacteria isolates exhibited antimicrobial properties in this study. These results indicated that Actinobacteria are still capable of producing highly bioactive metabolites and continued to provide high quality biological material for drug discovery. Nevertheless, to increase the chances of discovering bioactive isolates in future study, new targets could be used to detect more activities such as antimicrobial and cytotoxic activities [65], as the discovery of novel microbial natural products is encouraged not only by the quality of biological material but also by the innovation and sensitivity of the screening models used [1].
3.6. Detection of PKS and NRPS Genes in Actinobacteria Isolates
Many Actinobacterial isolates from marine environments contain polyketide synthetase (PKS) and nonribosomal polyketide synthetase (NRPS) pathways, the characteristics of secondary metabolite production [16]. In this study, eighty-seven isolates were screened for the presence of PKS-I, PKS-II, and NRPS gene sequences by specific amplification of chromosomal DNA with primer sets K1F-M6R, KSα-KSβ, and A3F-A7R (Table 1). The amplification products were examined by 1% agarose gel electrophoresis, and bands of 1200 to 1400 bp, 600 bp, and 700 to 800 bp were identified as products of PKS-I, PKS-II, and NRPS genes, respectively (Table 1). Of 87 isolates tested for the presence of PKS and NRPS genes, 52 isolates (59.7%) exhibited at least one type of biosynthetic sequences. PKS-II genes were the most frequent as it is detected in 42.5% (n = 37) of the isolates. PKS-I genes were detected in 19.5% of the isolates (n = 17), and NRPS genes were exhibited by only 5 isolates (5.7%). The only isolate that contained all of the biosynthetic genes (PKS-I, PKS-II, and NRPS) used is MUSC 134, a Nocardia species that is closely related to Nocardia Africana at 99.8% similarity. Isolates MUSC 119, MUSC 192, and MUSC 249 contained both of the PKS-I and PKS-II genes, whereas isolates MUSC 81 and MUSC 152 showed positive amplification of both PKS-I and NRPS genes products.
The percentage of bioactive isolates (10.3%) detected in this study is relatively low as compared to the percentage of isolates with at least one type of biosynthetic sequences (59.7%). This showed that the fermentation media (FM3) used may not be able to provide the nutrients and conditions needed to stimulate secondary metabolite production in many of the mangrove Actinobacteria isolates in this study. Therefore, the PCR detection of polyketide synthase types I and II and the nonribosomal peptide synthetase genes is vital to discover the potential of these isolates to produce valuable secondary metabolites. Some of the secondary metabolite biosynthetic gene clusters appear to be nonactive or silent under standard culture conditions and they may require some specific triggers to be activated [66]; these data exhibited that the nonbioactive isolates hold the genetic capability to produce useful secondary metabolites if they were cultivated under the appropriate conditions. As rare taxa such as the Nocardioides (isolate MUSC 201T) showed biosynthetic potential by carrying the PKS-II genes, shedding light on the possibility to serve as a source of natural products, but even this genus has not been shown to produce any natural products yet.
The absence of amplification products from some of the isolates may reveal the lack of PKS-I, PKS-II, and NRPS genes. However, the negative results could be caused by the types of degenerate primers used, which were not suitable for amplifying these genes [67], especially for the low detection rate of NRPS genes in this study. Moreover, it is reported that not all of the NRPS genes are involved in the biosynthesis of bioactive secondary metabolites, as these genes could be responsible in functionality of the bacteria such as quorum sensing [68] or iron metabolism [30]. The high rate of detection of PKS-I and PKS-II genes in the isolates tested was mostly Streptomyces, providing strong evidence for the high potential of Streptomyces to produce high number of biologically active metabolites. Therefore, the molecular screening of Actinobacteria isolates for genes encoding biosynthesis of bioactive compounds is still an effective and valuable approach for preselecting isolates for useful secondary metabolites production [29, 67, 69–73].
4. Conclusions
In conclusion, this study performed a comprehensive investigation into the diversity, phylogeny, and biosynthetic and antimicrobial potential of Actinobacteria isolated from tropical mangrove sediments in east coast of Peninsular Malaysia. A substantial diversity of Actinobacteria isolates was isolated from mangrove habitats, and it is apparent that isolates from genera such as Sinomonas, Microbacterium, and Streptomyces could merit novel species status. Many of the Actinobacteria isolates are producing bioactive secondary metabolites or possess detectable biosynthetic genes, indicating that mangrove habitats are a valuable source of discovery for novel Actinobacteria with promising potential to produce highly bioactive antimicrobial metabolites that could have important value in drug discovery programs.
Acknowledgments
This work was supported by the High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant no. H-50001-A000027) of University of Malaya awarded to Dr. Chan Kok-Gan, also MOHE FRGS Grant (Vote no. 2500110), and External Industry Grant (Vote no. GBA-808138) awarded to Dr. Learn-Han Lee.
Conflict of Interests
The authors declare that they have no conflict of interests.
References
- 1.Hong K, Gao A, Xie Q, et al. Correction: hong, K. et al. Actinomycetes for Marine Drug discovery isolated from mangrove soils and plants in China (Marine Drugs (2009) 7, (24–44)) Marine Drugs. 2009;7(4):495–496. doi: 10.3390/md7010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goshi K, Uchida T, Lezhava A, et al. Cloning and analysis of the telomere and terminal inverted repeat of the linear chromosome of Streptomyces griseus. Journal of Bacteriology. 2002;184(12):3411–3415. doi: 10.1128/JB.184.12.3411-3415.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo PCY, Lau SKP, Huang Y, Yuen K-Y. Genomic evidence for antibiotic resistance genes of actinomycetes as origins of antibiotic resistance genes in pathogenic bacteria simply because actinomycetes are more ancestral than pathogenic bacteria. Medical Hypotheses. 2006;67(6):1297–1304. doi: 10.1016/j.mehy.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 4.Arasu MV, Duraipandiyan V, Agastian P, Ignacimuthu S. Antimicrobial activity of Streptomyces spp. ERI-26 recovered from Western Ghats of Tamil Nadu. Journal de Mycologie Medicale. 2008;18(3):147–153. [Google Scholar]
- 5.Bérdy J. Bioactive microbial metabolites. Journal of Antibiotics. 2005;58(4):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 6.Fenical W, Baden D, Burg M, et al. From Monsoons to Microbes: Understanding the Ocean’s Role in Human Health. Washington, DC, USA: National Academy Press; 1999. [PubMed] [Google Scholar]
- 7.Lam KS. New aspects of natural products in drug discovery. Trends in Microbiology. 2007;15(6):279–289. doi: 10.1016/j.tim.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. Journal of Natural Products. 2007;70(3):461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 9.Jennerjahn TC, Ittekkot V. Relevance of mangroves for the production and deposition of organic matter along tropical continental margins. Naturwissenschaften. 2002;89(1):23–30. doi: 10.1007/s00114-001-0283-x. [DOI] [PubMed] [Google Scholar]
- 10.Yan B, Hong K, Yu Z. Archaeal communities in mangrove soil characterized by 16S rRNA gene clones. Journal of Microbiology. 2006;44(5):566–571. [PubMed] [Google Scholar]
- 11.Xu J, Wang Y, Xie S-J, Xiao J, Ruan J-S. Streptomyces xiamenensis sp. nov., isolated from mangrove sediment. International Journal of Systematic and Evolutionary Microbiology. 2009;59(3):472–476. doi: 10.1099/ijs.0.000497-0. [DOI] [PubMed] [Google Scholar]
- 12.Long H, Xiang W, Zhuang T, Lin P. Microorganism resource of mangrove ecosystems. Chinese Journal of Ecology. 2005;24(6):696–702. [Google Scholar]
- 13.Han L, Huang XS, Sattler I, Fu HZ, Grabley S, Lin WH. Two new constituents from mangrove Bruguiera gymnorrhiza . Journal of Asian Natural Products Research. 2007;9(3–5):327–331. doi: 10.1080/10286020600727574. [DOI] [PubMed] [Google Scholar]
- 14.Ara I, Kudo T, Matsumoto A, Takahashi Y, Omura S. Nonomuraea maheshkhaliensis sp. nov., a novel actinomycete isolated from mangrove rhizosphere mud. The Journal of General and Applied Microbiology. 2007;53(3):159–166. doi: 10.2323/jgam.53.159. [DOI] [PubMed] [Google Scholar]
- 15.Xie XC, Mei WL, Zhao YX, Hong K, Dai HF. A new degraded sesquiterpene from marine actinomycete Streptomyces sp. 0616208. Chinese Chemical Letters. 2006;17(11):1463–1465. [Google Scholar]
- 16.Salomon CE, Magarvey NA, Sherman DH. Merging the potential of microbial genetics with biological and chemical diversity: an even brighter future for marine natural product drug discovery. Natural Product Reports. 2004;21(1):105–121. doi: 10.1039/b301384g. [DOI] [PubMed] [Google Scholar]
- 17.Pisano AM, Sommer JM, Lopez MM. Application of pretreatments for the isolation of bioactive actinomycetes from marine sediments. Applied Microbiology and Biotechnology. 1986;25(3):285–288. [Google Scholar]
- 18.Takahashi Y, Matsumoto A, Seino A, Iwai Y, Omura S. Rare actinomycetes isolated from desert soils. Actinomycetologica. 1996;10(2):91–97. [Google Scholar]
- 19.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. International Journal of Systemic Bacteriology. 1966;16(3):313–340. [Google Scholar]
- 20.Küster E, Williams ST. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- 21.Atlas RM. Handbook of Microbiological Media. Boca Raton, Fla, USA: CRC Press; 1993. [Google Scholar]
- 22.MacFaddin JF. Biochemical Tests for Identification of Medical Bacteria. 3rd edition. Baltimore, Md, USA: Lippincott Williams & Wilkins; 2000. [Google Scholar]
- 23.Williams ST, Davies FL. Use of antibiotics for selective isolation and enumeration of actinomycetes in soi. Journal of General Microbiology. 1965;38(2):251–261. doi: 10.1099/00221287-38-2-251. [DOI] [PubMed] [Google Scholar]
- 24.Kelly KL. Inter-Society Color Council–National Bureau of Standards Color Name Charts Illustrated with Centroid Colors. Washington, DC, USA: US Government Printing Office; 1964. [Google Scholar]
- 25.Cerny G. Studies on the aminopeptidase test for the distinction of gram negative from gram positive bacteria. European Journal of Applied Microbiology. 1978;5(2):113–122. [Google Scholar]
- 26.Carrillo PG, Mardaraz C, Pitta-Alvarez SI, Giulietti AM. Isolation and selection of biosurfactant-producing bacteria. World Journal of Microbiology and Biotechnology. 1996;12(1):82–84. doi: 10.1007/BF00327807. [DOI] [PubMed] [Google Scholar]
- 27.Lemos ML, Toranzo AE, Barja JL. Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microbial Ecology. 1985;11(2):149–163. doi: 10.1007/BF02010487. [DOI] [PubMed] [Google Scholar]
- 28.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. Chichester, UK: Wiley; 1991. pp. 115–175. [Google Scholar]
- 29.Ginolhac A, Jarrin C, Gillet B, et al. Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Applied and Environmental Microbiology. 2004;70(9):5522–5527. doi: 10.1128/AEM.70.9.5522-5527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiology and Molecular Biology Reviews. 2002;66(2):223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakur D, Yadav A, Gogoi BK, Bora TC. Isolation and screening of Streptomyces in soil of protected forest areas from the states of Assam and Tripura, India, for antimicribial metabolites. Journal de Mycologie Medicale. 2007;17(4):242–249. [Google Scholar]
- 32.Garcia GD, Romero MF, Perez BJ, T Garcia D. Thiodepsipeptide isolated from a marine actinomycete WO9527730. Patent Number. 1999;(US5681813)
- 33.Grammer A. Antibiotic sensitivity and assay test. In: Collins CH, Lyne PN, editors. Microbiological Methods. London, UK: Butterworths; 1976. p. p. 235. [Google Scholar]
- 34.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim O, Cho Y, Lee K, et al. Introducing EzTaxon-e: a prokaryotic 16s rRNA gene sequence database with phylotypes that represent uncultured species. International Journal of Systematic and Evolutionary Microbiology. 2012;62(3):716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 36.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–789. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 39.Kimura MA. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 40.Cashion P, Holder-Franklin MA, McCully J. A rapid method for the base ratio determination of bacterial DNA. Analytical Biochemistry. 1977;81(2):461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- 41.de Ley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. European Journal of Biochemistry. 1970;12(1):133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 42.Huss VAR, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Systematic and Applied Microbiology. 1983;4(2):184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- 43.Meena B, Rajan LA, Vinithkumar NV, Kirubagaran R. Novel marine actinobacteria from emerald Andaman & Nicobar Islands: a prospective source for industrial and pharmaceutical byproducts. BMC Microbiology. 2013;13(1, article 145) doi: 10.1186/1471-2180-13-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho SH, Han J-H, Ko H-Y, Kim SB. Streptacidiphilus anmyonensis sp. nov., Streptacidiphilus rugosus sp. nov. and Streptacidiphilus melanogenes sp. nov., acidophilic actinobacteria isolated from Pinus soils. International Journal of Systematic and Evolutionary Microbiology. 2008;58(7):1566–1570. doi: 10.1099/ijs.0.65480-0. [DOI] [PubMed] [Google Scholar]
- 45.Huang Y, Cui Q, Wang L, et al. Streptacidiphilus jiangxiensis sp. nov., a novel actinomycete isolated from acidic rhizosphere soil in China. Antonie van Leeuwenhoek. 2004;86(2):159–165. doi: 10.1023/B:ANTO.0000036124.18820.ae. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Huang Y, Liu Z, Goodfellow M, Rodríguez C. Streptacidiphilus oryzae sp. nov., an actinomycete isolated from rice-field soil in Thailand. International Journal of Systematic and Evolutionary Microbiology. 2006;56(6):1257–1261. doi: 10.1099/ijs.0.64165-0. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y, Wei W, Wang X, Lai R. Proposal of Sinomonas flava gen. nov., sp. nov., and description of Sinomonas atrocyanea comb. nov. to accommodate Arthrobacter atrocyaneus . International Journal of Systematic and Evolutionary Microbiology. 2009;59(2):259–263. doi: 10.1099/ijs.0.000695-0. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Y, Chen XY, Zhang Y, Wang W, Xu J. Description of Sinomonas soli sp. nov., reclassification of Arthrobacter echigonensis and Arthrobacter albidus (Ding et al. 2009) as Sinomonas echigonensis comb. nov. and Sinomonas albida comb. nov., respectively, and emended description of the genus Sinomonas . International Journal of Systematic and Evolutionary Microbiology. 2012;62(2, part 4):764–769. doi: 10.1099/ijs.0.030361-0. [DOI] [PubMed] [Google Scholar]
- 49.Ding L, Hirose T, Yokota A. Four novel Arthrobacter species isolated from filtration substrate. International Journal of Systematic and Evolutionary Microbiology. 2009;59(4):856–862. doi: 10.1099/ijs.0.65301-0. [DOI] [PubMed] [Google Scholar]
- 50.Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by the genus Streptomyces? Archives of Microbiology. 2001;176(5):386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 51.Bentley SD, Chater KF, Cerdeño-Tárraga A-M, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417(6885):141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 52.Nimnoi P, Pongsilp N, Lumyong S. Endophytic actinomycetes isolated from Aquilaria crassna Pierre ex Lec and screening of plant growth promoters production. World Journal of Microbiology and Biotechnology. 2010;26(2):193–203. [Google Scholar]
- 53.Lee L-H, Cheah Y-K, Sidik SM, et al. Molecular characterization of Antarctic actinobacteria and screening for antimicrobial metabolite production. World Journal of Microbiology and Biotechnology. 2012;28(5):2125–2137. doi: 10.1007/s11274-012-1018-1. [DOI] [PubMed] [Google Scholar]
- 54.Cho CH, Lee J-S, An D-S, Whon TW, Kim S-G. Nocardioides panacisoli sp. nov., isolated from the soil of a ginseng field. International Journal of Systematic and Evolutionary Microbiology. 2010;60(2):387–392. doi: 10.1099/ijs.0.012690-0. [DOI] [PubMed] [Google Scholar]
- 55.Lee LH, Zainal N, Azman AS, Mutalib NS, Hong K, Chan KG. Mumia flava gen. nov., an actinobacterium of the family Nocardioidaceae . International Journal of Systematic and Evolutionary Microbiology. 2014;64(5):1461–1467. doi: 10.1099/ijs.0.058701-0. [DOI] [PubMed] [Google Scholar]
- 56.Wayne LG, Brenner DJ, Colwell RR, et al. Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. International Journal of Systematic Bacteriology. 1987;37(4):463–464. [Google Scholar]
- 57.Lee LH, Azman AS, Zainal N, et al. Microbacterium mangrovi sp. nov., an amylolytic actinobacterium isolated from Tanjung Lumpur mangrove forest. International Journal of Systematic and Evolutionary Microbiology. 2014 doi: 10.1099/ijs.0.062414-0. [DOI] [PubMed] [Google Scholar]
- 58.Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic Bacteriology. 1994;44(4):846–849. [Google Scholar]
- 59.Bull AT, Stach JEM, Ward AC, Goodfellow M. Marine actinobacteria: perspectives, challenges, future directions. Antonie van Leeuwenhoek. 2005;87(1):65–79. doi: 10.1007/s10482-004-6562-8. [DOI] [PubMed] [Google Scholar]
- 60.Bull AT, Stach JEM. Marine actinobacteria: new opportunities for natural product search and discovery. Trends in Microbiology. 2007;15(11):491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Bredholt H, Fjærvik E, Johnsen G, Zotchev SB. Actinomycetes from sediments in the Trondheim fjord, Norway: diversity and biological activity. Marine Drugs. 2008;6(1):12–24. [PMC free article] [PubMed] [Google Scholar]
- 62.Lee LH, Zainal N, Azman AS, et al. Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete inhibiting methicillin-resistant Staphylococcus aureus . International Journal of Systematic and Evolutionary Microbiology. 2014 doi: 10.1099/ijs.0.065045-0. [DOI] [PubMed] [Google Scholar]
- 63.Omura S, Ikeda H, Ishikawa J, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(21):12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Natural Product Reports. 2009;26(11):1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams PG. Panning for chemical gold: marine bacteria as a source of new therapeutics. Trends in Biotechnology. 2009;27(1):45–52. doi: 10.1016/j.tibtech.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Janso JE, Carter GT. Biosynthetic potential of phylogenetically unique endophytic actinomycetes from tropical plants. Applied and Environmental Microbiology. 2010;76(13):4377–4386. doi: 10.1128/AEM.02959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qin S, Li J, Chen H-H, et al. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna China. Applied and Environmental Microbiology. 2009;75(19):6176–6186. doi: 10.1128/AEM.01034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annual Review of Microbiology. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 69.Courtois S, Cappellano CM, Ball M, et al. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Applied and Environmental Microbiology. 2003;69(1):49–55. doi: 10.1128/AEM.69.1.49-55.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hornung A, Bertazzo M, Dziarnowski A, et al. A genomic screening approach to the structure-guided identification of drug candidates from natural sources. ChemBioChem. 2007;8(7):757–766. doi: 10.1002/cbic.200600375. [DOI] [PubMed] [Google Scholar]
- 71.Metsä-Ketelä M, Salo V, Halo L, et al. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiology Letters. 1999;180(1):1–6. doi: 10.1111/j.1574-6968.1999.tb08770.x. [DOI] [PubMed] [Google Scholar]
- 72.Schneemann I, Nagel K, Kajahn I, Labes A, Wiese J, Imhoff JF. Comprehensive investigation of marine actinobacteria associated with the sponge Halichondria panicea . Applied and Environmental Microbiology. 2010;76(11):3702–3714. doi: 10.1128/AEM.00780-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microbial Ecology. 2005;49(1):10–24. doi: 10.1007/s00248-004-0249-6. [DOI] [PubMed] [Google Scholar]


