Abstract
Once damaged, cardiac tissue does not readily repair and is therefore a primary target of regenerative therapies. One regenerative approach is the development of scaffolds that functionally mimic the cardiac extracellular matrix (ECM) to deliver stem cells or cardiac precursor populations to the heart. Technological advances in micro/nanotechnology, stem cell biology, biomaterials and tissue decellularization have propelled this promising approach forward. Surprisingly, technological advances in optical imaging methods have not been fully utilized in the field of cardiac regeneration. Here, we describe and provide examples to demonstrate how advanced imaging techniques could revolutionize how ECM-mimicking cardiac tissues are informed and evaluated.
Keywords: extracellular matrix, developing heart, multimodality imaging, tissue engineeringbiomaterials
ECM-informed cardiac tissue regeneration
Heart disease is the leading cause of death in the United States, with over 830 000 cases in 2011 [1]. The limited ability of the heart to regenerate has prompted efforts to drive stem cell differentiation to cardiomyocytes (or their precursors) and to deliver cells to the cardiac microenvironment for therapeutic applications. Efforts to augment differentiation and delivery have been explored independently, each having unique challenges. The ECM is a crucial component that could effectively address key challenges in both fields [2–4]. The current limitations to effectively employ the ECM are (i) limited knowledge of the spatial structure, component makeup and temporal dynamics of the ECM best suited for stem cell delivery, (ii) the inability to accurately assess the spatial structure and temporal dynamics of ECM-based, engineered tissues, and (iii) the difficulty in tracking cell behavior and cell–ECM interactions within these engineered tissues. The primary goal of this review is to describe how advanced imaging techniques can be employed to address these limitations (Box 1) and, in so doing, we will also summarize current knowledge of cardiac ECM composition during development and the state-of-the-art in ECM-based engineered cardiac tissues.
A blueprint for regeneration
ECM of the developing heart
The heart is a complex organ consisting of multiple chambers, one-way valves and a synchronized conduction system designed to generate sufficient force to move blood throughout the body. The embryonic development of the vertebrate heart is equally complex, transitioning from a tube containing beating cells through a phase of looping to the formation of distinct chambers and valves. This process utilizes cell proliferation, migration, rearrangement and differentiation. Given the wide array of structural and functional changes, it is not surprising that the research literature on cardiac ECM composition shows that the ECM is also changing, either as a response to, or as a precursor of, changes in the developing heart (Table 1). The extracellular matrix provides adhesion substrates, imparts structural support, stores and sequesters soluble factors, and transduces mechanical signals. Indeed, isolated cardiac cells require ECM to maintain or acquire function [5], and changes in cardiac ECM composition and function during development are crucial for directing tissue specification [6,7]. For this reason, defining the dynamics of ECM of this developing tissue could provide a critical design template, a blueprint, for constructing synthetic scaffolds for tissue repair. Although this blueprint concept is attractive, the current gap between what is known of ECM dynamics during cardiac development and the generation of constructs based on that knowledge is vast.
Table 1.
The spatiotemporal distribution of ECM (protein or RNA) in the developing heart from E11 to adulta,b,c
 Looping heart (E10.5) Looping heart (E10.5) |
 Remodeling heart (E12.5) Remodeling heart (E12.5) |
Maturation and septation (E12.5–E19.5) |
 Neonatal to adult Neonatal to adult |
Refs | ||||
|---|---|---|---|---|---|---|---|---|
| Collagen I | (E11) myo, endo AV filament | (E12) myo bundle | (E13) subepi parallel fibril | (E15) epi AT, V fibril | Subepi thick layer; myo reticulate; interstitium bundle; collagend V reticulate | [14,17,70–72] E11: [14] E12: [17] E13: [17] E15: [70] Adult: [17,71,72] |
||
| Colllagen III | (E12) peri, endo AV | (E13) widely distributed multiple orientation | (E15) BV, V fibril | (E16) epi | Subepi thick fiber; myo fine fiber BV reticulate | [17,70,71,73,74] E12: [73] E13: [17] E15: [70] E16: [74] Adult: [17,71] |
||
| Collagen IV | (E11) myo, endothelium BM fine fibril | BV, BM reticulate | [14,71] E11: [14] Adult: [71] |
|||||
| Fibronectin | (E11) myo, endo; epi, peri, endo AV | (E12) endo | (E14) epi right AV; myo V, OFT; endo BV, LV; mRNA V, AT | (E16) epi BV; peri | (E18) subepi fiber | BV, LV fine reticulate | [14,17,20,74–76] E11: [14,75] E12: [75] E14: [20,74,75] E16: [75] E18: [17] Adult: [20,76] |
|
| Laminin | (E11) myo, endothelium OFT | (E14) endo LV; V | (E15) whole heart punctate | Whole heart punctate; myo BM, BV fine fibril; LV fine fibrillar reticulate | [20,74,76–78] E11: [77] E14: [20,74] E15: [78] Adult: [20,76,78] |
|||
| Elastin | (E11) OFT, V fibrillar, parallel to OFT | (E12) myo OFT circumferential fiber; AV fibril | (E13) OFT | (E14) epi, subendo AT, BV bundle, fibril | E11–E14: [76] | |||
The instructive cues from these developmental stages are critical for building ECM-informed functional scaffolds. The spatiotemporal information is presented as time point, anatomical location (wall and/or chamber), and architecture.
The developing heart schematic was adapted, with permission, from [79].
OFT, outflow tract; AT, atrium; AV, atrioventricular; BM, basement membrane; endo, endocardium; epi, epicardium; LV, left ventricle; myo, myocardium; peri, pericardium; subepi, subepicardium; V, ventricle; BV, blood vessel.
Collagen types not specified.
ECM composition during development
Much of what is known of ECM composition with cardiac development corresponds to the most prevalent proteins. Collagen type I (ColI) and collagen type III (ColIII) are fibrillar proteins stabilized by hydrogen bonds, providing a framework and mechanical support of tissues. Collagen type IV (ColIV) and laminin are found in the basement membrane, forming networks that mediate cell adhesion, migration and differentiation. Fibronectin is a multi-domain dimer that interacts with multiple integrins, collagens, glycosaminoglycans and glycoproteins to mediate cell behavior. Elastin, the major component of elastic matrices, is critical for regulating elasticity. These ECM components are so important that rodents lacking genes encoding these proteins do not survive the early postnatal period (reviewed in [8]). Unfortunately, what is known of the spatiotemporal distribution of these primary ECM proteins is incomplete during the main period of prenatal heart growth (mouse embryonic day 11–day 18). The limited knowledge we have of the ECM in the heart during this period (Table 1) is primarily based on antibody labeling in essentially 2D histological sections of mouse or chick embryos. In the subsequent text, E indicates mouse embryonic day and the approximate conversion of chick developmental stages (Hamburger-Hamilton stages) to mouse embryonic days has been applied [9].
At stages prior to E11, the cardiac ECM consists mostly of ColI [10], ColIV [10] and ColIII [11]. Beginning at E11, the atrial and interventricular septa form [12] and epicardial cells migrate to cover the heart [13]. During this time, ColIV and laminin become localized in the ‘cardiac jelly’ layer of ECM between the endocardial and myocardial layers of the primitive heart [14]. ColIV and laminin may contribute to cell migration, particularly of mesenchymal cells in the endocardial cushions. Elastin and ColVI appear during this stage [15], whereas fibronectin is localized in the endocardial and myocardial tubes [14].
The atrial septum begins to form shortly after E12 and the outflow tract (OFT) is dividing into the pulmonary and systemic branches [12]. By E12.5, the four chambers of the heart are delineated as is septation of the OFT [16], with elastin reaching maximum expression during this time, mainly in the OFT [15]. The distribution pattern of the elastic matrices is progressively modified in association with the formation of the definitive four-chambered heart and the elastic matrices are localized in the epicardium and subendocardial space of the atria [15]. The fetal circulatory system is fully functional by E14.5 [12]. With continued development and maturation of the heart, ColI ultimately distributes circumferentially around the heart [17], increasing even more rapidly than the total heart weight [18], to provide the support needed to function under the pressures of gravity and activity following birth. Beyond E15.5, cardiomyocyte proliferation subsides and by postnatal day 5 most proliferation has ceased [19]. Elastin, except for association with blood vessels, is absent in later stages [15]; fibronectin, which is found alongside the endo- and epicardium (E14), is considerably decreased after birth [20]; whereas the presence of laminin along the basement membrane of myocardial cells is maintained even after birth [20].
Imaging technologies to address gaps in understanding of ECM dynamics with cardiac development
As summarized above, data exist identifying the ECM components (Table 1) present at some time points during cardiac development in chick and mouse embryos, but a clear profile of ECM composition, organization and functional contribution during heart formation is lacking. Different stages of heart development exhibit regional and compositional differences in ECM proteins, suggesting that cardiac precursors are likely to require a different ECM blueprint than fully differentiated cardiomyocytes, an important consideration for regenerative therapies. Thus the developmental range of time points that provides the ECM with the cues that are most essential for cardiomyocyte precursor differentiation, propagation and function must be defined. Subsequently, a comprehensive analysis of ECM content, organization and ECM–cell associations, in both two and three dimensions, will be needed. To date, most cardiac development ECM studies have consisted of 2D tissue sections, frequently with single antibody labeling; therefore, analysis of intact 3D tissues, with multiple labels, will be an important next step. Histology is still the gold standard for assessing cell and tissue morphology. Unfortunately, although this approach is powerful, the tissue is fixed, embedded, physically sliced and stained so that accurate 3D morphology cannot be acquired. Alternatively, one of the most widely utilized tools for high-resolution imaging of 3D biological structure is confocal laser scanning microscopy (CLSM). CLSM has improved optical sectioning, temporal and spatial resolution and signal to noise ratio (SNR) over traditional wide-field fluorescence approaches [21]. CLSM-generated optical sections can be 3D rendered to visualize compositional and structural relationships. Adaptations include reflectance confocal [22], which is useful for examining the ECM in thin samples.
There is great need for non-invasive high resolution imaging methods for in vitro and in vivo tissue that can preserve ECM integrity and limit histochemical artifacts. Nonlinear optical laser scanning methods utilize high power, wide-tunable pulsed excitation sources that yield nonlinear optical interactions. Multiphoton laser scanning microscopy (MPLSM) is a nonlinear optical sectioning technique allowing thick biological sections to be noninvasively imaged via simultaneous absorption of two or more near-infrared (NIR) photons resulting in improvements in depth, viability and SNR compared to traditional optical approaches, including CLSM [23]. The wavelength tunability of MPLSM allows for endogenous fluorophore excitation such as the intrinsic metabolic cofactors nicotinamide adenine dinucleotide (NADH) and flavoproteins [23]. Changes in these metabolites have been associated with changes in stem cell state including differentiation [24]. Thus, the autofluorescent metabolites offer a noninvasive means of tracking cell state with changes in ECM. Second harmonic generation (SHG) [25] is a nonlinear effect arising from the laser field passing through non-centrosymmetric ordered structures (such as collagen). SHG is sensitive to concentration, organization and orientation of the sample and can be used simultaneously with MPLSM, yet still be differentiated due to distinct emission signals.
The exciting consequence of these endogenous optical approaches is that they can be applied to fresh ex vivo tissue, such as mouse embryonic hearts, to noninvasively provide 3D information about the relationships between cells and the ECM. In one study, the SHG signals originating from the collagen matrix, along with fluorescence signals from the fibroblast nuclei and actin, showed the spatial reorganization of the collagen fibers in the 3D microenvironment [26]. Similarly, SHG and MPLSM have been utilized to compare spatial distribution of anisotropic collagen and isotropic elastin fibers [27] (Figure 1a–c). This live imaging methodology avoids artifacts accompanying chemical fixation, staining or physical treatments, which could produce potential inaccuracies in the spatiotemporal distribution of ECM proteins to define a construction blueprint. These nonlinear methods offer deeper penetration, higher sensitivity and reduced phototoxicity over other currently available methods that offer high spatial and temporal resolution. However, these techniques still are limited in depth due to scattering and refractive index changes in tissues and in resolution by the diffraction limit. Thus, other emerging methods may still be needed where greater spatial performance is critical, such as for nanometer scale resolution and millimeter depth imaging.
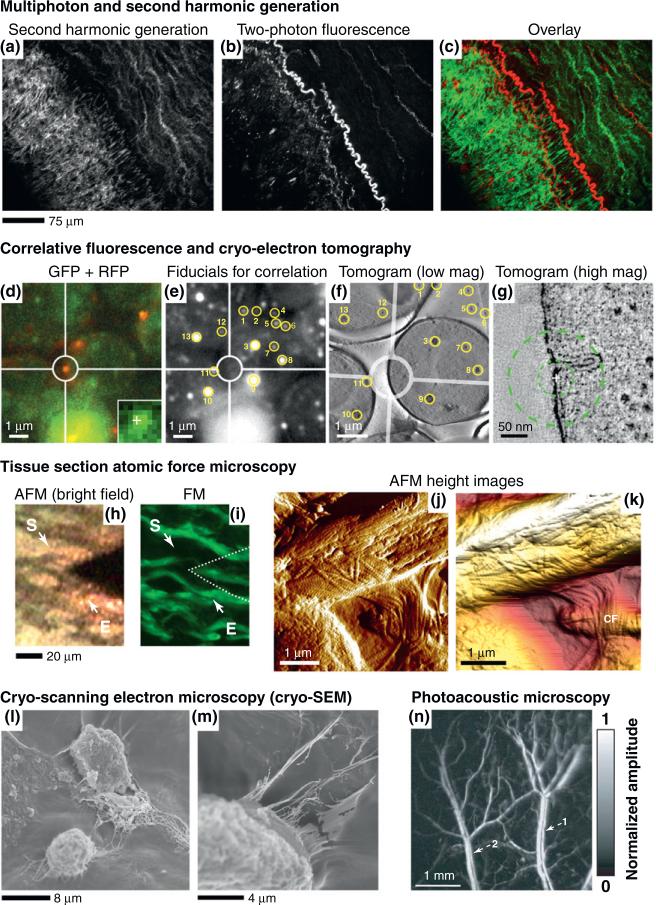
Figure 1.
Multimodality and high-resolution imaging techniques. (a) Second harmonic generation (SHG) image of collagen and (b) two-photon fluorescence image of elastin in an atherosclerotic artery. (c) Merged image of (a) collagen (green) and (b) elastin (red). Reproduced, with permission, from [27]. (d) The region of interest is highlighted by the circles and a cross in the inset marks a centroid position. (e) Fiducials selected for correlation with yellow circles and numbers. (f) Same fiducials as in (e) present from a tomogram of the cells of interest. (g) Slice of high magnification tomogram. The cross marks the centroid coordinates of the fluorescent patch transformed into tomogram coordinates. The inner circle corresponds to a probability radius of 50% (33 nm), outer circle to 80% (89 nm). Reproduced, with permission, from [45]. Imaging a sheep aorta region by (h) an atomic force microscopy (AFM) optical system and (i) fluorescence microscopy (FM). The location of the AFM tip is indicated by a dotted line. The start (S) and end (E) points of a series of 5 μm AFM scans are indicated by arrows. In (j) and (k), collagen fibrils (CF) are evident from amplitude scans collected. Reproduced, with permission, from [47]. (l) Cryo-scanning electron microscopy (cryo-SEM) of bone marrow-derived human mesenchymal stem cells (hMSCs) in 3D poly(ethylene glycol)-diacrylate hydrogel/collagen type I composite (unpublished work). (m) Note connections established by cells and ECM in a 3D environment. (n) Laser-scanning optical-resolution photoacoustic microscopy of vasculature within a Swiss Webster mouse ear, two pairs of parallel veins (1) and arteries (2). Reproduced, with permission, from [69].
Engineering cardiac tissues: building from the blueprint
The development of 3D ECM-mimicking biomaterials
For more than a decade, the elements of the ECM have been appreciated as essential cues in developing and maintaining appropriate 3D microenvironments for cardiac cell types and their precursors. Several approaches have been implemented to create such environments in vitro with a spectrum of potential benefits that include the ability to better understand the behavior of cardiac cell types, the improvement of means to guide the behavior of cardiac cell types, and the generation of tissues for repair of acute (i.e. myocardial infarction) or chronic (i.e. scarring and remodeling associated with age or disease) damage. These approaches include the use of native-ECM gels [28], decellularized ECM [3], endogenous ECM-based cell sheets [29], composites containing synthetic polymers with either native ECM proteins [30] or ECM-inspired chemical functionalities [31]. All approaches have merit, but perhaps the most control and flexibility is afforded by the last two, and so providing the greatest probability of ‘building the blueprint’. Plentiful examples exist to support the feasibility of incorporating defined, ECM-inspired, chemical functionality into non-functionalized materials. Such examples include conjugating cell-adhesion ligands to enhance cell attachment and proliferation [32], tethering extracellular ligands for maintaining pluripotent stem cells in an undifferentiated state [33], or delivering soluble factors often bound or sequestered by native or engineered ECM [34]. Given the complexity of the ECM and the dynamic features it displays a single ‘functionality’ may not suffice. Thus, more recent efforts to develop ECM-based 3D scaffolds employ material platforms capable of presenting multiple factors. In one example, matrix metalloproteinase (MMP)-sensitive motifs were presented in a poly(ethylene glycol) hydrogel to facilitate scaffold remodeling, while arginine-glycineaspartic acid-serine-proline (RGDSP) peptides of the same hydrogel promoted binding of integrins relevant in early cardiac development (α5β1 or αvβ1) to direct differentiation of pluripotent cardioprogenitors [35]. Similarly, a thermoresponsive poly(N-isopropylacrylamide-co-acrylic acid) hydrogel was used to modulate mechanical properties of a scaffold that also included MMP-sensitive motifs to facilitate cardiac stem cell transplantation [36].
Challenges in constructing 3D ECM-mimicking biomaterials
As technologies improve to incorporate multiple ECM-based signals in a single material, the challenge of evaluating all possible combinations of physical (stiffness, dimensionality, porosity, swelling), chemical (soluble factors, nutrients, oxygen), and biological (cell–matrix and cell–cell interactions) factors emerges. To address this challenge, 3D microarray platform approaches are under intense development, aiming to attain tight control of instructive signals, rapid modulation and miniaturization in a high-throughput manner [37]. For example, arrays of intact ECM proteins (i.e. ColI, ColIV, laminin and fibronectin) of various compositions were employed to identify the compositions that facilitated differentiation of specific cell types from embryonic stem cells [38]. As another example, a PEG-based hydrogel presenting arginine-glycine-aspartic acid (RGD) ligands for cell attachment and dithiothreitol (DTT) bridges for hydrolytic degradation was used as a well-defined and adaptable 3D array platform for high-throughput analysis of multiple cell–matrix interactions including cell adhesion, migration and differentiated function [39]. One important consideration when assessing the impact of multiple ECM-based material modifications on cell behavior is that the impact of multiple material modifications on material properties may not necessarily reflect the additive effects of each modification [40]. Copolymerizing two different types of macromonomers does not always yield the desired material: the final products can exhibit significant drift in composition due to different reactivity to one another from each macromonomer [41]. A modular molecular design that controls each factor independently without affecting one another is one strategy towards achieving multifunctionality [42].
ECM-based materials, whether in 3D microarray format or 3D tissue-scale format, must be analyzed in terms of material properties (i.e. pore size and distribution, fiber size and distribution, homogeneity of added functionality and activity of added functionality) and cell response to material feature(s) (i.e. viability, adhesion to material, differentiation and function). Despite great advances in high-throughput methods and synthetic chemistry to direct and/or understand cell–ECM interactions, visualization of these complex systems remains a challenge. Optical approaches as described above can be used to examine cell–ECM interactions but have limitations particularly with respect to nanometer scale spatial resolution. Fortunately, optical methods are amenable to multimodal imaging approaches where complementary imaging methods are combined or correlated, resulting in a combined set of attributes not collectable by a single modality.
Correlative fluorescence, cryo-electron tomography (cryo-ET) and tissue section atomic force microscopy (AFM)
Transmission electron microscopy (TEM) uses electrons instead of light to provide nanometer or greater scale resolution of fixed thin sections. The biological utility of TEM for visualizing 3D spatiotemporal arrangements and dynamics of constituent molecular machinery has been advanced through the use of cryo-TEM and cryo-ET [43]. However, specimen preparation artifacts, poor SNR and computationally-intensive image analysis still present significant technical challenges.
Another major limitation of high-resolution EM is that as the information from a small volume increases with higher magnification, the number of samples examined is reduced. In the case of cryo-ET, the cellular area covered in a single tomogram is approximately 2 μm2, about 1.5% of the peripheral area of a typical eukaryotic cell [43]. Correlative microscopy attempts to bridge the various temporal and resolution gaps of TEM to integrate structural information gathered from multiple levels of biological hierarchy into one common framework [44]. Because fluorescence microscopy (FM) is an ideal technique to localize ultrastructure and to observe cellular dynamics, a method integrating FM with 3D cryo-ET was recently used to image microtubule end structures using small fluorescent microspheres as fiducial markers that are visible by both FM and cryo-ET [45] (Figure 1d–g).
Multimodality imaging approaches can also be applied to AFM, a high-resolution non-destructive imaging modality that records interactions between a sharp tip (probe) and a sample surface on the nanometer scale. The combination of AFM and cryo-sectioning, which prevents protein denaturation and significant morphological changes, could serve as an alternative to cryo-ET. AFM can also be used to monitor force interaction such as modulus measurements of native ECM or ECM-based synthetic materials [46]. Recently, tissue section AFM has been used to visualize ferret left ventricle epicardium and, in correlation with FM, to examine the elastic lamellae in sheep aorta to confirm the chemical composition of these lamellae [47] (Figure 1h–k). Theoretically, tissue section AFM techniques could be applied to samples vulnerable to in vitro degradation or dehydration, permitting the quantitation of dynamics changes in tissue structures. Furthermore, new classes of AFM instruments (Bio-AFM), integrate fluorescence with AFM, enabling studies that track fluorescently-labeled cells in response to measured changes in ECM mechanical properties [48].
Despite the great promise of cryo-based TEM for better ultrastructure preservation, this technique is still very invasive and time consuming. Cryo-scanning electron microscopy (cryo-SEM) techniques that can monitor a sample's surface topography and composition in high resolution (Figure 1l,m) are of great interest to cardiac-ECM studies, but as with TEM, there is concern regarding preservation of structure. A new type of SEM has recently emerged, environmental SEM (ESEM), which can collect images of hydrated samples in a non-vacuum environment. Unlike traditional TEM or SEM, ESEM can capture images of an intact ECM without dehydration or coating. ESEM has been used for biomaterials analysis [49] and shows great promise in its ability to look at intact ECM with high resolution [50].
Emerging imaging innovation
The past 25 years have seen great advances in live cell imaging approaches where it is now possible to image at cell resolution specific players of interest, such as tagged ECM proteins and associated cells, in both time and space. In addition, these images can be collected in biologically relevant environments where temporal and spatial windows are preserved by rapid acquisition of intact 3D samples. Technological advances in genetic labeling, optics, electronics and computation allow the capture of important biological phenomena with fewer compromises. Thus, the imaging can be more than just a qualitative view of biological events but also a quantitative assessment of the progress and maturation of a protein or cell and its interaction with the microenvironment. Rapidly evolving technologies, such as spectral, fluorescence lifetime [51] and polarization [52] imaging can help characterize the microenvironment and the cellular interactions within. In addition, techniques are being developed to not only image the cell and tissue environments but provide optical tools that can directly probe and manipulate. Photomanipulation tools such as uncaging [53], photoactivation and fluorescence recovery after photobleaching (FRAP) [54] have been used to query phenomena from calcium channel activation [53] to protein signaling [54] and might be further adapted to assess sequestration, release and cellular uptake of soluble factors from the ECM of native tissues and ECM-based biomaterials.
The future also brings the promise of generating realistic 3D high-resolution views both in vitro and in vivo. New photochemistry [55] allows the creation of realistic 3D scenarios that mimic specific microenvironments, such as the ECM, or provide specific control, such as gradients for drug studies. There is significant potential for multimodality experiments, in addition to that described for correlative EM. Deeper imaging techniques such as optical coherence tomography [56], photoacoustic microscopy [57] (Figure 1n) and ultrasound imaging [58] can greatly improve our knowledge of intact tissue composition. Super resolution light microscopy (LM) [59] aims to overcome the Abbe diffraction limit and greatly improve spatial resolution while maintaining temporal resolution. For in vivo imaging, the goal is to develop optical strategies compatible with deeper imaging and imaging of intact specimens, including whole animals [60]. Long wavelength imaging [61] and adaptive optics [62] hold promise to overcome depth penetration limitations due to scattering and refractive index induced aberrations inherent in all tissues. Neuroscience and other fields are driving the development of optical endoscopes, where the microscope is replaced by an optical fiber that can remotely probe a biological site [63]. These technologies will become increasingly important for cardiac regeneration as ECM-based materials move from in vitro to in vivo environments.
Finally, bioimage informatics is critical for all imaging, both current and emerging [64]. Computational approaches are needed to quickly acquire these large heterogeneous datasets, convert them to standard analyzable data models [65], effectively mine the data [66], accurately identify and track objects [67] and provide image visualization [68]. As the cardiac ECM community utilizes more of these advanced imaging approaches, it will be imperative that a computational framework be developed that will not only enable the collection and analysis of these important cell-ECM datasets but also facilitate new discovery such as new computational models and algorithmic approaches to assess ECM organization and cell identification or data-mining with correlative genomic information. Multimodal imaging in conjunction with advanced computation could substantially advance the construction and evaluation of synthetic scaffolds for cardiac tissue repair.
Box 1. Building a blueprint for cardiac tissue regeneration.
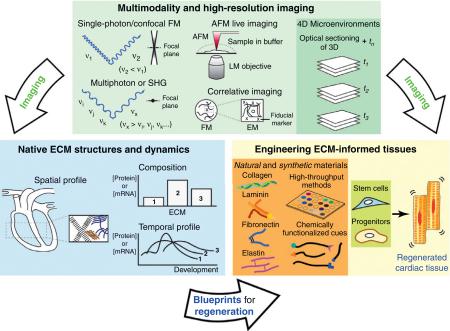
Multimodality and high-resolution imaging (Figure I): traditionally, single-photon and confocal FM have been utilized to investigate 3D tissue structures. However, the scattering and interference of incident beams primarily limit the application of optical imaging in visualizing extracellular microenvironments. Multiphoton or SHG significantly expands the ability of optical sectioning without employing exogenous markers. AFM combined with LM modality enables the exploration of nanometer-scale extracellular microenvironments. Correlative imaging utilizes a reference point (fiducial marker) to identify the region of interest under two different modalities (i.e. FM and EM). These imaging modalities in conjunction with time-resolved information specify 4D microenvironments.
Advanced imaging analysis could significantly expand the understanding of native ECM structures and dynamics by probing the spatiotemporal profiles and (relative) composition of ECM in cardiac tissue of interest. The information collected through advanced imaging will generate a blueprint for regenerating tissues.
The blueprint guides the design of ECM-informed tissues from natural or synthetic materials, which requires high-throughput screening to identify multiple factors directing cell proliferation, differentiation, and migration and a library of functionalities that provide instructive signals to cells. The directed cell–matrix interactions could regenerate functional cardiac tissues to repair acute or chronic damage, which is also subject to advanced imaging analysis to verify the properties of and performance of the regenerated cardiac tissues.
Figure I. Advanced imaging to define a blueprint for cardiac tissue regeneration.
Acknowledgments
The authors would like to thank Jose A. Santiago for preparing cryo–SEM images and Leanne M. Olds for help with graphic illustrations. This research was supported by the National Institutes of Health (BMO, HL100014) and the American Heart Association (BMO, IRG5570039).
References
- 1.Roger VL, et al. Heart Disease and Stroke Statistics-2011 Update. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santiago JA, et al. Heterogeneous differentiation of human mesenchymal stem cells in response to extended culture in extracellular matrices. Tissue Eng. Part A. 2009;15:3911–3922. doi: 10.1089/ten.tea.2008.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kouris NA, et al. A nondenatured, noncrosslinked collagen matrix to deliver stem cells to the heart. Regen. Med. 2011;6:569–582. doi: 10.2217/rme.11.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akhyari P, et al. Myocardial tissue engineering: the extracellular matrix. Eur. J. Cardiothorac. Surg. 2008;34:229–241. doi: 10.1016/j.ejcts.2008.03.062. [DOI] [PubMed] [Google Scholar]
- 5.Rajala K, et al. Cardiac differentiation of pluripotent stem cells. Stem Cells Int. 2011;2011:383709. doi: 10.4061/2011/383709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baudino TA, et al. Cell patterning: interaction of cardiac myocytes and fibroblasts in three-dimensional culture. Microsc. Microanal. 2008;14:117–125. doi: 10.1017/S1431927608080021. [DOI] [PubMed] [Google Scholar]
- 7.van Laake L, et al. Extracellular matrix formation after transplantation of human embryonic stem cell-derived cardiomyocytes. Cell. Mol. Life Sci. 2010;67:277–290. doi: 10.1007/s00018-009-0179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Dev. Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wessels A, Markwald R. Cardiac morphogenesis and dysmorphogenesis. I. Normal development. Methods Mol. Biol. 2000;136:239–259. doi: 10.1385/1-59259-065-9:239. [DOI] [PubMed] [Google Scholar]
- 10.Drake CJ, et al. Avian vasculogenesis and the distribution of collagens I, IV, laminin, and fibronectin in the heart primordia. J. Exp. Zool. 1990;255:309–322. doi: 10.1002/jez.1402550308. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, et al. Type III collagen is crucial for collagen I fibrillogenesis and for normal cardiovascular development. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1852–1856. doi: 10.1073/pnas.94.5.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rugh R. The Mouse: Its Reproduction and Development. Burgess Publishing; 1968. [Google Scholar]
- 13.Vega-Hernández M, et al. FGF10/FGFR2b signaling is essential for cardiac fibroblast development and growth of the myocardium. Development. 2011;138:3331–3340. doi: 10.1242/dev.064410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little CD, et al. Distribution of laminin, collagen type IV, collagen type I, and fibronectin in chicken cardiac jelly/basement membrane. Anat. Rec. 1989;224:417–425. doi: 10.1002/ar.1092240310. [DOI] [PubMed] [Google Scholar]
- 15.Hurle JM, et al. Elastic extracellular matrix of the embryonic chick heart: an immunohistological study using laser confocal microscopy. Dev. Dynam. 1994;200:321–332. doi: 10.1002/aja.1002000407. [DOI] [PubMed] [Google Scholar]
- 16.Cohen ED, et al. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008;135:789–798. doi: 10.1242/dev.016865. [DOI] [PubMed] [Google Scholar]
- 17.Tidball JG. Distribution of collagens and fibronectin in the subepicardium during avian cardiac development. Anat. Embryol. 1992;185:155–162. doi: 10.1007/BF00185916. [DOI] [PubMed] [Google Scholar]
- 18.Woessner JF, Jr, et al. Collagen development in heart and skin of the chick embryo. Biochim. Biophys. Acta Protein Struct. 1967;140:329–338. doi: 10.1016/0005-2795(67)90473-4. [DOI] [PubMed] [Google Scholar]
- 19.Savolainen SM, et al. Histology atlas of the developing mouse heart with emphasis on E11.5 to E18.5. Toxicol. Pathol. 2009;37:395–414. doi: 10.1177/0192623309335060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, et al. Expression of extracellular matrix components fibronectin and laminin in the human fetal heart. Cell Struct. Funct. 1999;24:19–26. doi: 10.1247/csf.24.19. [DOI] [PubMed] [Google Scholar]
- 21.White JG, et al. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J. Cell Biol. 1987;105:41–48. doi: 10.1083/jcb.105.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samatham R, et al. Optical properties of mutant versus wild-type mouse skin measured by reflectance-mode confocal scanning laser microscopy (rCSLM). J. Biomed. Opt. 2008;13:041309. doi: 10.1117/1.2953195. [DOI] [PubMed] [Google Scholar]
- 23.Zipfel WR, et al. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 24.König K, et al. Multiphoton fluorescence lifetime imaging of 3D-stem cell spheroids during differentiation. Microsc. Res. Tech. 2011;74:9–17. doi: 10.1002/jemt.20866. [DOI] [PubMed] [Google Scholar]
- 25.Campagnola PJ, Loew LM. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat. Biotechnol. 2003;21:1356–1360. doi: 10.1038/nbt894. [DOI] [PubMed] [Google Scholar]
- 26.Abraham T, et al. Collagen matrix remodeling in 3-dimensional cellular space resolved using second harmonic generation and multiphoton excitation fluorescence. J. Struct. Biol. 2010;169:36–44. doi: 10.1016/j.jsb.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Le TT. Label-free molecular imaging of atherosclerotic lesions using multimodal nonlinear optical microscopy. J. Biomed. Opt. 2007;12:054007. doi: 10.1117/1.2795437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan Y, et al. Hybrid gel composed of native heart matrix and collagen induces cardiac differentiation of human embryonic stem cells without supplemental growth factors. J. Cardiovasc. Translat. Res. 2011;4:605–615. doi: 10.1007/s12265-011-9304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekine H, et al. Cardiac cell sheet transplantation improves damaged heart function via superior cell survival in comparison with dissociated cell injection. Tissue Eng. Part A. 2011;17:2973–2980. doi: 10.1089/ten.tea.2010.0659. [DOI] [PubMed] [Google Scholar]
- 30.Hillel AT, et al. Photoactivated composite biomaterial for soft tissue restoration in rodents and in humans. Sci. Translat. Med. 2011;3:93ra67. doi: 10.1126/scitranslmed.3002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCall JD, et al. Affinity peptides protect transforming growth factor beta during encapsulation in poly(ethylene glycol) hydrogels. Biomacromolecules. 2011;12:1051–1057. doi: 10.1021/bm101379v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collier JH, Segura T. Evolving the use of peptides as components of biomaterials. Biomaterials. 2011;32:4198–4204. doi: 10.1016/j.biomaterials.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberti K, et al. Functional immobilization of signaling proteins enables control of stem cell fate. Nat. Meth. 2008;5:645–650. doi: 10.1038/nmeth.1222. [DOI] [PubMed] [Google Scholar]
- 34.Martino MM, et al. Engineering the growth factor microenvironment with fibronectin domains to promote wound and bone tissue healing. Sci. Translat. Med. 2011;3:100ra189. doi: 10.1126/scitranslmed.3002614. [DOI] [PubMed] [Google Scholar]
- 35.Kraehenbuehl TP, et al. Three-dimensional extracellular matrix-directed cardioprogenitor differentiation: systematic modulation of a synthetic cell-responsive PEG-hydrogel. Biomaterials. 2008;29:2757–2766. doi: 10.1016/j.biomaterials.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 36.Wall ST, et al. Biomimetic matrices for myocardial stabilization and stem cell transplantation. J. Biomed. Mater. Res. Part A. 2010;95A:1055–1066. doi: 10.1002/jbm.a.32904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobel S, Lutolf MP. Biomaterials meet microfluidics: building the next generation of artificial niches. Curr. Opin. Biotechnol. 2011;22:690–697. doi: 10.1016/j.copbio.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Yang F, et al. Combinatorial extracellular matrices for human embryonic stem cell differentiation in 3D. Biomacromolecules. 2010;11:1909–1914. doi: 10.1021/bm100357t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jongpaiboonkit L, et al. An adaptable hydrogel array format for 3-dimensional cell culture and analysis. Biomaterials. 2008;29:3346–3356. doi: 10.1016/j.biomaterials.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zustiak SP, et al. Influence of cell-adhesive peptide ligands on poly(ethylene glycol) hydrogel physical, mechanical and transport properties. Acta Biomat. 2010;6:3404–3414. doi: 10.1016/j.actbio.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apostolovic B, Klok H-A. Copolymerization behavior of N-(2-hydroxypropyl)methacrylamide and a methacrylated coiled-coil peptide derivative. Biomacromolecules. 2010;11:1891–1895. doi: 10.1021/bm100533g. [DOI] [PubMed] [Google Scholar]
- 42.Jung JP, et al. Multifactorial optimization of endothelial cell growth using modular synthetic extracellular matrices. Integr. Biol. 2011;3:185–196. doi: 10.1039/c0ib00112k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medalia O, Geiger B. Frontiers of microscopy-based research into cell-matrix adhesions. Curr. Opin. Cell Biol. 2010;22:659–668. doi: 10.1016/j.ceb.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Plitzko JM, et al. Correlative cryo-light microscopy and cryo-electron tomography: from cellular territories to molecular landscapes. Curr. Opin. Biotechnol. 2009;20:83–89. doi: 10.1016/j.copbio.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Kukulski W, et al. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raspanti M, et al. Structural aspects of the extracellular matrix of the tendon: an atomic force and scanning electron microscopy study. Arch. Histol. Cytol. 2002;65:37–43. doi: 10.1679/aohc.65.37. [DOI] [PubMed] [Google Scholar]
- 47.Graham HK, et al. Tissue section AFM: in situ ultrastructural imaging of native biomolecules. Matrix Biol. 2010;29:254–260. doi: 10.1016/j.matbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kassies R, et al. Combined AFM and confocal fluorescence microscope for applications in bio-nanotechnology. J. Microsc. 2005;217:109–116. doi: 10.1111/j.0022-2720.2005.01428.x. [DOI] [PubMed] [Google Scholar]
- 49.Chawla K, et al. Biodegradable and biocompatible synthetic saccharide–peptide hydrogels for three-dimensional stem cell culture. Biomacromolecules. 2011;12:560–567. doi: 10.1021/bm100980w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergmans L, et al. Microscopic observation of bacteria: review highlighting the use of environmental SEM. Int. Endod. J. 2005;38:775–788. doi: 10.1111/j.1365-2591.2005.00999.x. [DOI] [PubMed] [Google Scholar]
- 51.Bird DK, et al. Simultaneous two-photon spectral and lifetime fluorescence microscopy. Appl. Opt. 2004;43:5173–5182. doi: 10.1364/ao.43.005173. [DOI] [PubMed] [Google Scholar]
- 52.Tuer AE, et al. Nonlinear optical properties of type I collagen fibers studied by polarization dependent second harmonic generation microscopy. J. Phys. Chem. B. 2011;115:12759–12769. doi: 10.1021/jp206308k. [DOI] [PubMed] [Google Scholar]
- 53.Ishihara A, et al. Photoactivation of caged compounds in single living cells: an application to the study of cell locomotion. Biotechniques. 1997;23:268–274. doi: 10.2144/97232st01. [DOI] [PubMed] [Google Scholar]
- 54.Mavrakis M, et al. Fluorescence imaging techniques for studying Drosophila embryo development. Curr. Protoc. Cell Biol. Chapter. 2008;4:18. doi: 10.1002/0471143030.cb0418s39. [DOI] [PubMed] [Google Scholar]
- 55.Basu S, et al. Multiphoton excited fabrication of collagen matrixes cross-linked by a modified benzophenone dimer: bioactivity and enzymatic degradation. Biomacromolecules. 2005;6:1465–1474. doi: 10.1021/bm049258y. [DOI] [PubMed] [Google Scholar]
- 56.Skala MC, et al. Combined hyperspectral and spectral domain optical coherence tomography microscope for noninvasive hemodynamic imaging. Opt. Lett. 2009;34:289–291. doi: 10.1364/ol.34.000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maslov K, et al. Optical-resolution photoacoustic microscopy for in vivo imaging of single capillaries. Opt. Lett. 2008;33:929–931. doi: 10.1364/ol.33.000929. [DOI] [PubMed] [Google Scholar]
- 58.Insana MF, Hall TJ. A method for characterizing soft tissue microstructure using parametric ultrasound imaging. Prog. Clin. Biol. Res. 1991;363:241–256. [PubMed] [Google Scholar]
- 59.Lippincott-Schwartz J, Manley S. Putting super-resolution fluorescence microscopy to work. Nat. Methods. 2009;6:21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen PK, et al. Imaging: guiding the clinical translation of cardiac stem cell therapy. Circ. Res. 2011;109:962–979. doi: 10.1161/CIRCRESAHA.111.242909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balu M, et al. Effect of excitation wavelength on penetration depth in nonlinear optical microscopy of turbid media. J. Biomed. Opt. 2009;14:010508. doi: 10.1117/1.3081544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marsh P, et al. Practical implementation of adaptive optics in multiphoton microscopy. Opt. Expr. 2003;11:1123–1130. doi: 10.1364/oe.11.001123. [DOI] [PubMed] [Google Scholar]
- 63.Tang S, et al. Design and implementation of fiber-based multiphoton endoscopy with microelectromechanical systems scanning. J. Biomed. Opt. 2009;14:034005. doi: 10.1117/1.3127203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng H. Bioimage informatics: a new area of engineering biology. Bioinformatics. 2008;24:1827–1836. doi: 10.1093/bioinformatics/btn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Linkert M, et al. Metadata matters: access to image data in the real world. J. Cell Biol. 2010;189:777–782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moore J, et al. Open tools for storage and management of quantitative image data. Methods Cell Biol. 2008;85:555–570. doi: 10.1016/S0091-679X(08)85024-8. [DOI] [PubMed] [Google Scholar]
- 67.Vokes MS, Carpenter AE. Using CellProfiler for automatic identification and measurement of biological objects in images. Curr. Protoc. Mol. Biol. Chapter. 2008;14:17. doi: 10.1002/0471142727.mb1417s82. [DOI] [PubMed] [Google Scholar]
- 68.Rueden CT, Eliceiri KW. Visualization approaches for multidimensional biological image data. Biotechniques. 2007;43:33–36. doi: 10.2144/000112511. [DOI] [PubMed] [Google Scholar]
- 69.Xie Z, et al. Laser-scanning optical-resolution photoacoustic microscopy. Opt. Lett. 2009;34:1771–1773. doi: 10.1364/ol.34.001771. [DOI] [PubMed] [Google Scholar]
- 70.Hendrix MJC. Localization of collagen types in the embryonic heart and aorta using immunohistochemistry. Perspect. Cardiovasc. Res. 1981;5:213–225. [Google Scholar]
- 71.Speiser B, et al. The extracellular-matrix in human myocardium. 1. Collagens I, III, IV, and VI. Cardioscience. 1991;2:225–232. [PubMed] [Google Scholar]
- 72.Borg TK, et al. Changes in the distribution of fibronectin and collagen during development of the neonatal rat-heart. Coll. Relat. Res. 1982;2:211–218. doi: 10.1016/s0174-173x(82)80015-0. [DOI] [PubMed] [Google Scholar]
- 73.Niederreither K, et al. Coordinate patterns of expression of type I and III collagens during mouse development. Matrix Biol. 1995;14:705–713. doi: 10.1016/s0945-053x(05)80013-7. [DOI] [PubMed] [Google Scholar]
- 74.Samuel JL, et al. Expression of fibronectin during rat fetal and postnatal development: an in situ hybridisation and immunohistochemical study. Cardiovasc. Res. 1994;28:1653–1661. doi: 10.1093/cvr/28.11.1653. [DOI] [PubMed] [Google Scholar]
- 75.Bouchey D, et al. Distribution of connective tissue proteins during development and neovascularization of the epicardium. Cardiovasc. Res. 1996;31:E104–E115. [PubMed] [Google Scholar]
- 76.Speiser B, et al. The extracellular-matrix in human cardiac tissue. 2. Vimentin, laminin, and fibronectin. Cardioscience. 1992;3:41–49. [PubMed] [Google Scholar]
- 77.Davis LA, et al. Embryonic heart mesenchymal cell migration in laminin. Dev. Biol. 1989;133:37–43. doi: 10.1016/0012-1606(89)90294-7. [DOI] [PubMed] [Google Scholar]
- 78.Price RL, et al. Ultrastructural localization of laminin on in vivo embryonic, neonatal, and adult rat cardiac myocytes and in early rat embryos raised in whole-embryo culture. J. Histochem. Cytochem. 1992;40:1373–1381. doi: 10.1177/40.9.1506674. [DOI] [PubMed] [Google Scholar]
- 79.Harvey RP. Patterning the vertebrate heart. Nat. Rev. Genet. 2002;3:544–556. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]