Abstract
Domoic acid is an algal-derived seafood toxin that functions as a glutamate agonist and exerts excitotoxicity via overstimulation of glutamate receptors (AMPA, NMDA) in the central nervous system (CNS). At high (symptomatic) doses, domoic acid is well-known to cause seizures, brain lesions and memory loss; however, a significant knowledge gap exists regarding the health impacts of repeated low-level (asymptomatic) exposure. Here, we investigated the impacts of low-level repetitive domoic acid exposure on gene transcription and mitochondrial function in the vertebrate CNS using a zebrafish model in order to: 1) identify transcriptional biomarkers of exposure; and 2) examine potential pathophysiology that may occur in the absence of overt excitotoxic symptoms. We found that transcription of genes related to neurological function and development were significantly altered, and that asymptomatic exposure impaired mitochondrial function. Interestingly, the transcriptome response was highly-variable across the exposure duration (36 weeks), with little to no overlap of specific genes across the six exposure time points (2, 6, 12, 18, 24, and 36 weeks). Moreover, there were no apparent similarities at any time point with the gene transcriptome profile exhibited by the glud1 mouse model of chronic moderate excess glutamate release. These results suggest that although the fundamental mechanisms of toxicity may be similar, gene transcriptome responses to domoic acid exposure do not extrapolate well between different exposure durations. However, the observed impairment of mitochondrial function based on respiration rates and mitochondrial protein content suggests that repetitive low-level exposure does have fundamental cellular level impacts that could contribute to chronic health consequences.
Keywords: domoic acid, chronic toxin exposure, microarray, subclinical effects
Introduction
Domoic acid is a small neurotoxic molecule that is naturally-produced by phytoplankton and accumulates in seafood products during harmful algal blooms (HABs) (Wright et al., 1989). At high doses, it is responsible for the human neurotoxic illness known as amnesic shellfish poisoning (ASP), with symptoms that include seizures, memory loss, coma, and death (Quilliam & Wright, 1989). The toxin was first identified in 1987 when over 100 people became ill and four died after consuming domoic acid-contaminated mussels (Perl et al., 1990). Domoic acid has since been shown to severely impact seabirds and marine mammals via transfer through planktivorous prey (Scholin et al., 2000; Work et al., 1993). For humans, risks of exposure to high levels of domoic acid are managed by testing shellfish and regulating harvests based on toxin loads (≥20 μg domoic acid/g = harvest closure); however, there are no regulatory guidelines in place for protection from long-term low-level repetitive exposure (< 20 μg domoic acid/g = harvest open) due to a lack of knowledge regarding the health impacts of chronic or episodic low level domoic acid exposure (Lefebvre & Robertson, 2010).
The knowledge gap regarding health impacts of low-level repetitive exposure to naturally-produced algal toxins, such as domoic acid, is particularly alarming in light of the increasing emergence, magnitude and frequency of domoic acid-producing HABs globally – a pattern that is predicted to worsen with continued climate change (Van Dolah, 2000). Of particular concern are coastal and tribal communities that rely heavily on seafood products as a major food source. For example, in the Pacific Northwest region of the US, the Quinault Tribe has a strong cultural reliance on razor clams for subsistence and ceremonial harvests (Lefebvre & Robertson, 2010). These clams are known to retain low levels of domoic acid for up to a year after a HAB and are eaten on a year-round basis in these communities, thereby presenting a risk of low level repetitive exposure to the consumer (Wekell et al., 1994). Further risks to the developing fetus or nursing offspring have also been identified in naturally-exposed marine mammals and in laboratory studies with rodent and primate models (Gulland et al., 2002; Levin et al., 2006).
Domoic acid functions as a glutamate agonist and exerts excitotoxicity via overstimulation of glutamate receptors (AMPA, NMDA) in the central nervous system (CNS) (Berman & Murray, 1997). Domoic acid-mediated increases in glutamatergic activation may alter cellular transcriptional responses that if gene product formation follows transcriptional changes, would lead to changes in synapse structure and function, dendrite and nerve terminal integrity, and may produce neuronal injury. Changes in synaptic function and loss of select glutamatergic neurons, or their synapses, occurring during neurodevelopment or in the aged may be particularly deleterious for normal CNS function (Morrison & Hof, 1997). Potential health risks of chronically elevated glutamatergic activity, like those that might result from chronic/episodic DA exposure, are also supported by the studies of a transgenic mouse model selectively overexpressing neuronal glutamate dehydrogenase (Glud1). Bao et al. (2009) and Wang et al. (2010) demonstrated that chronic moderately-elevated synaptic glutamate in the Glud1 mice leads to reductions in glutamate synapses in specific (vulnerable) brain regions, with reductions in glutamate synapses increasing with advancing age (Bao et al., 2009). Notably, these decreases in glutamate synapses were associated with upregulation of cellular pathways associated with nervous system development, neuronal growth, and synaptic transmission, as well as oxidative stress, cell injury, and inflammation, indicating apparent compensatory responses to stress that promoted growth of neuronal processes (Wang et al., 2010).
In the present study, we investigated the impacts of low-level repetitive domoic acid exposure on gene transcripton and mitochondrial function in the vertebrate CNS using the zebrafish (Danio rerio) laboratory model in order to 1) identify transcriptional biomarkers of exposure and 2) examine potential pathophysiological impacts that may occur in the absence of overt excitotoxic behaviors. We examined temporal patterns in the transcriptome response over a 36-week domoic acid exposure period to determine whether asymptomatic exposure elicited transcriptional changes in genes related to neuronal damage and compensatory cellular responses in the CNS.
Materials and methods
Zebrafish & chronic domoic acid exposures
Wild-type zebrafish (Danio rerio, AB strain), approximately 5 months of age, were obtained from Oregon State University (Sinnhuber Aquatic Research Laboratory Corvallis, OR). Fish were maintained at the Northwest Fisheries Science Center (NWFSC, Seattle, WA) in a ZebTec stand-alone recirculating and continuously-monitored zebrafish rack system with UV sterilizer (Techniplast, Exton, PA). Water temperature was maintained at 26°C, and fish were kept on a 12:12 hr light:dark cycle and fed daily with BioVita fish feed (Bio-Oregon, Longview, WA). Starting at ~7 months of age, zebrafish were repeatedly exposed to asymptomatic doses of domoic acid via intracoelomic injection (10 μL volume delivered with 33-gauge needle in custom-made auto-injector from Hamilton®) for 36 weeks. Toxin and vehicle (PBS) injections were given once a week (dose= 0.31 ± 0.03 mg domoic acid/g fish) for the first 6 weeks, then once every 2 weeks (dose = 0.18 ± 0.02 mg domoic acid/g fish) for the duration of the exposure period. After each injection, fish were observed for 30–45 minutes to note the instance of any neurobehavioral symptoms (e.g., circle- or spiral-swimming). Domoic acid doses of less than half of the previously-reported EC50 for zebrafish (0.86 mg domoic acid/g fish; (Lefebvre et al., 2009)) were chosen to ensure asymptomatic exposures. All domoic acid dose concentrations were verified via HPLC as described in Lefebvre et al. (2009). Fish (n= 9–10 per treatment group per time period) were harvested at six time points, including 2, 6, 12, 18, 24 and 36 weeks exposure for whole brain transcriptome analyses. Additionally, whole fish (n≈10 per treatment) at the 36-week exposure time point were preserved for histopathological analysis.
Histopathological examination
Zebrafish allocated for histopathological examination (n≈10 per treatment) at the 36-week exposure time point were examined grossly for any externally visible abnormalities, placed in an ice bath and exsanguinated via a tail bleed to obtain serum for another study. Fish were then carefully opened with a midline incision of the abdomen, and the entire fish was preserved in 20 mL Dietrich’s fixative (Fournie et al., 2000) at an ~1:20 (v/v) tissue to fixative ratio and fixed for three days on an orbital shaker. After fixation, the fish were rinsed in 2–3 changes of water and placed in 70% ethanol. Fish were bisected along the sagittal plane using a razor blade, cutting parallel to and on the left side of the spinal cord, before they were processed. The bisected fish were loaded into cassettes for processing, using a Shandon Hypercenter XP tissue processing center. The tissue processing protocol closely followed that specified by the Zebrafish International Research Center, University of Oregon, Eugene, OR (http://zfin.org/zirc/disMan/diseaseManual.php). Processed tissues were infiltrated with a 50:50 ratio of Fisher Paraplast PLUS tissue embedding medium and Surgipath Formula ‘R’ infiltrating and embedding paraffin. Tissues were then embedded in Surgipath Formula ‘R’ infiltrating and embedding paraffin, using a Shandon Embedding Center.
Fixed, paraffin-embedded fish were cut in the sagittal plane with a high-profile disposable blade in multiple, stepped-sections (~20 μm apart) at 5–7 μm thickness. Multiple sections of gills, eyes, brain, spinal cord and all internal organs were taken to ensure complete histopathological evaluation. Sections were routinely stained by hematoxylin and eosin, using the protocols described in “Diseases of Zebrafish in Research Facilities” (Zebrafish International Research Center). Sections were examined by light microscopy on a Nikon Optiphot microscope, using 4×, 10×, 20×, 40×, and 100× planachromat objectives; selected sections of the central nervous system were photographed by digital photomicroscopy on a Nikon Eclipse E600 microscope, using a SPOT™ camera and SPOT™ software. All major organs were examined for pathological changes, with a focus on alterations in components of the central nervous system. Any observed anomalies were scored by type, distribution within the tissue (e.g., focal/multifocal/diffuse) and severity, using a semiquantitative, ordinal scoring system detailed in Fairgrieve et al. (2005). Potential lesions/anomalies of interest were shared as needed with other fish histopathologists experienced in the examination of zebrafish (Dr. Trace Peterson and Dr. Michael Kent of Oregon State University) for confirmation and verification.
Brain dissections & RNA extraction
Zebrafish were euthanized via decapitation and whole brains were dissected from the fish as described in Lefebvre et al. (2009). Global transcriptome expression was quantified by microarray in three RNA replicates, each consisting of RNA from 3 pooled brains, for exposed and control treatments. The test group size (n= 9–10 per treatment per time point) was chosen based on a previous study (Lefebvre et al., 2009). Total RNA was isolated from zebrafish brains using the miRNeasy Mini Kit (Qiagen Inc., Valencia, CA) according to the vendor’s defined method, and stored at −70°C. RNA quantity (ng/μL) was determined by measuring the OD260 with a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA); RNA purity was assessed by measuring OD260/280 and OD260/230 ratios. RNA integrity (quality) was characterized using the Agilent RNA 6000 Nano Kit with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Only total RNA samples with appropriate size distribution, quantity, and OD260/280 as well as OD260/230 ratios of 1.8–2.1 were used for microarray-based analysis. Individual brain RNA samples were split, and half was used for microarray analyses and half retained for RT-PCR confirmation.
Global transcriptome profiling in brain
The RNA samples were labeled and prepared for hybridization onto a Zebrafish (V3) Gene Expression Microarray (Agilent Technologies, Inc. Santa Clara, CA) using the manufacturer’s established protocols. Hybridization and washing of these arrays was accomplished using HS 400 Pro hybridization and wash stations (Tecan Systems, Inc., San Jose, CA) and scanned using an Agilent DNA Microarray Scanner (Agilent Technologies, Inc. Santa Clara, CA), according to the manufacturer’s established standard protocol. In total, 36 microarrays were used, including 18 for the control group and 18 for the chronic exposure group. Separate pools of RNA, each consisting of three individuals, were hybridized to each array resulting in three biological replicates (a total of nine individuals) per treatment group at each time point.
Microarray analyses
Raw microarray data were generated with the Agilent Feature Extraction image analysis software (Agilent), and further processed and analyzed with tools from Bioconductor (Gentleman et al., 2004). Data were normalized using the BioconductorAgi4644Pre-Process package (Smyth, 2004); additionally, the normexp option for background adjustment and quantile normalization was used for the between array normalization step. Using the normalized data, genes with significant evidence for differential expression were identified using the limma package. The limma methodology calculates a P-value for each gene using a modified t-test in conjunction with an empirical Bayes method to moderate the standard errors of the estimated log-fold changes. This method of detecting differentially expressed genes draws strength across genes for more robust and accurate detection of differentially expressed genes; such an adjustment has been shown to avoid an excess of false positives when identifying differentially expressed genes (Allison et al., 2006). Using the P-values from limma, the Bioconductor package p.adjust (Benjamini & Hochberg, 1995) was used to estimate the false discovery rate associated with the list of differentially expressed genes. This methodology allows us to address the multiple testing problem without resorting to an excessively conservative approach that controls the family-wise error, such as a Bonferroni correction. Comparative analyses on bio-functions of the differentially transcribed genes were conducted using Ingenuity PathwayAnalysis (IPA) software (Ingenuity Systems, Redwood City, CA, USA; Build Version 242990, Content Version 12710793 (Release date 2012-05-07)). IPA uses a Fischer’s exact test to calculate the P-values. All microarray data are available through the NCBI Gene Expression Omnibus database (accession #: GSE34716).
Quantitative RT-PCR analyses
Six neurodevelopment-related genes belonging to Biological Functions categories; Nervous system development and function, Neurological disease, Cell death, Cellular function and maintenance, and Cellular growth and proliferation were further validated by RT-PCR in samples from all time points (6, 12, 18, 24 and 36 weeks). Total RNA was isolated from individual zebrafish brains (n= 9 per treatment per time point). RT-PCR was performed on pooled samples as described for microarray analyses (i.e., n=3 brains per pooled sample, yielding n=3 independent samples per treatment per time point). Briefly, reverse transcription was performed according to the manufacturer’s established protocol using total RNA and the SuperScriptH III First-Strand Synthesis System (Invitrogen, Carlsbad, CA.). For gene expression measurements, 2 μL of cDNA were included in a PCR reaction (12 μL final volume) that consisted of the ABI inventoried TaqManH Gene Expression Assays mix (Applied Biosystems Inc., Foster City, CA), or customized forward and reverse primers, probes and TaqMan Gene Expression Master Mix (Applied Biosystems Inc., Foster City, CA). The PCR primers and the dual-labeled probes for the genes were designed using ABI Primer Express v.1.5 software (Applied Biosystems Inc., Foster City, CA). Amplification and detection of PCR amplicons were performed with the ABI PRISM 7900 system (Applied Biosystems Inc., Foster City, CA) with the following PCR reaction profile: 1 cycle of 95°C for 10 min, 40 cycles of 95°C for 30 sec, and 60°C for 1 min. Beta-actin 1 amplification plots derived from serial dilutions of an established reference sample were used to create a linear regression formula in order to calculate expression levels, and Beta-actin 1 gene expression levels were utilized as an internal control to normalize the data.
Western blotting of oxidative damage & mitochondrial proteins
In situ mitochondrial respiration was measured in whole brains of control and exposed zebrafish at 18 weeks of chronic asymptomatic exposure. Fish were euthanized via decapitation and whole brains were dissected and either placed in ice-cold respiration buffer for mitochondrial respiration (n=6 per group) or homogenized for western blotting (n=11 per group) of protein carbonyls and electron transport chain proteins as described previously (Siegel et al., 2012). Briefly, homogenates were diluted 1:25 in lysis buffer containing protease inhibitors, combined 1:1 with Laemli buffer and separated on a 4–20% gradient gel. Proteins were transferred to nitrocellulose membrane and Ponceau stained to visualize protein loading and immunoblotted as follows: complex II 30kDa subunit (1 hr 1:1K Mitosciences #203 in 5% NFDM), complex IV subunit IV (1 hr 1:1K Mitosciences #407 in 5% NFDM), and protein carbonyls (according to Millipore kit S7150). Secondary Ab from Cell Signaling (#7076, 1:10K in 1% NFDM) was conjugated to horseradish peroxidase. Blots were developed in Western C chemiluminescent developer (BioRad, Hercules, CA) and visualized with a ChemiDoc imaging system. Band densities were quantified with Quantity One software and normalized to total lane Ponceau staining.
Quantification of mitochondrial respiration
Mitochondrial respiration was measured in whole brain homogenates in respiration buffer (210 mM sucrose, 0.5 mM EGTA, 3 mM MgCl2, 10 mM KH2PO4, 20 mM HEPES, 20 mM taurine, 50 mM K-MES, 1 g/L BSA, pH=7.1) using a clark-type electrode in an Oxygraph 2K (Oroboros Instruments, Austria). Multiple respiration states were measured using the following substrate inhibitor combinations: proton leak (state 4; 10mM glutamate/5mM pyruvate/2mM malate, without ADP), state 3 (state 4 conditions plus 1mM ADP with and without 10 mM succinate), fully uncoupled (ETS, 1 μM CCCP) or complex IV (0.5mM N,N,N′,N′-tetramethyl-ρ-phenylenediamine (TMPD), 2mM ascorbate, 2.5μm antimycin A). Potassium cyanide (2μM) was used to confirm complex IV-specific activity. The amount of oxygen consumed was calculated by assuming the O2 solubility in media to be 0.920 and by calibrating initial oxygen concentration in the buffer for each experiment and correcting for pressure, temperature and instrumental oxygen consumption as described by Gnaiger et al. (2009).
Results
Chronic low-level domoic acid exposure did not cause neurobehavioral excitotoxicity or visible histopathological lesions
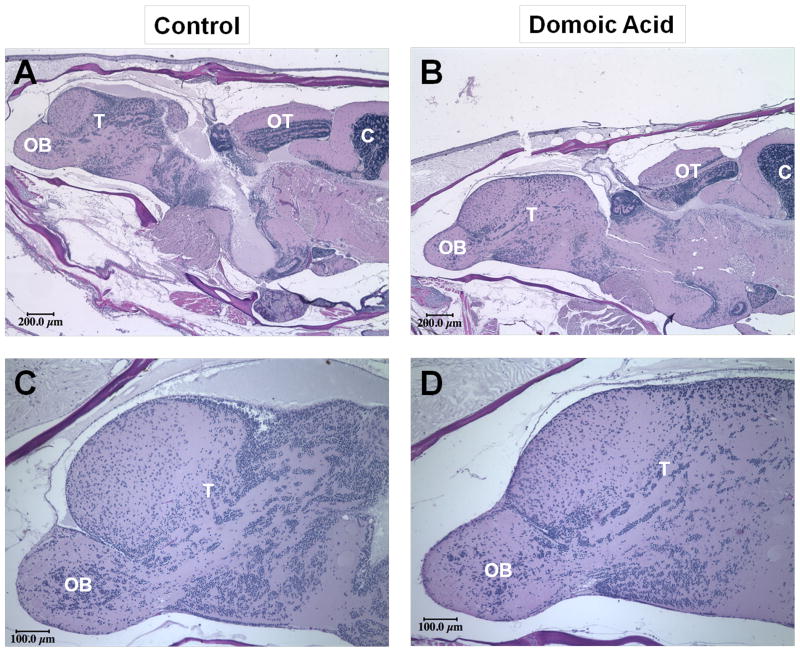
The domoic acid exposure regimen used here did not produce any visible signs of neurobehavioral excitotoxicity (i.e., circle- or spiral-swimming) when observed over a 30–45 minute period following each injection, consistent with an asymptomatic exposure regimen (data not shown). Because domoic acid is well-known to produce excitotoxic neuron death and histopathological lesions in glutamate-rich brain areas, we examined whether there was evidence of lesions in sagitally-sectioned brains of control and exposed zebrafish from the 36-week exposure time point. Semi-quantitative scoring of the brain and spinal cord revealed no visible histological differences between control and exposed fish (Fig. 1). All other major organ systems were also examined and appeared normal (data not shown). Consistent with our histopathological analysis, microarray analysis of gfap gene expression, a marker of neuroinflammation and astrocyte activation, revealed no significant changes (Table 1).
Figure 1.
Representative H&E-stained sagittal sections of control (A, C) and domoic acid-exposed (B, D) zebrafish at 36 weeks. Whole brain images (A, B) include olfactory bulb (OB), telencephalon (T), optic tectum (OT) and anterior cerebellum (C) (4×); panels (C) and (D) depict olfactory bulb (OB) and telencephalon (T) (10×). The telencephalon phylogenetically develops into the brain region that includes the hippocampus in higher-order organisms.
Table 1.
Gene transcription data for gfap (glial fibrillary acidic protein; log2(fold-change), p value), a marker of astrocyte activation and neuroinflammation, as measured by microarray. Transcriptional changes are shown for animals exposed to domoic acid relative to the time-matched vehicle controls. Note that all array probes for this gene are non-significant, highlighting the absence of gross neuroinflammation and corroborating the histopathological assessment presented in Figure 1.
| NCBI GenBank ID | Probe ID |
gfap Log2(fold-change) (p value)
|
|||||
|---|---|---|---|---|---|---|---|
| 2 weeks | 6 weeks | 12 weeks | 18 weeks | 24 weeks | 36 weeks | ||
| NM_131373 | A_15_P107505 | 0.33 (0.19) | 0.02 (0.93) | 0.14 (0.56) | 0.45 (0.07) | 0.13 (0.59) | −0.14 (0.57) |
| NM_131373 | A_15_P131871 | 0.02 (0.77) | −0.03 (0.65) | 0.03 (0.64) | 0.05 (0.47) | −0.03 (0.67) | −0.07 (0.30) |
| AY397679 | A_15_P551122 | 0.00 (0.90) | 0.01 (0.78) | −0.01 (0.71) | −0.02 (0.30) | 0.00 (0.88) | −0.01 (0.62) |
| NM_131373 | A_15_P658936 | 0.05 (0.57) | −0.01 (0.91) | 0.04 (0.59) | 0.19 (0.02) | 0.03 (0.73) | −0.05 (0.55) |
Chronic low-level domoic acid exposure led to temporally variable gene transcription responses
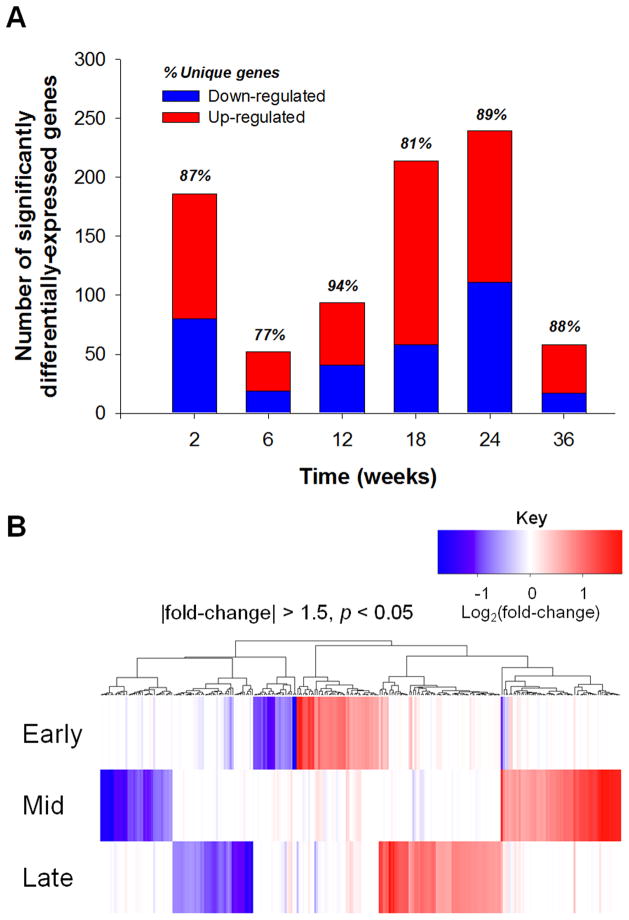
The number of significantly-differentially transcribed genes (±1.5-fold-change compared to control, p<0.05) ranged from 52–239 at each time point, with the majority of genes having increased transcription (60–64%) (Fig. 2A). Notably, the vast majority (77–94%) of differentially-transcribed genes were unique to a particular time point, and very few individual genes were differentially-transcribed across multiple time points (Fig. 2A). In order to characterize broad temporal patterns in the transcriptome response to chronic domoic acid exposure, we performed Gene Set Analysis (GSA) on grouped time points (Early = 2 and 6 weeks; Mid = 12 and 18 weeks; Late = 24 and 36 weeks); time points were grouped in order to achieve improved statistical power for GSA. The heatmap in Figure 2B illustrates these grouped time points, underscoring the temporally-dynamic gene transcription profile over the duration of domoic acid exposure, suggesting that the gene transcriptome response to chronic domoic acid exposure depends largely on the exposure duration.
Figure 2.
(A) Number of genes significantly differentially-regulated throughout duration of chronic exposure to asymptomatic domoic acid compared to time-matched vehicle controls. The proportion of up- and down-regulated genes is shown by red and blue bars, respectively. Percentage of genes unique to each time point is listed above each bar. (B) Heatmap depicting clustered gene transcription responses (±1.5-fold, p<0.05, 265 probes) to chronic domoic acid exposure across exposure duration; Early = 2 and 6 week time points, Mid = 12 and 18 week time points, Late = 24 and 36 week time points.
While GSA revealed a broad range of significantly-altered Biological Processes & Molecular Functions (as defined by Gene Ontology), no clear temporal patterns emerged from this analysis (data not shown). That is, we were unable to detect any transcriptional effect aligned with our expectation that early response (2 and 6 weeks exposure duration) would transition into a consistent response profile later in the chronic exposure paradigm (12, 18, 24 and 36 weeks).
In order to gain a more function-based understanding of pathways and biological functions affected by chronic domoic acid exposure, we also performed Ingenuity Pathway Analysis (IPA) of gene expression at each individual exposure duration time point. Our primary hypothesis entering the study was that chronic asymptomatic domoic acid exposure would alter the transcription of genes associated with cellular death/damage and neurological function/development. Ingenuity Pathway Analysis identified 9 – 70 significant Biological Function categories at each time point, with categories defined here as containing ≥3 genes (data not shown). The Bio Functions most relevant to our hypothesis, i.e., Nervous system development and function, Neurological disease, Cell death, Cellular growth and proliferation, and Cellular function and maintenance, are shown in Figure 3 (full gene list in Table 2). Notably, these IPA Bio Functions were significantly-altered by domoic acid exposure at five to six of the six exposure duration time points (Fig. 3). However, while there is overlap of individual genes across Bio Functions within each time point (~73–97% overlap between Bio Function genes within individual time points), there is very little overlap of individual genes across individual time points (e.g., only ~2 genes within a Bio Function category appear at two or more time points). This further supports the finding that the transcriptome response to chronic domoic acid is highly temporally variable and dynamic.
Figure 3.
Significantly altered (p<0.05) Biological Functions (from Ingenuity Pathway Analysis) associated with cellular and neurological health, function and development. Bubble diameter corresponds to –log(p value) for functional categories and time points possessing ≥3 genes; # indicates not significant and/or <3 genes within the category. Scale provides reference for relative bubble diameter.
Table 2.
All genes falling into the Bio Function categories shown in Figure 3 at all time points.
| Category | Genes | |
|---|---|---|
| Cell Death (92) | Up-regulated: | ADIPOQ, AGGF1, APOE, APP, ARG1, ATG3 (includes EG:171415), B2M, B4GALNT1, BCL2L1, BNIP3, RFWD2, CCNI, CCT7, CD53, CD74, CDC73 (includes EG:214498), CHFR, CORO1A, COX5A, includes EG:12858), CTSB, CTSD, CTSS, CYLD, DRAM1, DUSP2, ELMO1, EPO, FKBP5, GAS6, GMFG, GRN, HAND2, HBZ, HDAC5, AGT, HLA-DRB1, IRF1 (includes EG:16362), IRF8, JAG1, LCP1, LECT2, LGALS3, LGALS3BP, MCL1, MCM10 (includes EG:307126), MEF2C, MT2A, NFKBIA, NTRK3, NUDCD3, PPP1CA, PRKCD, PSME2, RBBP4, SOCS1, SOCS3, TAP2, TFDP1, TOP1, TRIM2, UCP2, USP2, VDAC2, WAS, WEE1 |
| Down-regulated: | AKT1, ANXA11, ATP2A1, AURKA, CALCB, CAV1, CCDC47, CGA, CHKA, CLCN3, DYRK1B, FAH, GH1, GRIA2, LCK, LIFR, NOD1, P4HB, POMC, PPID, PPM1L, PTGES, RFXANK, SH3BP5, SIRT2, TASP1, TSG101 | |
| Cellular Function and Maintenance (81) | Up-regulated: | ADIPOQ, AGT, APOE, APP, ATP6V1B2, B2M, B4GALNT1, BCL2L1, BNIP3, CACNB3, CD74, CORO1A, COX5A (includes EG:12858), CTSD, CTSS, CYLD, DGKA, DRAM1, DUSP2, ELMO1, EPO, GAS6, GPR12, GRN, HDAC5, HIVEP2, IRF1 (includes EG:16362), IRF8, JAG1, LCP1, LECT2, LGALS3, MCL1, Mcpt4, MGAT5, MT2A, NEO1, NFKBIA, NIN, NTRK3, PPP1CA, PRKCD, PSMB10, PSME2, RBBP4, RHCG, SLC9A3 (includes EG:105243), SOCS1, SOCS3, STK35, Tlr12, TOP1, TPM2, UCP2, UCP3, WAS |
| Down-regulated: | AKT1, ATP2A1, BCL11A, CALCB, CAV1, CGA, CLCN3, CREB3L2, CUBN, CYP4A11, GH1, GNB1, GORASP1, LCK, LIFR, NEFM, NOD1, PAK7, POMC, PPID, PPP1CA, SASH3, SLC25A47, SPC25 (includes EG:100144563), TSG101, ULK1 | |
| Cellular Growth and Proliferation (49) | Up-regulated: | ADIPOQ, AGT, APOE, APP, B2M, B4GALNT1, CD74, CDC73 (includes EG:214498), CORO1A, CTSB, CTSD, DUSP2, EPO, GAS6, GRN, HLA-DRB1, IRF1 (includes EG:16362), IRF8, JAG1, MCL1, MKNK1, MT2A, NFKBIA, NR1D1, PRKCD, PSMB10, PSMB9, RAD17 (includes EG:19356), SOCS1, SOCS3, TPM2, WAS |
| Down-regulated: | AKT1, CAV1, CD151, CGA, CHKA, DYRK1A, DYRK1B, GH1, IK, LCK, LIFR, PKP3, POMC, PTGES, RAB5C, SIRT2, TSG101 | |
| Nervous System Development and Function (41) | Up-regulated: | AGT, APOE, APP, B4GALNT1, BCL2L1, CACNB3, CTSF, EPO, FEV, FGFRL1, GAS6, GJA8, GNGT1, MT2A, NFKBIA, NR1D1, NRXN2, NTRK3, PAK4, PDE6G, RPE65, TRIM2 |
| Down-regulated: | AKT1, ALS2CR8, BCL11A, CAV1, CGA, CLCN3, DYRK1A, FZR1, GH1, GRIA2, LIFR, NEFM, PAK7, POMC, PPP1CA, RBP4, RTN4RL1, SIRT2, ULK1 | |
| Neurological Disease (62) | Up-regulated: | ADIPOQ, ALDH7A1, APOE, APP, ARG1, ATP1B1, ATP6V1B2, B2M, B4GALNT1, BCL2L1, CD74, CORO1A, CTSB, CTSD, CTSF, CTSS, ELMO1, EPO, GNGT1, GRN, HLA-DRB1, HNRNPU, IDH2, IRF1 (includes EG:16362), MGAT5, MKNK1, MT2A, NFKBIA, PCSK5, PDE6H, PSMB9, PSME1, RARS2, RPE65, SLC1A4, SOCS3, SRD5A1, TAP2, TPM2 |
| Down-regulated: | ACOX1, AKT1, ANKRD11, ATP2A1, BEST1, CAV1, CDC73 (includes EG:214498), CGA, CLCN3, CLEC3B, GH1, GRIA2, IMPDH1, MYH7, NEFM, PER2, PPID, PTGES, RNASET2, SEPP1, SRM, TASP1, TEAD1 | |
Real-time quantitative PCR confirms microarray gene transcription results
Six genes (gria2a, nrxn2a, appa, nfkbiab, bcl2L1, and pak7) were used for RT-PCR confirmation at all time points (2, 6, 12, 18, 24, 36 weeks). Eighty percent of the genes across all 36 comparisons agreed in fold-change direction.
Chronic low-level domoic acid exposure led to oxidative damage & mitochondrial dysfunction
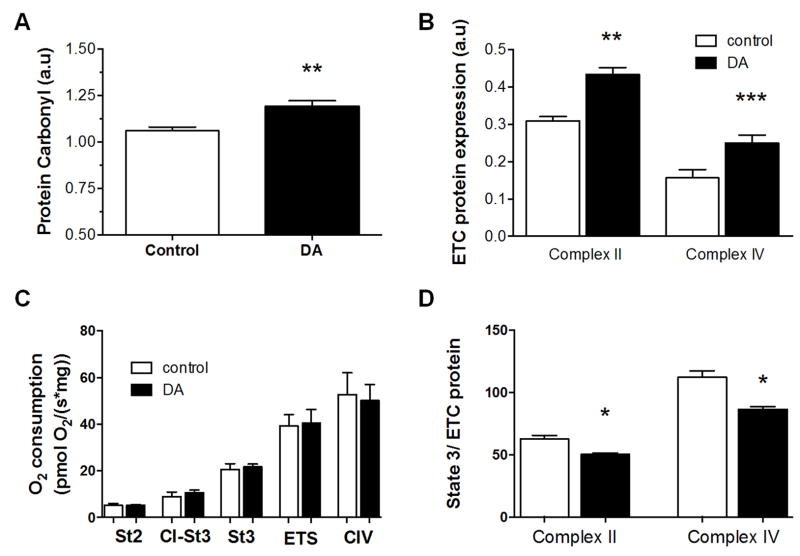
To test whether chronic domoic acid exposure resulted in changes in cell function, we measured oxidative damage to proteins, mitochondrial protein content, and mitochondrial respiration rates in the brains of zebrafish following 18 weeks of exposure to asymptomatic doses of domoic acid. Protein carbonyls (Fig. 4A), a measure of oxidative damage, were significantly elevated in whole brains. Electron transport chain (ETC) complex lI and complex lV subunit expression were significantly higher in the domoic acid exposed zebrafish brains (Fig. 4B). Despite the increase in mitochondrial proteins, there were no significant differences in mitochondrial respiration per unit tissue (Fig. 4C). However, maximum mitochondrial respiration rates normalized to complex IV and complex II protein expression (Fig. 4D) were significantly decreased in the brains of domoic acid-exposed fish compared to saline-exposed controls. These results suggest that chronic low-level exposure to domoic acid leads to mitochondrial dysfunction that is partially offset by increases in mitochondrial content.
Figure 4.
Comparison of oxidative damage and mitochondrial content and function in brains of zebrafish exposed for 18 weeks to domoic acid and time-matched vehicle controls. (A) Protein carbonyl levels were significantly higher in exposed fish compared to controls. (B) Expression of complex II (30 kD subunit) and complex IV (subunit IV) were both significantly higher in exposed fish compared to controls. (C) Oxygen flux per unit brain tissue at five different measures of mitochondrial respiration (St4 = state 4, C1-St3 = Complex I State 3, St3 = Complex I & II State 3, ETS = Fully uncoupled respiration, and CIV = Complex IV) was not significantly different between control and exposed fish. (D) Maximal state 3 respiration (ADP, plus succinate and glutamate/malate) was normalized to complex ll and complex IV protein content. Flux per content was reduced in exposed fish compared to controls. n=6–11 for control and exposed treatments, mean ± SEM. *-P<0.05, **-P<0.01, ***-P<0.0001 relative to controls.
Chronic low-level domoic acid exposure led to unique transcriptome responses compared to an elevated glutamate release exposure model
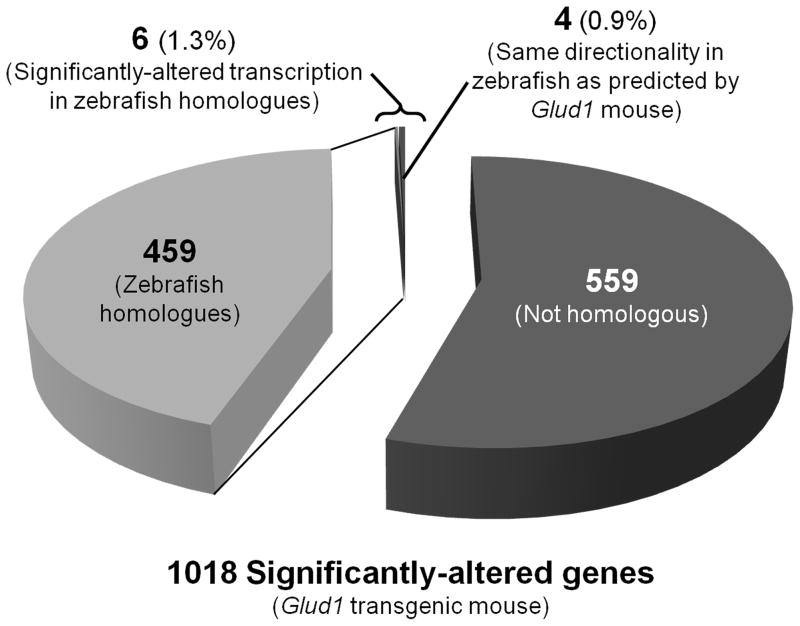
To test our hypothesis that chronic asymptomatic domoic acid exposure in zebrafish produces brain transcriptome-level changes consistent with chronic excess synaptic glutamate release, we compared the gene transcriptome changes in zebrafish with reported gene transcriptome changes in the hippocampi of Glud1 transgenic mice (Wang et al., 2010). This mouse model was developed in order to study the neurological consequences of excess glutamatergic stimulation, and constitutively over-expresses a copy of the glud1 gene, leading to moderate over-production of glutamate and increased (~30%) release of glutamate during synaptic transmission (Bao et al., 2009). For our analysis, we compared the significance and directionality of zebrafish/mouse homologues, as well as Biological Functions identified by IPA. We identified 459 zebrafish homologues of the 1018 Glud1 genes that were differentially-expressed compared to wild-type. Interestingly, only six of those zebrafish homologues were significantly differentially-regulated with chronic domoic acid exposure in zebrafish at any of the exposure duration time points; of those six genes, four were altered in the same direction as their Glud1 mouse counterparts (Fig. 5). Two Biological Functions, as identified by IPA, were found in common between the Glud1 mouse model analysis and our targeted analysis of the zebrafish brain transcriptome: Nervous System Function & Development; Cell Death. Genes differentially-transcribed at multiple domoic acid-exposure duration time points in adult zebrafish whole brain do not correspond to genes differentially-transcribed in the nine month-old glud1 mouse hippocampus.
Figure 5.
Comparison of transcriptome responses in zebrafish whole brain and Glud1 transgenic mouse hippocampus. Number of genes for each wedge is indicated. Percent of zebrafish homologues is listed parenthetically. Of the 459 zebrafish homologues identified, only 6 were significantly altered; of those 6 genes, 4 were regulated in the same direction as in the Glud1 hippocampus.
Discussion
In order to elucidate the neurological consequences of chronic exposure to low-level (asymptomatic) domoic acid doses, we conducted a transcriptome profiling analysis on the brains of zebrafish exposed for 2 – 36 weeks. We hypothesized that: 1) chronic asymptomatic domoic acid exposure would lead to a consistent gene transcription profile in the brains of exposed zebrafish; and 2) this profile would be similar to the gene transcription profile in the hippocampi of Glud1 transgenic mice, a model of chronic moderate glutamatergic over-stimulation (Bao et al., 2009; Wang et al., 2010). We found that genes and biological functions related to neurological function and development were significantly altered; however, the transcriptome response was highly-variable across exposure duration, with little to no overlap of specific genes across the six exposure duration time points. Additionally, while biological function categories were similar between exposed zebrafish and the Glud1 mouse model, less than 1% of individual genes overlapped between models.
Chronic low-level domoic acid exposure does not lead to a consistent transcriptome profile
We expected to identify early changes in gene transcription following 2 – 6 weeks of domoic acid exposure, leading to a consistent response at the later chronic time points. Instead, we found that the response was extremely variable and dynamic across all exposure duration time points. The number of differentially-transcribed genes unique to each time point ranged from 77–94% (Fig. 2A), with only three genes significantly-regulated at more than two time points. This shows that prolonged asymptomatic domoic acid exposure produced a continuum of diverse gene transcription responses.
This study is highly unique in its ability to evaluate the temporal nature of the transcriptome response because it assessed six time points across a duration that spanned a time period equivalent to one-third of the organism’s life. Prior studies have typically investigated gene transcription changes following only an acute exposure to symptomatic doses of domoic acid (Ananth et al., 2001; Ryan et al., 2005). Additionally, our previous work has shown that whole-brain transcriptional response to symptomatic doses of domoic acid differs markedly from the transcriptional response to asymptomatic exposure (Lefebvre et al., 2009). Our current results suggest that studies utilizing only a single domoic acid exposure duration may observe transcription profiles unique only to that particular time point, and may not extrapolate to other exposure durations.
Chronic low-level domoic acid exposure alters transcription of genes related to nervous system function & development
We hypothesized that chronic asymptomatic domoic acid exposure would lead to transcriptional changes in genes related to neurological homeostatic function and development based on 1) evidence that perturbation to the glutamate system is linked to several neurological disorders (Choi, 1988); and 2) a model of differential neural insult whereby subtle insults, similar to chronic asymptomatic domoic acid exposure, lead to ‘alerted’ microglia, which contribute to a neuro-supportive environment by release of neurotrophic factors (Hanisch & Kettenmann, 2007). This is in contrast to the cascade of more overt damage caused by full activation of microglia, which can ultimately lead to astrocyte activation and neuroinflammation. Notably, gfap (glial fibrillary acidic protein), a marker of astrocyte activation and neuroinflammation, was not significantly up-regulated at any of the time points (Table 1), highlighting the absence of overt neuroinflammation consistent with the unremarkable histopathology reported in Figure 1.
Ingenuity Pathway Analysis (IPA) of our datasets revealed several significantly altered transcriptional responses in genes categorized in Biological Functions relevant to our hypotheses (Fig. 3). Within the IPA Bio Function categories Nervous system development & function and Cell death, we examined six genes of interest, including gria2a, nrxn2a, pak7, appa, nfkbiab, and bcl2L1. At the 2-week time point, the AMPA glutamate receptor encoded by gria2 was down-regulated (Table 2). Notably, this was the only glutamate-related gene significantly altered by domoic acid exposure at any time point. Down-regulation of this receptor has been shown to occur following seizure activity (Huang et al., 2002), suggesting that the down-regulation of gria2a at 2 weeks is a short-term compensatory response to elevated glutamatergic activity.
Neurexin 2a gene (nrxn2a), which encodes a synaptic organization cell adhesion molecule, was up-regulated at 2 weeks (Table 2). Neurexins have been linked to Alzheimer’s Disease (Gauthier et al., 2011), and may play a role in maintaining mature neural networks (Rozic-Kotliroff & Zisapel, 2007); up-regulation at this time point suggests an alteration of synaptic signaling and/or neural disruption. A member of the PAK family of Ser/Thr protein kinases encoded by pak7, was down-regulated at the 36-week time point (Table 2). Pak7 is involved in neurite outgrowth (Dan et al., 2002) and its disruption suggests that synaptic trafficking may be altered (Strochlic et al., 2012) following prolonged exposure to domoic acid.
Transcriptional responses of a trio of genes involved in neuroprotective, anti-inflammatory, and apoptotic responses, appa, nfkbiab, and bcl2L1, were significantly up-regulated following 24 weeks of domoic acid exposure (Table 2). Amyloid precursor protein (APP) is a membrane protein that is metabolized to β-amyloid – the main component of Alzheimer’s Disease (AD) plaques (Klunk & Abraham, 1988). While the function of APP in the non-AD brain is unknown, it has been suggested to play a neuroprotective role under normal physiological conditions (Wentzell et al., 2012). Nfkbia, up-regulated 2.8-fold, encodes a cellular protein that inhibits the NF-kB transcription factor (Arenzana-Seisdedos et al., 1995), which contributes to a pro-inflammatory cascade when activated. A member of the Bcl-2 family, bcl2L1, was also up-regulated; the proteins in this family play a role in the regulation of apoptosis and are localized to the mitochondrial membrane (Plas & Thompson, 2002).
Chronic low-level domoic acid exposure causes oxidative damage, impairs mitochondrial function & may trigger compensatory mitochondrial biogenesis
As a glutamate agonist, domoic acid overexcites glutamatergic receptors and ultimately causes excess Ca2+ influx at acute doses, making mitochondria an obvious target for toxic insult due to their central role in Ca2+ homeostasis (Nicholls, 2009; Nijjar & Nijjar, 2000). In the present study, differential transcription of the bcl2L1 gene, involved in mitochondrial regulation of apoptosis described above, provides evidence of mitochondrial involvement in chronic asymptomatic domoic acid toxicity. Previous studies in mice have further demonstrated that oxidative stress-induced mitochondrial dysfunction is the underlying mechanism of domoic acid-mediated cognitive deficits after acute exposure (Lu et al., 2012). Elevated protein carbonyl levels in chronically-exposed zebrafish found in the present study suggest that oxidative stress also plays a role in toxicity. Additionally, enzymatic assays of the mitochondrial electron transport chain (complexes l-V) in isolated rat mitochondria showed marked concentration-dependent impairment in cardiac energetics following exposure to domoic acid (Vranyac-Tramoundanas et al., 2008); this is consistent with our findings of reduced mitochondrial-specific respiration in the brains of chronically-exposed zebrafish (Fig. 4D). Our findings of increased oxidative damage (Fig. 4A) coupled with reduced mitochondrial respiration (Fig. 4D) suggest that low-level repetitive domoic acid exposure also causes oxidative stress-related mitochondrial dysfunction. Interestingly, mitochondrial content was significantly higher in exposed zebrafish (Fig. 4B), suggesting that a compensatory mitochondrial biogenesis response may be initiated to meet energy demands in a living organism. Evidence for improvement in domoic acid-induced cognitive deficits by stimulation of estrogen receptor-α-mediated mitochondrial biogenesis signaling has been reported previously (Lu et al., 2012).
Chronic low-level domoic acid exposure leads to unique transcriptome profiles compared to the Glud1 transgenic mouse model
We anticipated that the whole-brain transcriptome profile in zebrafish chronically-exposed to asymptomatic domoic acid doses would be similar to the hippocampal transcriptome response in the Glud1 transgenic mouse model (Wang et al., 2010). This transgenic line transcribes extra copies of Glud1 (glutamate dehydrogenase 1) under the control of the promoter for neuron-specific enolase (Nse) in neurons. This contributes to increased glutamate release in striatal and hippocampal neurons compared to wild-type mice (Bao et al., 2009).
We compared transcriptome results from the brains of chronically-exposed zebrafish to hippocampi from nine-month old Glud1 mice (Wang et al., 2010), and identified transcriptional changes for genes from two IPA Bio Functions groups in both the present study and the Glud1 mouse study: 1) Nervous System Function & Development; and 2) Cell Death. However, individual gene transcription responses in zebrafish were different from those reported in the Glud1 mouse hippocampus (Fig. 5). Out of 459 zebrafish homologues to the 1,018 differentially-transcribed Glud1 mouse genes, only six genes (1.3% of zebrafish homologues) were significantly differentially-transcribed following domoic acid exposure, and only four (0.9% of homologues) were altered in the same direction as the Glud1 genes. This indicates that episodic domoic acid exposure leads to a toxicological response similar to one produced by chronically moderately-elevated glutamatergic activity at a biological function/pathway level, but not at the level of individual genes.
Conclusion
Chronic asymptomatic domoic acid exposure altered the transcription of genes in several IPA-identified Bio Functions related to nervous system function and neurodevelopment. The transcriptome response was highly dynamic over the 36 week exposure period, involving a varying list of differentially-transcribed genes and revealing that gene transcription responses are unique at different exposure durations. Finally, oxidative stress-related mitochondrial dysfunction appears to play a role in chronic low-level exposure similar to that shown in previous acute exposure studies. These results highlight the temporal variability of the biological response to chronic perturbation of the glutamate system, and demonstrate the potential for subclinical pathology with low-level environmental toxin exposure.
Highlights.
Low-level seafood toxin exposure alters gene transcription in the CNS.
Transcriptional responses to low-level exposure are highly dynamic over time.
Transcriptome alterations occur in genes related to nervous system function.
Low-level seafood toxin exposure impairs mitochondrial function.
Low-level seafood toxin exposure induces compensatory mitochondrial biogenesis.
Acknowledgments
Funding
This work was supported by an Ecology and Oceanography of Harmful Algal Blooms (ECOHAB) grant; the National Oceanic and Atmospheric Administration’s Oceans and Human Health Initiative; the National Institutes of Health [ES021930]; the National Institute of Environmental Health Sciences [P30ES007033]; and the National Science Foundation [DGE-0809125-006 to E.M.H. and OCE-1314088]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Oceanic and Atmospheric Administration.
The authors would like to thank Patricia Stapleton (University of Washington) for sample processing and RT-PCR analyses, Heather Weidenhoft for maintenance of test fish, Rudolph Stuppard and Dr. Shane Kruse (University of Washington) for help with mitochondrial content and function assays, and Dr. Walt Dickhoff and Dr. Linda Rhodes for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Emma M Hiolski, Email: ehiolski@ucsc.edu.
Preston S Kendrick, Email: psk39@u.washington.edu.
Elizabeth R Frame, Email: Elizabeth.Frame@kingcounty.gov.
Mark S Myers, Email: mark.s.myers@noaa.gov.
Theo K Bammler, Email: tbammler@u.washington.edu.
Richard P Beyer, Email: dbeyer@u.washington.edu.
Federico M Farin, Email: freddy@u.washington.edu.
Hui-wen Wilkerson, Email: jasminew@u.washington.edu.
Donald R Smith, Email: drsmith@ucsc.edu.
David J Marcinek, Email: dmarc@uw.edu.
Kathi A Lefebvre, Email: kathi.lefebvre@noaa.gov.
References
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature reviews Genetics. 2006;7(1):55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Ananth C, Thameem Dheen S, Gopalakrishnakone P, Kaur C. Domoic acid-induced neuronal damage in the rat hippocampus: changes in apoptosis related genes (bcl-2, bax, caspase-3) and microglial response. Journal of neuroscience research. 2001;66(2):177–90. doi: 10.1002/jnr.1210. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11592113. [DOI] [PubMed] [Google Scholar]
- Arenzana-Seisdedos F, Thompson J, Rodriguez MS, Bachelerie F, Thomas D, Hay RT. Inducible nuclear expression of newly synthesized I kappa B alpha negatively regulates DNA-binding and transcriptional activities of NF-kappa B. Molecular and cellular biology. 1995;15(5):2689–96. doi: 10.1128/mcb.15.5.2689. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=230499&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Pal R, Hascup KN, Wang Y, Wang WT, Xu W, Hui D, Agbas A, Wang X, Michaelis ML, Choi IY, Belousov AB, Gerhardt GA, Michaelis EK. Transgenic expression of Glud1 (glutamate dehydrogenase 1) in neurons: in vivo model of enhanced glutamate release, altered synaptic plasticity, and selective neuronal vulnerability. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29(44):13929–44. doi: 10.1523/JNEUROSCI.4413-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Mulitple Testing. Journal of the Royal Statistical Society. 1995;57 (1):289–300. [Google Scholar]
- Berman FW, Murray TF. Domoic acid neurotoxicity in cultured cerebellar granule neurons is mediated predominantly by NMDA receptors that are activated as a consequence of excitatory amino acid release. Journal of neurochemistry. 1997;69 (2):693–703. doi: 10.1046/j.1471-4159.1997.69020693.x. [DOI] [PubMed] [Google Scholar]
- Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988;1(8):623–34. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- Dan C, Nath N, Liberto M, Minden A. PAK5, a New Brain-Specific Kinase, Promotes Neurite Outgrowth in N1E-115 Cells. Molecular and Cellular Biology. 2002;22 (2):567–577. doi: 10.1128/MCB.22.2.567-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairgrieve WT, Masada CL, McAuley WC, Peterson ME, Myers MS, Strom MS. Accumulation and clearance of orally administered erythromycin and its derivative, azithromycin, in juvenile fall chinook salmon Oncorhynchus tshawytscha. Diseases of Aquatic Organisms. 2005;64:99–106. doi: 10.3354/dao064099. [DOI] [PubMed] [Google Scholar]
- Fournie J, Krol R, Hawkins W. Fixation of fish tissues. In: Ostrander G, editor. The Laboratory Fish, The handbook of Experimental Animals. New York, New York: Academic Press; 2000. pp. 569–578. [Google Scholar]
- Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, Lapointe M, Spiegelman D, Noreau A, Lafrenière RG, Fathalli F, Joober R, Krebs MO, DeLisi LE, Mottron L, Fombonne E, Michaud JL, Rouleau GA. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Human genetics. 2011;130(4):563–73. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5(10) doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnaiger E. Capacity of oxidative phosphorylation in human skeletal muscle: New perspectives of mitochondrial physiology. The International Journal of Biochemistry & Cell Biology. 2009;41:1837–1845. doi: 10.1016/j.biocel.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Gulland FM, Haulena M, Fauguier D, Langlois G, Lander ME, Zabka T, Duerr R. Domoic acid toxicity in Californian sea lions (Zalophus californianus): clinical signs, treatment and survival. The Veterinary Record. 2002;150(15):475–80. doi: 10.1136/vr.150.15.475. [DOI] [PubMed] [Google Scholar]
- Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nature neuroscience. 2007;10(11):1387–94. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- Huang Y, Doherty JJ, Dingledine R. Altered Histone Acetylation at Glutamate Receptor 2 and Brain-Derived Neurotrophic Factor Genes Is an Early Event Triggered by Status Epilepticus. The Journal of Neuroscience. 2002;22(19):8422–8428. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Abraham DJ. Filamentous proteins in Alzheimer’s disease: new insights through molecular biology. Psychiatric developments. 1988;6(2):121–52. [PubMed] [Google Scholar]
- Lefebvre KA, Robertson A. Domoic acid and human exposure risks: A review. Toxicon : official journal of the International Society on Toxinology. 2010;56(2):218–230. doi: 10.1016/j.toxicon.2009.05.034. [DOI] [PubMed] [Google Scholar]
- Lefebvre KA, Tilton SC, Bammler TK, Beyer RP, Srinouanprachan S, Stapleton PL, Farin FM, Gallagher EP. Gene expression profiles in zebrafish brain after acute exposure to domoic acid at symptomatic and asymptomatic doses. Toxicological sciences: an official journal of the Society of Toxicology. 2009;107(1):65–77. doi: 10.1093/toxsci/kfn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin ED, Pang WG, Harrison J, Williams P, Petro A, Ramsdell JS. Persistent neurobehavioral effects of early postnatal domoic acid exposure in rats. Neurotoxicology and teratology. 2006;28(6):673–80. doi: 10.1016/j.ntt.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Lu J, Wu D, Zheng Y, Hu B, Cheng W, Zhang Z. Purple sweet potato color attenuates domoic acid-induced cognitive deficits by promoting estrogen receptor-α-mediated mitochondrial biogenesis signaling in mice. Free Radical Biology and Medicine. 2012;52(3):646–659. doi: 10.1016/j.freeradbiomed.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Morrison JH, Hof PR. Life and Death of Neurons in the Aging Brain. Science. 1997;278(412):412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- Nicholls DG. Mitochondrial calcium function and dysfunction in the central nervous system. Biochimica et biophysica acta. 2009;1787(11):1416–1424. doi: 10.1016/j.bbabio.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijjar M, Nijjar S. Domoic acid-induced neurodegeneration resulting in memory loss is mediated by Ca2+ overload and inhibition of Ca2+ + calmodulin-stimulated adenylate cyclase in rat brain (review) International Journal of Molecular Medicine. 2000;6(4):377–466. doi: 10.3892/ijmm.6.4.377. [DOI] [PubMed] [Google Scholar]
- Perl TM, Bedard L, Kosatsky T, Hogkin JC, Todd ECD, Remis RS. An outbreak of toxic encephalopathy caused by eating mussels contaminated with domoic acid. New England Journal of Medicine. 1990;322:1775–80. doi: 10.1056/NEJM199006213222504. [DOI] [PubMed] [Google Scholar]
- Plas DR, Thompson CT. Cell metabolism in the regulation of programmed cell death. Trends in Endocrinology & Metabolism. 2002;13(2):75–8. doi: 10.1016/s1043-2760(01)00528-8. [DOI] [PubMed] [Google Scholar]
- Quilliam M, Wright J. The amnesic shellfish poisoning mystery. Analytical chemistry. 1989;61(18):1053A–106A. doi: 10.1021/ac00193a002. [DOI] [PubMed] [Google Scholar]
- Rozic-Kotliroff G, Zisapel N. Ca2+-dependent splicing of neurexin IIa. Biochemical and biophysical research communications. 2007;352:226–230. doi: 10.1016/j.bbrc.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Ryan JC, Morey JS, Ramsdell JS, Van Dolah FM. Acute phase gene expression in mice exposed to the marine neurotoxin domoic acid. Neuroscience. 2005;136(4):1121–32. doi: 10.1016/j.neuroscience.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Scholin CA, Gulland F, Doucette GJ, Benson S, Busman M, Chavez FP, Cordaro J, DeLong R, De Vogelaere A, Harvey J, Haulena M, Lefebvre K, Lipscomb T, Loscutoff S, Lowenstine LJ, Marin R, Miller PE, Van Dolah FM. Mortality of sea lions along the central California coast linked to a toxic diatom bloom. Nature. 2000;403(6765):80–4. doi: 10.1038/47481. [DOI] [PubMed] [Google Scholar]
- Siegel MP, Wilbur T, Mathis M, Shankland EG, Trieu A, Harper M, Marcinek DJ. Impaired adaptability of in vivo mitochondrial energetics to acute oxidative insult in aged skeletal muscle. Mechanisms of Ageing and Development. 2012;133(9–10):620–628. doi: 10.1016/j.mad.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Strochlic TI, Concilio S, Viaud J, Eberwine RA, Wong LE, Minden A, Turk BE, Plomann M, Peterson JR. Identification of neuronal substrates implicates Pak5 in synaptic vesicle trafficking. PNAS. 2012;109(11):4116–4121. doi: 10.1073/pnas.1116560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dolah FM. Marine algal toxins: origins, health effects, and their increased occurrence. Environmental health perspectives. 2000;108(Suppl March):133–41. doi: 10.1289/ehp.00108s1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranyac-Tramoundanas A, Harrison JC, Clarkson AN, Kapoor M, Winburn IC, Kerr DS, Sammut Ia. Domoic acid impairment of cardiac energetics. Toxicological sciences: an official journal of the Society of Toxicology. 2008;105(2):395–407. doi: 10.1093/toxsci/kfn132. [DOI] [PubMed] [Google Scholar]
- Wang X, Bao X, Pal R, Agbas A, Michaelis EK. Transcriptomic responses in mouse brain exposed to chronic excess of the neurotransmitter glutamate. BMC Genomics. 2010;11(360) doi: 10.1186/1471-2164-11-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wekell J, Gauglitz E, Jr, Barnett H, Hatfield C, Simons D, Ayres D. Occurrence of domoic acid in Washington state razor clams (Siliqua patula) during 1991–1993. Natural Toxins. 1994;2(4):197–205. doi: 10.1002/nt.2620020408. [DOI] [PubMed] [Google Scholar]
- Wentzell JS, Bolkan BJ, Carmine-Simmen K, Swanson TL, Musashe DT, Kretzschmar D. Amyloid precursor proteins are protective in Drosophila models of progressive neurodegeneration. Neurobiology of Disease. 2012;46(1):78–87. doi: 10.1016/j.nbd.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Work TM, Barr B, Beale AM, Fritz L, Quilliam MA, Wright JLC. Epidemiology of Domoic Acid Poisoning in Brown Pelicans (Pelecanus occidentalis) and Brandt’s Cormorants (Phalacrocorax penicillatus) in California. Journal of Zoo and Wildlife Medicine. 1993;24(1):54–62. [Google Scholar]
- Wright JLC, Boyd RK, de Freitas ASW, Falk M, Foxall R, Jamieson WD, Laycock MV, McCulloch AW, McInnes AG, Odense P, Pathak VP, Quilliam MA, Ragan MA, Sim PG, Thibault P, Walter JA. Identification of domoic acid, a neuroexcitatory amino acid, in toxic mussels from eastern Prince Edward Island. Canadian Journal of Chemistry. 1989;67:481–490. [Google Scholar]