Abstract
Endogenously and externally generated mechanical forces influence diverse cellular activities, a phenomenon defined as mechanotransduction. Deformation of protein domains by application of stress, previously documented to alter macromolecular interactions in vitro, could mediate these effects. We engineered a photon-emitting system responsive to unfolding of two repeat domains of the actin filament (F-actin) crosslinker protein filamin A (FLNA) that binds multiple partners involved in cell signaling reactions and validated the system using F-actin networks subjected to myosin-based contraction. Expressed in cultured cells, the sensor-containing FLNA construct reproducibly reported FLNA domain unfolding strikingly localized to dynamic, actively protruding, leading cell edges. The unfolding signal depends upon coherence of F-actin-FLNA networks and is enhanced by stimulating cell contractility. The results establish protein domain distortion as a bona fide mechanism for mechanotransduction in vivo.
Introduction
The actin cytoskeleton provides shape and mechanical stability to cells, and bidirectional signals between the cytoskeleton and membrane receptors and adhesion proteins regulate cell shape and movement. While chemical signals downstream of receptor ligation, such as ATP hydrolysis and phosphorylation can mediate cytoskeletal rearrangements, force-dependent conformational changes of load-bearing molecules can generate mechanotransduction, where mechanical forces are converted to biochemical signals 1, 2, 3, 4, 5, 6. However, little information is available concerning the spatio-temporal regulation of such molecules in cells primarily due to a limitation of reagents capable of detecting force-induced conformational change in situ 6. Obtaining such information is potentially important, because mechanical strain has medical consequences such as pathological vascular and cardiac hypertrophy 7.
Filamin A (FLNA), the most widely expressed and abundant member of the filamin family of F-actin cross-linking proteins, is a critical mechanosensing element 8, 9 that is essential for mechanoprotection, stretch-activated channel regulation, and the integration of distant parts of the cell to maintain the mechanical coherence of the cytoplasm 10, 11, 12. FLNA is a homodimer of elongated subunits, each composed of an N-terminal actin-binding domain (ABD) followed by 24 Ig-like repeats segmented into 2 rod domains by ‘hinges” (Fig. 1a) 13, 14. The second rod domain (repeats 16–23 or rod-2) contains the majority of binding sites for numerous and diverse cellular partners including signaling molecules and plasma membrane embedded proteins 13, 15. Structural studies of rod-2 have revealed that its partner-binding sites on repeats 19 and 21 are hidden by a β sheet designated strand A of repeats 18 and 20, respectively. In contrast to rod 1, the rod 2 Ig repeats overlap to form a more compact structure 16, 17, 18, 19, 20. This architecture provides a basis for physiologically relevant forces to induce a conformational change in FLNA rod-2 that can expose these cryptic binding sites, particular the integrin binding site on repeat 21 8, 9, 15, 21. In vitro studies have experimentally documented such changes 8, 21.
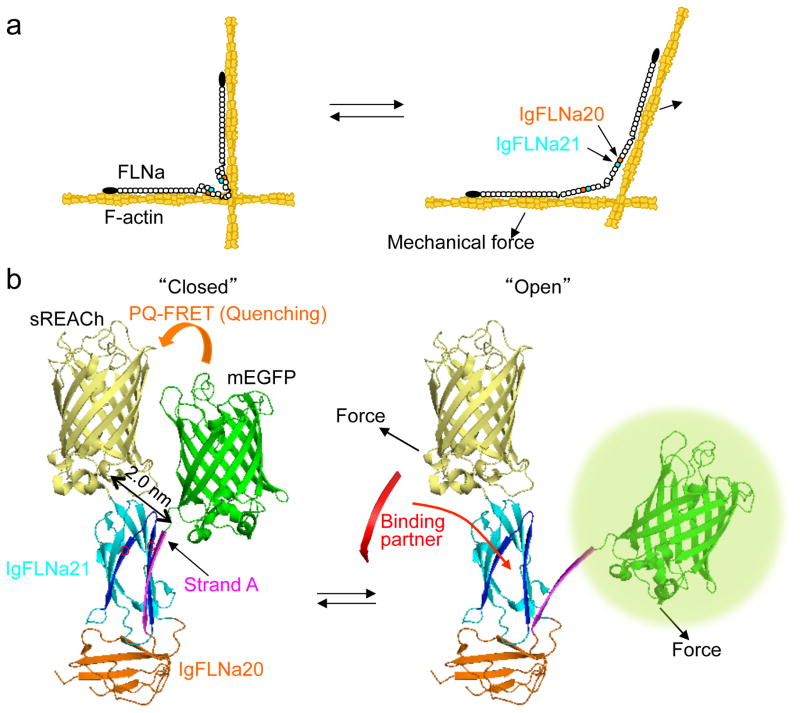
Figure 1. The FLNA repeat 21-conformation sensor.
(a) Model of FLNA crosslinking F-actin showing how its molecular conformation is altered by mechanical forces. FLNA interacts with F-actin through its N-terminal ABD and repeat 10 of rod-1 segment. This posits the rod-2 domain and the self-association site at the vertex of the crosslink where actin filament displacement can linearize the rod 2 domain. (b) Structure of FLNA repeats 20–21 (Protein Data Bank 2J3S) showing how strand A of repeat 20 auto-inhibits the partner binding site on repeat 21 and how mechanical force displaces repeat 20 strand A to expose the 21 partner binding site. The hypothetical location of mEGFP (donor) and sREACh (quencher) on the open and closed conformation of repeats 20–21 is shown. In the non-stressed state (left), the two fluorophores are in close proximity and the fluorescence of mEGFP is quenched by sREACh (or lifetime of mEGFP is shortened). When force opens the cryptic binding site (right), the probes move apart (PQ-FRET efficiency decreases and the fluorescence intensity of mEGFP increases or its lifetime becomes longer).
Here we engineered a novel photoquenching fluorescence resonance energy transfer (PQ-FRET)-based FLNA sensor cassette that reports on this conformational change in cells (Fig. 1b). We demonstrate that full-length FLNA containing the FRET pair cassette is responsive to contractile force exerted by myosin embedded in actin networks in vitro. In living cells, time-lapse ratio imaging reveals the FLNA sensor reports both spatial and temporal changes in FLNA conformation. Moreover, we show that ligands for FLNA repeat 21 induce molecular unfolding of that domain both in vitro and in vivo. These data confirm that FLNA-mediated mechanotransduction occurs in motile cells.
RESULTS
Construction and characterization of FLNA PQ-FRET probes
To detect FLNA’s conformation changes in cells, we constructed and inserted into FLNA’s rod-2 segment a genetically encoded PQ-FRET-based sensor. This sensor was composed of monomeric enhanced GFP (mEGFP) and a FRET fluorescence quencher, sREACh (Fig. 1) that were separated by FLNA Ig-repeats 20 and 21. We reasoned that “opening” or dissociation of Ig-repeat 20’s strand A from its binding position on Ig-repeat 21 would increase the fluorescence emission of the sensor by moving sREACh away from mEGFP (Fig. 1b). sREACh is a nonfluorescent yellow fluorescent protein (YFP) mutant with high absorbance that well overlaps with the emitted fluorescence of mEGFP, making sREACh an efficient quencher (Supplementary Figs. 1 and 2) 22, 23.
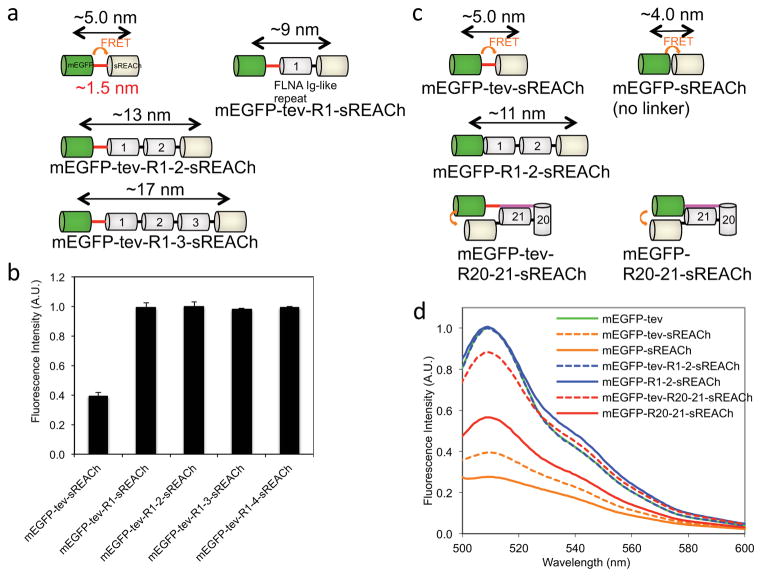
To test this idea, we expressed, purified and assessed PQ- FRET pair constructs mEGFP and sREACH separated by various FLNA Ig repeats. Attachment of sREACh to the C-terminus of mEGFP diminishes the fluorescence emission of mEGFP by 73 ± 3% (Fig. 2d) while the insertion of one or more linear FLNA repeats between this FRET pair prevented quenching (Fig. 2b). Critically, an insertion of FLNA repeats 20–21 between the sensor pair reconstituted quenching: mEGFP fluorescence intensity was diminished by 44 ± 5%, indicating that the 20–21 repeats position their ends and hence, the probes near one another (Figs. 1 and 2d). We next added a TEV protease site after mEGFP to test if cleavage could relieve quenching (Fig. 2 and Supplementary Fig. 1). Insertion of the cleavage site after mEGFP in the PQ-FRET probes slightly diminished the ability of the sREACh to quench, presumably by increasing the gap between them. However, mEGFP fluorescence further increased after cleavage of this linker sequence (Supplementary Fig. 1). These data demonstrate that the PQ-FRET cassette linked with repeats 20–21 can detect conformational changes while those linked with repeats 1–2, or lacking a linker, can serve as control standards that emit minimum or maximum mEGFP fluorescence, respectively. These sensor cassettes were Ca2+ insensitive and responded proportionally to pH, validating them as intracellular sensors for different ionic environments (Supplementary Fig. 3). Control probes with repeats 1–2, or without a linker, when they are embedded into full-length FLNA molecule, showed no change in FRET in vitro and in cells as described below.
Figure 2. Effect of insertions between mEGFP and sREACh on their FRET.
(a and c) Schematic representation of fluorescence protein probes tested in this experiments. TEV protease recognition sequence (ENLYFQ) is indicated in red line. The strand A of repeat 20 is indicated in magenta. (b) Addition of a single FLNA Ig repeat between mEGFP and sREACh is sufficient to obliterate their FRET. Excitation was set at 458 nm and emissions of mEGFP were measured at 511 nm. Error bars represent SD (n=3). (d) Removal of TEV protease recognition site (tev) increases FRET efficiency of mEGFP-tev-sREACh and EGFP-tev-R20–21-sREACh, but not that of mEGFP-tev-R1–2-sREACh. The emission spectrum is shown for excitation at 458 nm. Fluorescence intensities in panels b and d were normalized to protein concentration and are given in arbitrary units (AU).
FLNA binds F-actin through a high affinity actin-binding domain and a low affinity rod-1 segment (repeats 1–15) allowing force imposed on F-actin networks to concentrate at the rod-2 segment (Fig. 1) 15, 16. Guided by this model, we inserted the sensor cassettes at two different locations of full-length FLNA molecules in the hope that force would efficiently transmit to the sensor while preserving the integrity and functionality of native FLNA molecules (Supplementary Fig. 4). For example, the 20–21 sensor cassette was linked between repeat 17 and 22 of FLNA (repeats 18–21 were replaced with mEGFP-20–21-sREACh) because the: 1) probe contains four domains each of which has the similar size to each Ig repeat (two fluorophores and two Ig repeats, Supplementary Fig. 4b) and; 2) N-terminal of repeat 19 is close to C-terminal of repeat 20 (Supplementary Fig. 4c) and this configuration is similar to the probe (Supplementary Fig. 4b). Since this probe responded best to mechanical strain in cells, all sensor cassettes were inserted at this position unless otherwise stated. The FLNA conformation sensor having repeats 20–21 inserted between the FRET pair was designated as FLNA-CS(20–21).
FRET-FLIM imaging of FLNA conformation sensor in living cells
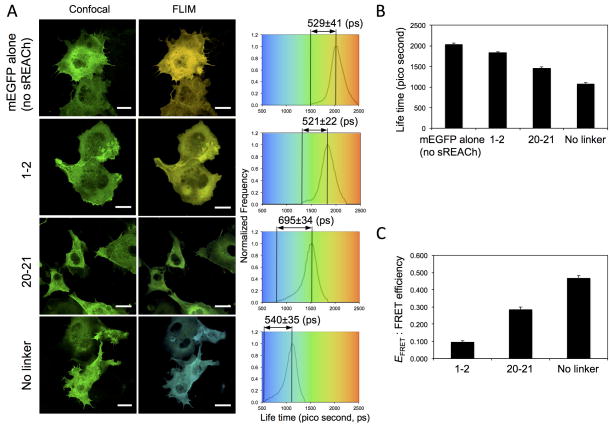
Since FRET shortens the mEGFP donor lifetime in concentration-independent manner 24, we anticipated that fluorescence lifetime imaging microscopy (FLIM) -FRET would be useful to visualize conformational changes of the FLNA probe in living cells. The probes were successfully expressed in COS7 cells and the lifetime of mEGFP changed as expected (Fig. 3). The wider distribution of lifetime between minimum and mean lifetime with FLNA-CS(20–21) suggests that there is more alterations in lifetime of the donor in the confirmation sensitive probe compared to the lifetime of the donor itself and those of the control probes in living cells. However, the long acquisition times (2~3 min) required in FLIM measurements, limit its utility for monitoring dynamic events during cell protrusion and migration.
Figure 3. FLIM-FRET study of FLNA-CS expressed in live COS-7 cells.
(a) COS-7 cells were transfected with pFLAG-FLNA-CS with (repeats 1–2 or 20–21) or without linker between mEGFP and sREACh (Supplementary Fig. 4, without mCherry), or without sREACh and were used to acquire mEGFP intensity (confocal) and lifetime (FLIM) images. Bar=20 μm. The lifetime distributions are shown for representative cells expressing FLNA-CS. The results of the fluorescence lifetime analysis are summarized in panels (b) and (c). Error bars represent SD (n=3). FRET efficiency (EFRET) was calculated from EFRET = 1−τDA/τD where τ is the lifetime of the donor (mEGFP) in the presence of quencher (sREACh) and τD is the average lifetime of the donor in the absence of quencher.
Construction of an intensity-based FLNA PQ-FRET sensor
To capture the rapid dynamics of the biosensor activities in high resolution, we used intensity-based ratiometric FRET 25, 26. The signal from the PQ-FRET probe depends on not only conformational changes of the probe but also its local concentration. Therefore, we attached mCherry to the N- or C-terminal end of FLNA (Supplementary Fig. 4) as an internal control to normalize localized probe concentration because of its excellent stability and ease of quantitative detection without interfering with mEGFP in our microscopic system (Supplementary Fig. 2). The ratio of the two fluorophores (mEGFP/mCherry) thus reflects the liberation of the quenching group or conformational changes of the sensor. These probes retained F-actin gelation activity, the intrinsic fluorescence characteristics of native FLNA (Supplementary Fig. 5), and distributed with endogenous FLNA in cytosol without altering cell morphology (Supplementary Figs. 6 and 7). In addition negligible cross talk (bleed through) occurred between the mEGFP and mCherry signals, and the fluorescence of sREACh is low (Supplementary Figs. 2 and 6), simplifying image analysis (Supplementary Table 1).
Unfolding of purified FLNA-CS(20–21) by myosin in vitro
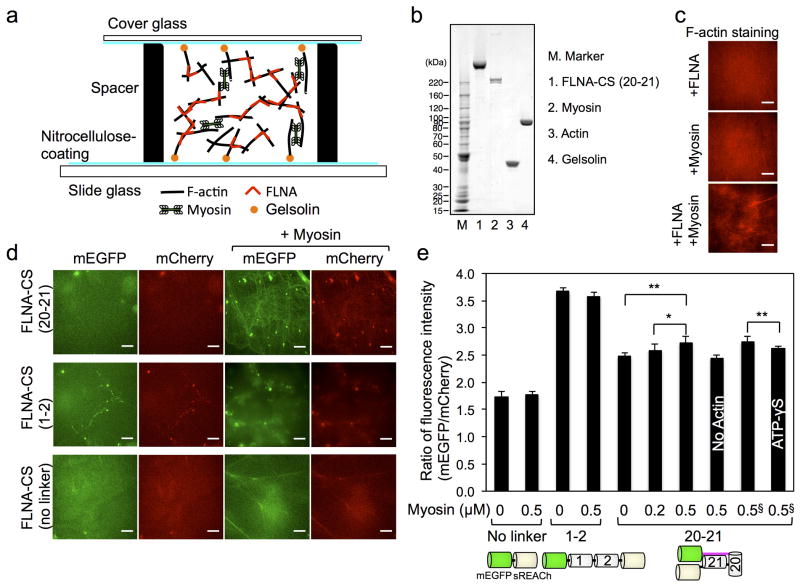
We reconstituted homogeneous actomyosin networks crosslinked by recombinant FLNA-CSs in sealed chambers constructed from a gelsolin-coated coverslip and glass slide (Fig. 4a) 27. We measured fluorescent intensities of mEGFP and mCherry using spinning disk microscopy and plotted their ratio (Fig. 4d, e). The donor fluorescence intensity of FLNA-CS(20–21), embedded in F-actin networks, increased when myosin was added. This result indicates a lowering of donor energy transfer as the FRET pairs move further apart due to increasing myosin-based contractility, whereas control protein constructs were insensitive (Fig. 4e). To inhibit myosin contraction, we used a non-hydrolyzable ATP analogue ATP-γS, because more specific myosin inhibitors interfere with our FRET measurements 28, 29. Replacement of ATP with its non-hydrolyzable analogue reduced donor fluorescent intensity of FLNA-CS(20–21) (Fig. 4e), indicating that the FRET change is not simply due to myosin-mediated actin cross-linking. These results validate the probe as a mechan osensor.
Figure 4. Myosin-dependent conformational changes of purified FLNA-CS in actin networks.
(a) Actin networks crosslinked with FLNA-CS are internally stressed by myosin motors. Actin filaments are anchored to glass surface through gelsolin coated on the surface. (b) Coomassie blue stain of SDS-PAGE gel of purified proteins used in this study. (c) 10 μM F-Actin, crosslinked with 0.1 μM FLNA or not, was internally stressed by 0.5 μM myosin. F-actin is stained with Alexa Fluor® 568 phalloidin. (d) Fluorescence images of mCherry-FLNA-CS (0.1 μM) embedded in actin filaments (10 μM) in the presence or absence of myosin (0.5 μM). Ratio of fluorescence intensities of mCherry and mEGFP was calculated and plotted in panel e. Scale bar = 10μm. (e) Error bars represent SD, n ≥ 4 independent experiments. §These two experiments were independently performed and compared. Statistical significance was determined by a two-tailed t test (*P<0.05, **P<0.005).
Conformational changes of FLNA-CS by a high affinity ligand
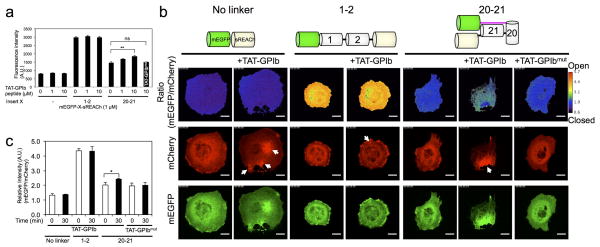
A high affinity FLNA peptide ligand derived from the GPIbα cytoplasmic tail of the von-Willebrand Factor receptor has been reported to bind FLNA’s repeat 21 in the absence of force 19, 21, 30, 31. We therefore determined if this peptide could induce the conformational change in the PQ-FRET sensor in FLNA-CS in vitro and in vivo. As expected, the GPIbα peptide markedly increased the fluorescent intensity of mEGFP linked to sREACh within the repeat 20–21 construct in a dose-dependent manner, whereas a β-integrin peptide emitted a much weaker signal (Supplementary Table 2 and Fig. 8). Time-lapse ratio images (mEGFP/mCherry) using FLNA-CS expressed in COS-7 and HEK-293 cells demonstrated that a cell-permeable GPIbα peptide (TAT-GPIbα) increased the relative fluorescent intensity of mEGFP when FLNA repeats 20–21 were inserted between the quenching pair. In contrast, FLNA-CS linked with repeats 1–2 or lacking a linker were insensitive to TAT-GPIbα peptide, and therefore, used as a standard for maximum and minimum ratio values in the scale, respectively (Fig. 5 and Supplementary Fig. 9a and movies 1 and 2). As a control, we tested a GPIbα peptide mutated at critical amino acid residues for FLNA-binding (Supplementary Table 2). This peptide did not unfold the auto-inhibition site on repeat 21 in vitro or in vivo (Fig. 5).
Figure 5. A cell-permeable high affinity ligand (TAT-GPIbα) for repeat 21 of FLNA induces conformational changes in the FLNA PQ-FRET probe (FLNA-CS) expressed in COS-7 cells.
(a) Effect of increasing amounts of TAT-GPIbα peptide (0 – 10 μM) or TAT-GPIbα mutant peptide (10 μM) on the fluorescence intensity of purified mEGFP-sREACh constructs containing no linker, linked together with FLNA repeats 1–2, or linked together with repeats 20–21 (1.0 μM each). Fluorescence was measured in a microplate reader with excitation/emission wavelengths of 458/511 nm (mEGFP). (b) Fluorescence images of mCherry and mEGFP of FLNA-CS expressed in COS-7 cells before and after treatment with TAT-peptides. Ratio images and Ratio color scale (mEGFP/mCherry) were generated using Ratio Plus plugin (ImageJ). The cells were treated with 100 μM cell-permeable TAT-GPIbα peptide or TAT-GPIbα mutant peptide. Arrows highlight foci induced by the cell-permeable TAT-GPIbα peptide. Note that the ratio values at the foci are not quantitative because the fluorescence intensity was saturated. Scale bar = 20μm. The corresponding movies are shown in Supplementary movie 1. (c) The ratio of fluorescence intensities of mEGFP versus mCherry are plotted before and after treatment with the cell-permeable peptides. Error bars represent SD (n=3). *P<0.05, **P<0.005; ns, not significant by a two-tailed t test.
Spatio-temporal regulation of FLNA-CS unfolding in cells
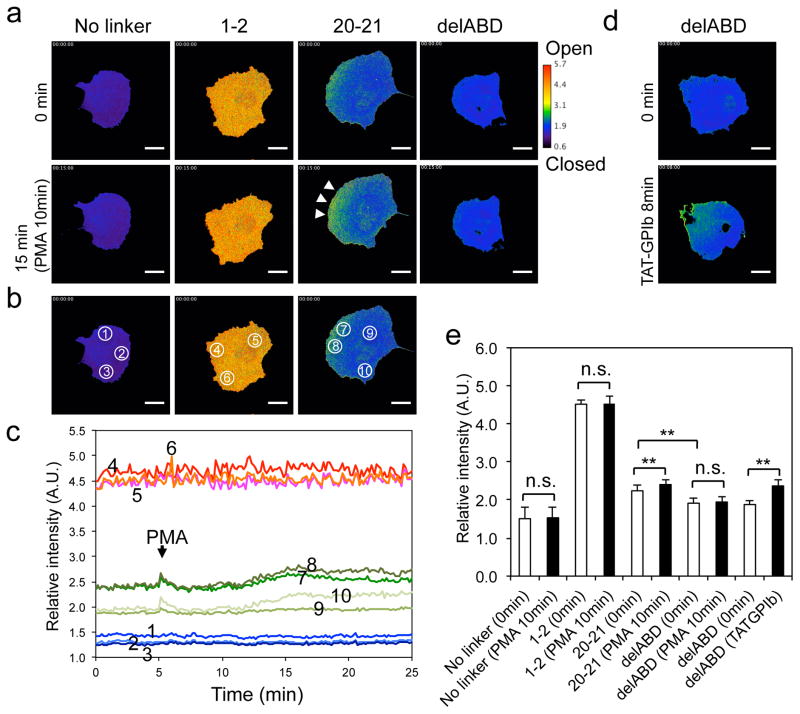
We reacted COS-7 and HEK-293 cells transfected with the three FLNA-CS probes with phorbol 12-myristate 13-acetate (PMA) to induce cell spreading and migration 32, 33 These cell lines were chosen because of their ease of transfection. The large size of COS-7 cells facilitates visualizing the molecular dynamics of a given protein in the cytosol. Since fluorescence intensity decreases when a lower-power objective lens is used, and different lenses may impact on mEGFP/mCherry ratios, we used the same high-power (×60) objective lens throughout our experiments. To monitor multiple cells, we used HEK-293 cells, which are relatively small. We found that the rod-2 domain of FLNA opens in specific cellular locations following cell stimulation with PMA (Fig. 6 and Supplementary movies 3–5). Quantitative measurements of relative intensities in different regions of cells as a function of time revealed that maximal openning of repeat 21 occurs 5~10 min after the addition of PMA and that unfolding occurs predominantly at the cell edge; the relative intensity of the FLNA-CS (20–21) probe increased by 11% at the cell edge (2.40 ± 0.03 to 2.64 ± 0.08, mean ± S. D (circled regions 7 and 8 in Fig. 6b, c). No ratio change was observed in cells transfected with constructs containing no linker or repeats 1–2 between the FRET pair (Fig. 6 and Supplementary Fig. 9 and movies 3 and 4), endorsing the methodology.
Figure 6. Conformational changes in the FLNA-CS(20–21) probe expressed in COS-7 cells require binding to the actin cytoskeleton.
(a) Spatial conformational changes in FLNA-CSs expressed in COS-7 cells following PMA treatment. Scale bar 20 μm. The corresponding movies are shown in Supplementary movie 3. (b–e) Quantification of spatial conformational changes in the FLNA-CSs probe expressed in COS-7 cells. Intensities of different regions (circled in panel b) of the ratio images were quantified and plotted as a function of time (c). COS-7 cells expressing FLNA-CS(20–21) lacking its ABD before and after treatment with cell permeable TAT-GPIbα peptide (d). The corresponding movies are shown in Supplementary movie 6. Average intensities of three independent ratio images (whole cells) before and after PMA stimulation were quantitated and plotted (e). Error bars represent SD (n=3). **P<0.005; ns, not significant by a two-tailed t test.
FLNA-CS(20–21) lacking the ABD, which retains sensitivity to TAT-GPIbα-binding, diffusely localizes in the cytoplasm, and the intensity of its ratio image was lower than that of full-length FLNA-CS(20–21) and was not changed by PMA stimulation (Fig. 6). In elongated cells, unfolded FLNA-CS(20–21) localizes mainly at the cell ends (Supplementary Fig. 10 and Supplementary movie 7). Depolymerization of actin filaments with latrunculin A closed the mechanosensing site of FLNA localized at these regions (Supplementary Fig. 10 and Supplementary movie 7).
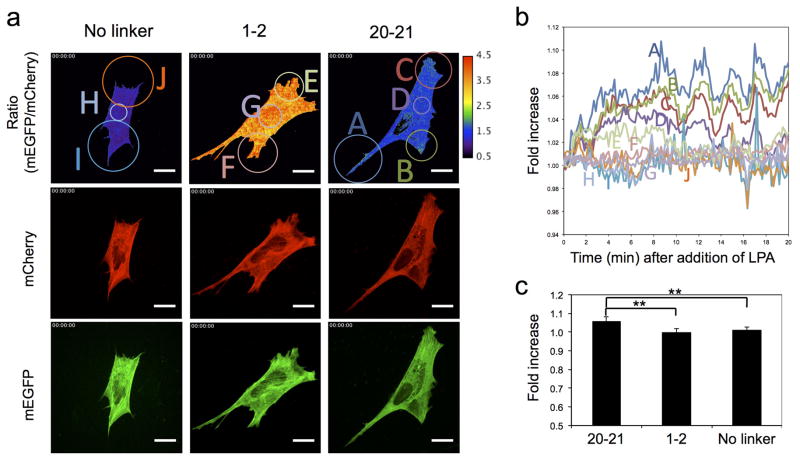
We transfected our probes into mouse embryonic fibroblast (MEF) cells and stimulated them with lysophosphatidic acid (LPA) to activate the well-established signaling pathway to myosin contraction downstream involving Rho/Rho-kinase 34. To optimize transfection efficiency, the plasmid vector was switched, and a different transfection reagent used (see Methods). Consistent with the above results, no ratio change was observed in MEF cells transfected with constructs containing no linker or repeats 1–2 between the FRET pair (Figure 7 and Supplementary movie 7). Upon LPA stimulation, MEF cells transfected with the FLNA-CS(20–21) probe contracted, and the FRET ratio (mEGFP/mCherry) increased. Relative changes of ratio in different regions of cells as a function of time showed that maximal FLNA probe opening of repeat 21 occurred ~10 min after the addition of LPA and that unfolding was restricted to protruding regions where the relative intensity of the FLNA-CS (20–21) probe increased by 6~10% (Figure 7 and Supplementary movie 7). We used the Rho-kinase inhibitor, Y27632, to suppress myosin contraction 35. Pretreatment of MEF cells with Y27632 inhibited LPA-induced opening of FLNA-CS(20–21) (Supplementary Fig. 11 and Supplementary movie 9).
Figure 7. LPA-induced myosin contraction increases conformational changes in the FLNA-CS(20–21) probe expressed in MEF cells.
(a) Spatial conformational changes in FLNA-CSs expressed in MEF cells following LPA (1 μM) treatment. Scale bar 20 μm. The corresponding movies are shown in Supplementary movie 8. (b and c) Quantification of spatial conformational changes in the FLNA-CSs probe expressed in MEF cells stimulated with LPA. Intensities of different regions (circled in panel a) of the ratio images were quantified, normalized (relative to before stimulation), and plotted as a function of time (b). Changes of average intensities of ten independent ratio images before and after (15 min) LPA stimulation were quantitated and plotted (c). Error bars represent SD (n=10). **P<0.005 by a two-tailed t test.
Finally, we investigated spatio-temporal changes of FLNA-CS(20–21) in MEF cells undergoing shape change and migration after starvation and stimulation with serum (Supplementary movie 10). Opening of FLNA-CS(20–21) was observed in the protruding regions of these cells, and opening of FLNA reversed as protrusive activity ceased. Although filamin is bound in stress fibers and mature adhesion sites, molecular opening was not observed in these areas. Maximum probe closing was observed when cells rounded due to lost of adhesion or intracellular contraction (left upper panel in Supplementary movie 10). Although open FLNA has a higher affinity for β-integrin tail than closed molecules 8, 21, we did not observe FLNA accumulation at focal adhesion sites or stress fibers and instead, found a broad distribution of the open probe in actively protruding cytoplasm.
DISCUSSION
Mechanosensing molecules function to transmit and transduce force to regulate key biological processes such as focal adhesion formation, differentiation, growth, and mechanosensing abnormalities and loss of mechanoprotection are implicated in disease processes 36, 37, 38, 39. Since mechanotransduction is mediated by changes in the structure of specific molecules in cells, spatio-temporal information on such conformational changes in cells and tissue is crucial. However, only limited methodology is available to monitor mechanotransduction events in general and the only currently available techniques to accomplish this task in vivo is FRET 40., Furthermore, insufficient structural information concerning the limited number of molecules identified as mechanosensor elements limits construction of useful FRET probes 6, 8, 41, 42, 43. In contrast, much information predicts that mechanotransduction in FLNA is triggered by conformational changes that expose the cryptic binding site of repeat 21. Hence, our FRET pair was designed to unquench as the conformation of FLNA opens and reports positive fluorescence as opposed to conventional FRET where fluorescence would decrease. Therefore, when most of the FLNA molecules are dormant (closed), the signal from the probe is low, alleviating signal to noise complications in detection within a high background. Because of this low background, we were able to detect a ~11% increase of relative intensity of mEGFP in cells expressing FLNA-CS(20–21) after PMA stimulation.
Although a FLNA-based probe has been previously used to reveal mechanotransduction, it has not been applied to cell dynamics and movement 44,45. This probe contains a conventional FRET pair (Cerulean/Venus) inserted at position 1000 in the FLNA molecule. This positions it within repeat 8 and between two F-actin-binding sites (N-terminal ABD and repeat 10 16,46). For this probe to report, repeat 8 must be unfolded. Unfolding of an Ig repeat requires higher forces (>30 pN) than those which open the repeat domain pairing between repeat 20 and 21 (2~5 pN) 21, 47, 48. It is not clear if such repeat unfolding occurs in living cells. In addition, the biological meaning of unfolding repeat 8, if any, has not been documented. The atomic structures of FLNA’s repeats 20–21 are well defined, and force-induced ligand binding to repeat 21 has been demonstrated 8, 17, 21. In this study, using our mechanosensor probe, we documented temporal and spatial conformational changes in this domain in living cells.
Myosin-dependent opening of FLNA-CS(20–21) was observed only in protruding regions of cells, and not in stress fibers or focal adhesions. Opening in focal adhesions was predicted, because the open form of FLNA has a higher affinity for the β-integrin tail 8, 21. Since previous studies showed that FLNA competes with talin and is a negative regulator of integrin activation 49, 50, our findings suggest that open FLNA does not translocate to focal adhesion, but rather binds to integrins in protruding cell regions, which are subsequently delivered to talins at focal adhesion. In addition to β-integrins, FLNA repeat 21 also interacts with migfilin and a large number of possible partners (SEK1, JNK1, etc) whose interactions have been delimited to larger regions of the FLNA molecule that include repeat 21 15, 49, 51, 52.
Since the affinity of β-integrin tail for the CD pocket in repeat 21, when covered with strand A of repeat 20 in an unstressed state, is insufficient to dissociate strand A, force can regulate this interaction. High affinity ligands, such as the GPIbα tail can also bind FLNA’s repeat 21 in the absence of force, and induce a conformational change detected by FLNA-CS(20–21) 19, 21, 31. Hence, PMA-stimulated unfolding of the sensor could be induced by ligand-binding or by force. However, expression of FLNA-CS(20–21) without its N-terminal ABD significantly reduced the basal level of fluorescent signal in cells and made the sensor unresponsive to PMA stimulation. In addition, the conversion of FLNA-CS(20–21) to the open form in MEF cells is LPA-dependent and suppressed with a Rho-kinase inhibitor. These data indicate that ligand binding to repeat 21 requires myosin-mediated contractile force transmitted through the actin cytoskeleton. The loss of signal from labeled FLNA lacking its actin-binding domain further implies that FLNA linked to actin filaments is pre-stressed, although, as suggested previously, its repeat 21 is not fully unfolded 53. Although stress fibers are force-generating and tension-bearing structures 54, FLNA-CS(20–21) was not converted to its open conformation in stress fibers, suggesting that three dimensional orthogonal structure of FLNA/F-actin networks optimizes this conformational change in FLNA.
Whereas FLNA molecules reside throughout the cytoplasm, in particular in F-actin-rich regions, the open FLNA configuration recognized by our probe localized strikingly at the dynamic protruding leading edges of activated cells. This finding is consistent with much known information about the structure of FLNA-F-actin complexes in different circumstances 16, 55. FLNA molecules in complexes composed of low FLNA-actin ratios in vitro (such as those we used to validate activation of the probe under myosin-induced contractile tension) and characteristic of the leading edge of motile cells reside at the vertices of orthogonal cross-linked actin networks (as depicted in Fig. 4). Exposed to force induced by myosin-based F-actin contraction or conceivably hydrostatic pressure 56, such networks would be expected to deform in a manner that would open FLNA’s C-terminal domains. The resulting accessibility of those domains to FLNA’s many binding partners relevant to cell signaling is consistent with the leading cell edge being a region where many if not most intracellular signaling reactions occur. In contrast, at high FLNA-actin ratios in vitro or when the FLNA is bound to F-actin in stress fiber bundles, at the contracted back ends of polarized cells or in dense complexes of cell adhesion structures, FLNA molecules would predictably be compressed, and their C-terminal partner-binding domains closed.
PQ-FRET should be applicable to study any biomaterial capable of signal emission in response to inter- or intra-molecular dissociation 57. It may be useful for high-throughput screening for molecules or drugs that regulate mechanotransduction and protein conformational changes as demonstrated here with the synthetic peptide derived from the GPIbα tail.
In conclusion, using a novel and defined FLNA-based mechanosensing probe, we found that opening of FLNA’s integrin binding site occurs at actively protruding regions of moving cells but not at retracting regions, focal adhesions or stress fibers. These results indicate that mechanical forces exerted in cortical orthogonal actin networks to FLNA recruits signaling molecules to protruding regions.
METHODS
Synthetic peptides and chemicals
Synthetic peptides (Supplementary Table 2) were purchased from Peptide 2.0 (>95% purity) and dialyzed against PBS using Micro Float-A-Lyzer (Spectrumlabs). Other chemicals were purchased from Sigma unless otherwise specified.
Plasmid construction
pFASTBAC-HTb (Invitrogen, San Diego, CA, USA) vector for protein expression in insect cells was modified by insertion of cDNA encoding EGFP, sREACh, and FLNA domains using conventional molecular biology techniques. To engineer pFASTBAC-HTb-mEGFP-tev and pFASTBAC-HTb-mEGFP, cDNA encoding mEGFP was amplified by PCR using pEGFP-C1 (Clontech) mutated at A206K as the template, the forward primer, CATGCCATGGTGAGCAAGGGCGAG, containing a NcoI site, and the reverse primers, CGGGATCCCTGAAAATACAGGTTTTCGATCCCGGCGGCGG, containing a BamHI site and encoding TEV cleavage site (ENLYFQ/G) or CGGGATCCGATCCCGGCGGCGGTC, containing a BamHI site without encoding TEV cleavage site. The amplified fragment was purified, NcoI/BamHI-digested, and ligated into NcoI/BamHI sites in the pFASTBAC-HTb vector. To generate pFASTBAC-HTb-mEGFP-tev--sREACh and pFASTBAC-HTb-mEGFP--sREACh, cDNA encoding sREACh was amplified by PCR using pCAG-sREACh-Actin (Addgene, 23) as the template, the forward primer, CGCGTCGACAAGGGCGAGGAGCTGTTC containing a SalI site, and the reverse primer, GCTCTAGATCAGATCCCGGCGGCGGTC, containing a XbaI site. The amplified fragment was purified, SalI/XbaI-digested, and ligated into SalI/XbaI sites in pFASTBAC-HTb-mEGFP-tev and pFASTBAC-HTb-mEGFP vectors. To engineer pFASTBAC-HTb-mEGFP-tev-sREACh and pFASTBAC-HTb-mEGFP-sREACh, cDNA encoding sREACh was amplified by PCR using pCAG-sREACh-Actin (Addgene) as the template, the forward primer, CGGGATCCGTGAGCAAGGGCGAGGAG, containing BamHI site, and the reverse primer, GCTCTAGATCAGATCCCGGCGGCGGTC, containing a XbaI site. To insert cDNA enconding FLNA Ig domains into pFASTBAC-HTb-mEGFP-tev and pFASTBAC-HTb-mEGFP vectors, FLNA Ig domains were amplified by PCR using primers containing BamHI site (forward primer) and SalI site (reverse primer) and the amplified fragments were ligated into BamHI/SalI sites in the vectors.
pmCherry-FLNA was engineered using pmCherry-C1 (Clontech). Briefly, the N-terminal FLNA domain (1–154 aa) was subcloned into BglII/EcoRI sites of pmCherry-C1 by PCR using pEGFP-FLNA as the template, the forward primers CGGGATCCATGAGTAGCTCCCACTCTC, and the reverse primer GGAATTCAGGGCATGGAGATGGAG. pmCherry-FLNA was generated by inserting a fragment of FLNA cDNA digestested out from pFASTBAC-FLNA vector with SalI and XbaI into pmCherry-FLNA(1–154). pFASTBAC-HTb-mCherry-FLNA was engineered from pFASTBAC-HTb. Briefly, cDNA encoding mCherry was amplified by PCR using pmCherry-C1 as the template, the forward primer, GAAGATCTATGGTGAGCAAGGGCGAGG, containing a BglII site, the reverse primer, CGGGATCCGAGTCCGGACTTGTACAGCTCGTCCA, containing a BamHI site. The amplified fragments were purified, BglII/BamHI-digested, and ligated into BamHI sites in the pFASTBAC-HTb vector to generate pFASTBAC-HTb-mCherry. The N-terminal FLNA domain (1–154 aa) was subcloned into BamHI/EcoRI sites of pFASTBAC-HTb-mCherry by PCR using pEGFP-FLNA as the template, the forward primers CGGGATCCATGAGTAGCTCCCACTCTC, and the reverse primer GGAATTCACGTCACTTGGAAGGGGC. pFASTBAC-HTb-mCherry-FLNA was generated by inserting a fragment of FLNA cDNA digestested out from pFASTBAC-FLNA vector with SalI and XbaI into pFASTBAC-HTb-mCherry-FLNA(1–154). mCherry was also attached at the C-terminal of full-length FLNA as follows. First, cDNA encoding FLNA repeat 24 was amplified by PCR using pFASTBAC-FLNA as the template, the forward primer, CGGGATCCGCTGACGCCAGCAAGGTG, the reverse primer, CTGGAGCTC GGC GGGGCACCACAACGCGG, containing a SacI site (and NotI and XbaI sites). The amplified fragments were purified, EcoNI/SacI-digested, and ligated into EcoNI/SacI sites in the pBluescript II (+)-FLNA(ClaI/XbaI) vector that was generated by subcloning 3′ fragment of FLNA cDNA digestested out from pFASTBAC-FLNA vector with ClaI and XbaI. cDNA encoding mCherry was amplified by PCR using pmCherry-C1 as the template, the forward primer, GAATGCGGCCGCAATGGTGAGCAAGGGCGAGG, containing a NotI site, the reverse primer, GCTCTAGACTACTTGTACAGCTCGTCCATG, containing a XbaI site. The amplified fragments were purified, NotI/XbaI-digested, and ligated into NotI/XbaI sites in the pBluescript II (+)-FLNA(ClaI/XbaI)-vector containing NotI and XbaI sites to generate pBluescript II (+)-FLNA(ClaI/XbaI)-mCherry vector.
To engineer full-length FLNA constructs, FLNA repeats 18–21 were deleted and simultaneously SpeI and AflII sites were introduced between repeats 17 and 22, hinge-1 and repeat 16, or strand A of repeat 16 and repeat 23 using QuickChange® Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) in accordance with the manufacturer’s protocol. Briefly, the following primers were designed CACAGGTGACGACACTAGTCTTAAGGGCGACGCCCGCCG and CGGCGGGCGTCGCCCTTAAGACTAGTGTCGTCACCTGTG, CCAGCAGACTTGGACTAGTCTTAAGGCCCCGGAGAGGC and GCCTCTCCGGGGCCTTAAGACTAGTCCAAGTCTGCTGG, or TGTCAATGGGCTGGATACTAGTCTTAAGGGGGACCCAGGCTT and AAGCCTGGGTCCCCCTTAAGACTAGTATCCAGCCCATTGACA. PCR was done on pBluescript II (+)-FLNA(ClaI/XbaI). To introduce SpeI site at 5′ terminus of mEGFP of pFASTBAC-HTb-mEGFP, the forward primer, CATGCCATGGCAACTAGTATGGTGAGCAAGGGCG, containing a NcoI site was used instead of CATGCCATGGTGAGCAAGGGCGAG described above. Likewise, AflII sites at 3′ terminus of sREACh of pFASTBAC-HTb-mEGFP-tev—sREACh was introduced using the reverse primer, GCTCTAGACTTAAGCCCGGCGGCGGTCACTCT, containing XbaI site instead of GCTCTAGATCAGATCCCGGCGGCGGTC as described above. The cDNA encoding mEGFP-sREACh with or without FLNA Ig repeats were digested with SpeI and AflII and ligated into pBluescript II (+)-FLNA(ClaI/XbaI)-R17-SpeI/AflII-R22, pBluescript II (+)-FLNA(ClaI/XbaI)-H1-SpeI/AflII-R16, or pBluescript II (+)-FLNA(ClaI/XbaI)-R16stA-SpeI/AflII-R23. For some construct, mCherry was attached to the C-terminal of FLNA by inserting EcoNI/XbaI-digested fragment of pBluescript II (+)-FLNA(ClaI/XbaI)-mCherry into these constructs. The FLNA fragment containing the mEGFP-sREACh reporters were digested out with ClaI and XbaI and ligated into pFASTBAC-HTb-mCherry-FLNA, pmCherry-FLNA, and pFLAG-FLNA. Likewise, the mEGFP-sREACh reporters were inserted at position 1000 as previously described 45.
To switch the CMV promoter of pFLAG-FLNA constructs to EF1α promoter, pBApo-EF1α Neo DNA (Takara) was digested with XbaI and calf intestinal alkaline phosphatase and ligated with fragments derived from the pFLAG-FLNA constructs digested with NheI and XbaI.
pFASTBAC-HTb-Halo-mCherry was constructed by PCR. Briefly, cDNA encoding Halo-tag was amplified by PCR using pFN21A HaloTag® CMV Flexi® Vector (Promega) as the template, the forward primer, CATGCCATGGCAGAAATCGGTACTGG, containing a NcoI site, and the reverse primer, GCGGATCCTCCACCGGAAATCTCCAGAGTAGACAGCCAGCGCGCGATC, containing BamHI site. The amplified fragments were purified, NcoI/BamHI-digested, and ligated into NcoI/BamHI sites in the pFASTBAC-HTb vector to generate pFASTBAC-HTb-Halo. Next, cDNA encoding mCherry was amplified by PCR using pmCherry-C1 as the template, the forward primer, GAAGATCTATGGTGAGCAAGGGCGAGG, containing a BglII site, and the reverse primer, CGGGATCCGAGTCCGGACTTGTACAGCTCGTCCA, containing BamHI site. The amplified fragments were purified, BglII/BamHI-digested, and ligated into BamHI sites in the pFASTBAC-HTb-Halo vector to generate pFASTBAC-HTb-Halo-mCherry. To gerate pFASTBAC-HTb-Halo-mCherry-16–23-Avi, the cDNA fragment encoding IgFLNa16–23-Avi was digested out from pFASTBAC-HTb-16–23-Avi using BamHI and HindIII, and ligated into pFASTBAC-HTb-Halo-mCherry digested with BamHI and HindIII. The cDNA encoding the force sensor was inserted into pFASTBAC-HTb-Halo-mCherry-16–23-Avi using BspMI digestion of pBluescript II (+)-FLNA(ClaI/XbaI)-R17-EGFP-sREACh-R22.
Expression and purification of Recombinant protein
Recombinant proteins were expressed using a Baculovirus Expression System (Invitrogen) in sf9 insect cells and purified58. A virus was generated with the BAC-TO-BAC system (Invitrogen). Sf9 cells were cultured in Sf-900 II SFM (Invitrogen) supplemented with 50 units ml−1 penicillin and 50 μg ml−1 streptomycin and used for virus amplification and protein expression. Forty-eight hours post infection, the cells were collected by centrifugation, washed with PBS, and stored at −80°C. The expressed proteins were purified by Ni-NTA affinity and gel filtration chromatography (Superose 12 and Superdex 200, GE Healthcare)58. In brief, the proteins were extracted from the Sf9 cells in lysis solution (20 mM sodium phosphate, pH 8.0, 100 mM NaCl, 20 mM imidazole,1 mM β-mercaptoethanol, 2 mM PMSF, 10 μg ml−1 aprotinin, and 10 μg ml−1 leupeptin) at 4°C. The extracts were centrifuged at 20,000 g for 30 min at 4°C and loaded onto a Ni-NTA column (1 ml; QIAGEN). The column was washed with washing solution I (20 mM sodium phosphate, pH 8.0, 20 mM imidazole, 1 mM β-mercaptoethanol, 300 mM NaCl, 0.1% Triton X-100) followed by washing solution II (20 mM sodium phosphate, pH 8.0, 20 mM imidazole, 100 mM NaCl, 1 mM β-mercaptoethanol) and bound recombinant proteins were eluted with 20 mM sodium phosphate, pH 8.0, 200 mM imidazole, 1 mM β-mercaptoethanol. Purified proteins were concentrated using an Amicon Ultra-15 (Millipore) and gel-filtered on Superose 12 and Superdex 200 (10/300; GE healthcare) columns pre-equilibrated with PBS. Full-length FLNA proteins containing the mEGFP-sREACh reporters were purified by ion-exchange and gel filtration chromatography16. In brief, protein extracts were prepared by suspending the Sf9 cells in a solution containing 10 mM Tris-HCl, pH 8.5, 100 mM NaCl, 1% Triton X-100, 10 mM EGTA, 1 mM EDTA, 5 mM 2-mercaptoethanol, and a mixture of protease inhibitors (10 μg ml−1 each chymostatin, leupeptin, and pepstatin and 1 mM phenylmethylsulfonyl fluoride). The cell lysates were centrifuged at 20,000 × g at 4 °C and the supernatant fluid was loaded onto a HiTrap Q column (5-ml column, GE Healthcare) pre-equilibrated with buffer solution QA8.5 (10 mM Tris-HCl, pH 8.5, 1 mM EDTA, 1 mM EGTA, 5 mM 2-mercaptoethanol, 0.02% Triton X-100). Proteins were eluted with a 100-ml linear gradient of 200–400 mM NaCl in the equilibrating buffer at a flow rate of 4 ml min−1. Fractions containing recombinant FLNA-CS (eluting around 300 mM NaCl) were pooled and diluted 1:2 with QA8.5 solution containing the protease inhibitor mixture. Proteins in the pooled fractions were absorbed again onto a Hi- Trap Q column, and the column was washed with QA6.5 solution (10 mM PIPES-NaOH, pH 6.5, 1 mM EDTA, 1 mM EGTA, 5 mM 2-mercaptoethanol, 0.02% Triton X-100) containing 100 mM NaCl. Recombinant FLNa was eluted with a linear salt gradient (100 ml) from 100 to 400 mM NaCl in buffer QA6.5 at a flow rate of 4 ml min−1. Fractions containing FLNA-CS (eluting around 200 mM NaCl) were pooled and concentrated using Ultrafree-15 (Biomax-50K, Milli- pore). FLNA-CS was further purified by Superose 6 (GE Healthcare) column chromatography with S6 solution (10 mM Tris- HCl, pH 7.4, 120 mM NaCl, 0.5 mM EGTA, 1 mM dithiothreitol). Isolated FLNA-CS was concentrated using Ultrafree-15, frozen in liquid nitrogen, and stored at −80 °C. 58,16 Protein concentration was measured by absorption at 280 nm using parameter calculated by ProtParam tool (http://au.expasy.org/tools/protparam.html) or 660nm protein assay kit (Pierce). Recombinant protein was stored at −80°C and thawed and clarified before use.
F-actin gel point assay
Gelation was determined using a miniature falling ball viscometer 59. Actin polymerization was initiated by the addition of 24 μM unpolymerized actin in the presence or absence of different concentrations of FLNA-CS in buffer B (20 mM Tris-HCl, 100 mM KCl, 2 mM MgCl2, 0.5 mM EGTA, 0.5 mM ATP). The mixtures were immediately drawn into vertically positioned 100-μl capillary tubes (Fisher) and incubated at 25 °C for 60 min unless indicated otherwise. The time required for a 0.7-mm diameter stainless steel ball to fall 5 cm was measured. 59
Fluorometry
Fluorescence intensities of purified proteins were measured by a fluorescence microplate reader (SpectraMax Gemini EM, Molecular Devices).
Cell culture and expression of FLNA-CS in mammalian cells
HEK-293 and COS-7 cells were trypsinized with 0.05% trypsin/EDTA for 5 min and centrifuged in PBS. The cells were resuspended in growth medium and placed on a Lab-Tek™ II chamber slide (Cat# 154534, Thermo Scientific: for scanning-disk microscopy) and Delta T culture dishes (Cat# 04200415C, Bioptechs Inc., Butler, PA: for FLIM) coated with 0.1% (w/v) gelatin solution (G1393, Sigma-Aldrich) or 10 μg ml−1 fibronectin (F0895, Sigma-Aldrich) for overnight in a 5% CO2 incubator at 37°C. The cells were transfected with TransIT-LT1 (Mirus Bio) in accordance with manufacturers’ instructions 24 or 48 hr before imaging. Three hours before imaging, growth medium were replaced with growth medium supplemented with 25 mM HEPES without phenol red. MEF cells were cultured on a Lab-Tek™ II chamber slide coated with 0.1% (w/v) gelatin. The growth medium was replaced with Opti-MEM (Life technologies) and the cells were transfected with TransIT-X2 (Mirus Bio). 24 hours post-transfection, the medium was replaced with growth medium supplemented with 25 mM HEPES without serum and phenol red. In some experiments, transfected cells were treated with a pharmacological activator or inhibitor as indicated in figure legends.
Fluorescence lifetime microscopy (FLIM)
FLIM was performed using a Becker & Hickl time correlated single photon counting (TCSPC) system with a Hamamatsu hybrid detector (model HPM-100-50; Hamamatsu Photonics) attached to the non-descanned detection (NDD) light path on a Zeiss 710 laser-scanning confocal/multi-photon system with an upright Zeiss Examiner Z.1 stand. Two-photon excitation was achieved using a Chameleon Vision II laser (Coherent) to pump a mode-locked frequency-doubled Ti:Sapphire laser that provided 140-femtosecond pulses at a 80-Mhz repetition rate with an output power of 3 W at the peak of the tuning curve (800 nm).
EGFP and sREACh fluorophores were used as a FRET pair for all of the FLIM–FRET measurements. The two-photon laser was set to 920nm for excitation of the donor (EGFP). Fluorescence emission of EGFP fusion proteins was collected using a bandpass filter (500–550nm) to limit detection to only the donor. Laser power was adjusted to 14% to give a mean photon count rate of the order 104 photons s−1.
For imaging live cells by FLIM, cells were plated in Delta T culture dish coated with 0.1% (w/v) gelatin solution. The dish and Zeiss W Plan-Apochromat 63x/1.0NA objective were heated using feedback control Bioptechs objective and stage heater.
For fixed cells, transfected cells were cultured on a cover glass coated with 0.1% (w/v) gelation and fixed with 4% formaldehyde (EM grade, Electron Microscopy Sciences) in PBS for 20 min at 37°C. The cells were rinsed with 20 mM Tris-HCl, pH 7.4, 150 mM NaCl and mounted in PermaFluor (Thermo Scientific). Fluorescence from fixed samples was collected with a Zeiss Plan-Apochromat 63x/1.40 NA Oil objective.
Spinning-disk confocal microscopy and image acquisition
Cells expressing PQ-FRET sensors were imaged using a spinning-disk confocal microscope: spinning-disk head (CSU-X1, Yokogawa), microscope stand (IX81, Olympus), 488nm and 561nm laser combiner (ALC, Andor Technology), EMCCD camera (iXon 897, Andor Technology), laser based focusing system (ZDC1, Olympus), objective (60×/1.2 NA UPLSAPO 60XW water immersion lens, Olympus), image acquisition software (MetaMorph 7.7.7.0, Molecular Devices), stage top humidified chamber (INU-ZILCS-F1, Tokai Hit), whole microscope 37°C chamber (custom designed by HMS Cell Biology machine shop), Semrock barrier filters (525/40, 607/36), Semrock dicroic mirror (405/488/568/647). Wavelengths of the excitation laser beam were 488 and 561 nm and fluorescence for mEGFP and mCherry was detected using 488/568 nm Yokogawa emission filter (Em01-R488/568-15, Semrock). Images were collected using the maximum field of view with 512 × 512 image size. Timelapse images were collected with the frame acquisition rate ranged from 1.0 to 30 s per frame (indicated in the video legend).
Immunofluorescence microscopy
Cells were plated on a gelatin-coated chamber slide, transfected with PQ-FRET sensors, fixed with 4% formaldehyde in CB (10mM MES pH 6.1, 138mM KCl, 3mM MgCl2, 2mM EGTA) containing 0.32M sucrose for 20 min, rinse in TBS (20mM Tris-HCl pH7.4, 150mM NaCl), permeabilized with 0.5% Triton X-100 in TBS for 10min, rinse in TBS-0.1%Tx (TBS containing 0.1% Triton X-100), blocked in 2% BSA in TBS-0.1%Tx, and incubated with Alexa568-phalloidin (Invitrogen, 1:200 dilution) or primary antibodies (mixture of mAb3–14 (5μg ml−1) 16 and mAb1678 (5μg ml−1), Millipore) for 1 hr. After several washes with TBS-0.1%Tx, the cells were incubated with secondary antibodies (Invitrogen, 5μg ml−1), washed with TBS-0.1%Tx, and mounted with mounting media (Spring Bioscience). Cells were imaged using a spinning-disk confocal microscope as described above with a 60×/1.42 NA oil immersion objective (PLAPON 60XO, Olympus).
FLNA-actin-myosin network
To prepare nitrocellulose-coated slide glass and cover glass, the glasses were covered with 0.1% (v/v) collodion in amylacetate and air-dried. The surfaces were coated with human recombinant gelsolin (50 μg ml−1) in PBS for 1 min and blocked with 1% bovine serum albumin in blocking buffer (25 mM KCl, 20 mM Tris-HCl, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT, pH 7.4) for 30 min. The coated surfaces were washed with the blocking buffer five times. Reconstituted networks were prepared by mixing 10 μM actin, 0.1 μM FLNA, 0.5 or 0.2 μM myosin (porcine cardiac myosin, Sigma M0531), 5 μM phalloidin, and 1x polymerization buffer (20 mM Tris_HCl, pH7.4, 50 mM NaCl, 0.5 mM EGTA, 4 mM MgCl2, 1 mM CaCl2, 5 mM DTT, 1 mM ATP; 1 mM γS-ATP was used for negative control) and sandwiched between the coated-slide glass and cover glass with a 100 μm spacer (illustrated in Fig. 4). Each sample was allowed to polymerize for one hour and fluorescence intensity was measured by spinning-disk confocal microscopy. Fluorescence intensities of mEGFP and mCherry were measured by NIH ImageJ and their ratio was calculated.
Data analysis of PQ-FRET images
Datasets were analyzed using ImageJ 1.46j software (National Institutes of Health). A mEGFP/mCherry ratio image was created using ImageJ plugin “Ratio Plus” and intensities of the ratio images were measured by ImageJ (Measure<Analyze or “Intensity v Time Monitor”<”Stacks - T-functions” plugins). The single steps for data processing are outlined in Supplementary Table 1.
Statistical analysis
Excel and Prism were used to access statistical significance.
Supplementary Material
Acknowledgments
We thank Lisa Cameron, Lai Ding, and Daniel Tom for technical assistance. This work is supported by the HUSEC Seed Fund for Interdisciplinary Science and a National Institutes of Health grant (HL19749).
Footnotes
Contributions
F.N., J.H.H. and T.P.S. conceived the project. F.N. and M.S. carried out the experiments and analysed the data. F.N., J.H.H. and T.P.S. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Brown AE, Discher DE. Conformational changes and signaling in cell and matrix physics. Current biology: CB. 2009;19:R781–789. doi: 10.1016/j.cub.2009.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Developmental cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heisenberg CP, Bellaiche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Guillot C, Lecuit T. Mechanics of epithelial tissue homeostasis and morphogenesis. Science (New York, NY. 2013;340:1185–1189. doi: 10.1126/science.1235249. [DOI] [PubMed] [Google Scholar]
- 6.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyndt F, et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–49. doi: 10.1161/CIRCULATIONAHA.106.622621. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478:260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razinia Z, Makela T, Ylanne J, Calderwood DA. Filamins in mechanosensing and signaling. Annual review of biophysics. 2012;41:227–246. doi: 10.1146/annurev-biophys-050511-102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kainulainen T, Pender A, D’Addario M, Feng Y, Lekic P, McCulloch CA. Cell death and mechanoprotection by filamin a in connective tissues after challenge by applied tensile forces. The Journal of biological chemistry. 2002;277:21998–22009. doi: 10.1074/jbc.M200715200. [DOI] [PubMed] [Google Scholar]
- 11.Sharif-Naeini R, et al. Polycystin-1 and -2 dosage regulates pressure sensing. Cell. 2009;139:587–596. doi: 10.1016/j.cell.2009.08.045. [DOI] [PubMed] [Google Scholar]
- 12.Luo W, et al. Analysis of the local organization and dynamics of cellular actin networks. J Cell Biol. 2013;202:1057–1073. doi: 10.1083/jcb.201210123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stossel TP, et al. Filamins as integrators of cell mechanics and signalling. Nature reviews Molecular cell biology. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 14.Gorlin JB, et al. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura F, Stossel TP, Hartwig JH. The filamins: Organizers of cell structure and function. Cell adhesion & migration. 2011;5 doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lad Y, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. The EMBO journal. 2007;26:3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heikkinen OK, et al. Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin. The Journal of biological chemistry. 2009;284:25450–25458. doi: 10.1074/jbc.M109.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruskamo S, et al. The C-terminal rod 2 fragment of filamin A forms a compact structure that can be extended. The Biochemical journal. 2012;446:261–269. doi: 10.1042/BJ20120361. [DOI] [PubMed] [Google Scholar]
- 20.Tossavainen H, et al. Model of a six immunoglobulin-like domain fragment of filamin A (16–21) built using residual dipolar couplings. Journal of the American Chemical Society. 2012;134:6660–6672. doi: 10.1021/ja2114882. [DOI] [PubMed] [Google Scholar]
- 21.Rognoni L, Stigler J, Pelz B, Ylanne J, Rief M. Dynamic force sensing of filamin revealed in single-molecule experiments. Proc Natl Acad Sci U S A. 2012;109:19679–19684. doi: 10.1073/pnas.1211274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganesan S, Ameer-Beg SM, Ng TT, Vojnovic B, Wouters FS. A dark yellow fluorescent protein (YFP)-based Resonance Energy-Accepting Chromoprotein (REACh) for Forster resonance energy transfer with GFP. Proc Natl Acad Sci U S A. 2006;103:4089–4094. doi: 10.1073/pnas.0509922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakoshi H, Lee SJ, Yasuda R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain cell biology. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Shyy JY, Chien S. Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing. Annual review of biomedical engineering. 2008;10:1–38. doi: 10.1146/annurev.bioeng.010308.161731. [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki N, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 27.Koenderink GH, et al. An active biopolymer network controlled by molecular motors. Proc Natl Acad Sci U S A. 2009;106:15192–15197. doi: 10.1073/pnas.0903974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bond LM, Tumbarello DA, Kendrick-Jones J, Buss F. Small-molecule inhibitors of myosin proteins. Future medicinal chemistry. 2013;5:41–52. doi: 10.4155/fmc.12.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dantzig JA, Walker JW, Trentham DR, Goldman YE. Relaxation of muscle fibers with adenosine 5′-[gamma-thio]triphosphate (ATP[gamma S]) and by laser photolysis of caged ATP[gamma S]: evidence for Ca2+-dependent affinity of rapidly detaching zero-force cross-bridges. Proc Natl Acad Sci U S A. 1988;85:6716–6720. doi: 10.1073/pnas.85.18.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ithychanda SS, et al. Identification and characterization of multiple similar ligand-binding repeats in filamin: implication on filamin-mediated receptor clustering and crosstalk. The Journal of biological chemistry. 2009;284:35113–35121. doi: 10.1074/jbc.M109.060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ylilauri M, Pentikainen OT. MMGBSA as a tool to understand the binding affinities of filamin-peptide interactions. Journal of chemical information and modeling. 2013;53:2626–2633. doi: 10.1021/ci4002475. [DOI] [PubMed] [Google Scholar]
- 32.Kaneider NC, Djanani A, Fischer-Colbrie R, Wiedermann CJ. Sphingosine kinase-dependent directional migration of leukocytes in response to phorbol ester. Biochemical and biophysical research communications. 2002;297:806–810. doi: 10.1016/s0006-291x(02)02304-5. [DOI] [PubMed] [Google Scholar]
- 33.Sumagin R, Robin AZ, Nusrat A, Parkos CA. Activation of PKCbetaII by PMA facilitates enhanced epithelial wound repair through increased cell spreading and migration. PLoS ONE. 2013;8:e55775. doi: 10.1371/journal.pone.0055775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukata Y, Amano M, Kaibuchi K. Rho-Rho-kinase pathway in smooth muscle contraction and cytoskeletal reorganization of non-muscle cells. Trends in pharmacological sciences. 2001;22:32–39. doi: 10.1016/s0165-6147(00)01596-0. [DOI] [PubMed] [Google Scholar]
- 35.Uehata M, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 36.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 37.Discher D, et al. Biomechanics: cell research and applications for the next decade. Ann Biomed Eng. 2009;37:847–859. doi: 10.1007/s10439-009-9661-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janmey PA, Wells RG, Assoian RK, McCulloch CA. From tissue mechanics to transcription factors. Differentiation; research in biological diversity. 2013;86:112–120. doi: 10.1016/j.diff.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Wang N. FRET and mechanobiology. Integrative biology: quantitative biosciences from nano to macro. 2009;1:565–573. doi: 10.1039/b913093b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science (New York, NY. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nature cell biology. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 43.Borghi N, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng F, Suchyna TM, Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J. 2008;275:3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng F, Suchyna TM, Lazakovitch E, Gronostajski RM, Sachs F. Real Time FRET Based Detection of Mechanical Stress in Cytoskeletal and Extracellular Matrix Proteins. Cellular and molecular bioengineering. 2011;4:148–159. doi: 10.1007/s12195-010-0140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suphamungmee W, Nakamura F, Hartwig JH, Lehman W. Electron microscopy and 3D reconstruction reveals filamin Ig domain binding to F-actin. Journal of molecular biology. 2012;424:248–256. doi: 10.1016/j.jmb.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, Zhu X, Cong P, Sheetz MP, Nakamura F, Yan J. Differential mechanical stability of filamin A rod segments. Biophysical journal. 2011;101:1231–1237. doi: 10.1016/j.bpj.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Chandrasekar S, Sheetz MP, Stossel TP, Nakamura F, Yan J. Mechanical perturbation of filamin A immunoglobulin repeats 20–21 reveals potential non-equilibrium mechanochemical partner binding function. Scientific reports. 2013;3:1642. doi: 10.1038/srep01642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiema T, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Das M, Ithychanda SS, Qin J, Plow EF. Migfilin and filamin as regulators of integrin activation in endothelial cells and neutrophils. PLoS ONE. 2011;6:e26355. doi: 10.1371/journal.pone.0026355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tu Y, Wu S, Shi X, Chen K, Wu C. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell. 2003;113:37–47. doi: 10.1016/s0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 52.Takafuta T, Saeki M, Fujimoto TT, Fujimura K, Shapiro SS. A new member of the LIM protein family binds to filamin B and localizes at stress fibers. The Journal of biological chemistry. 2003;278:12175–12181. doi: 10.1074/jbc.M209339200. [DOI] [PubMed] [Google Scholar]
- 53.Gardel ML, Nakamura F, Hartwig JH, Crocker JC, Stossel TP, Weitz DA. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc Natl Acad Sci U S A. 2006;103:1762–1767. doi: 10.1073/pnas.0504777103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burridge K, Wittchen ES. The tension mounts: stress fibers as force-generating mechanotransducers. J Cell Biol. 2013;200:9–19. doi: 10.1083/jcb.201210090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartwig JH, Stossel TP. Structure of macrophage actin-binding protein molecules in solution and interacting with actin filaments. Journal of molecular biology. 1981;145:563–581. doi: 10.1016/0022-2836(81)90545-3. [DOI] [PubMed] [Google Scholar]
- 56.Oster G. Biophysics of the leading lamella. Cell motility and the cytoskeleton. 1988;10:164–171. doi: 10.1002/cm.970100121. [DOI] [PubMed] [Google Scholar]
- 57.Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annual review of cell and developmental biology. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 58.Nakamura F, et al. Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS ONE. 2009;4:e4928. doi: 10.1371/journal.pone.0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakamura F, Osborn E, Janmey PA, Stossel TP. Comparison of filamin A-induced cross-linking and Arp2/3 complex-mediated branching on the mechanics of actin filaments. The Journal of biological chemistry. 2002;277:9148–9154. doi: 10.1074/jbc.M111297200. [DOI] [PubMed] [Google Scholar]
- 60.Kardash E, Bandemer J, Raz E. Imaging protein activity in live embryos using fluorescence resonance energy transfer biosensors. Nature protocols. 2011;6:1835–1846. doi: 10.1038/nprot.2011.395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.