Background: The role of microRNA (miRNA)-23b/27b/24 in metastasis is controversial.
Results: Ectopic expression of the miRNA-23b/27b/24 cluster enhances metastasis in vivo by direct inhibition of prosaposin (PSAP).
Conclusion: PSAP is a metastasis suppressor that is targeted by miRNA-23b/27b/24 in breast cancer.
Significance: This finding represents a novel miRNA-target interaction for therapeutic intervention of breast cancer metastasis.
Keywords: Breast Cancer, Invasion, Metastasis, MicroRNA (miRNA), Tumor Metastases
Abstract
MicroRNAs (miRNAs) have been shown to function as key regulators of tumor progression and metastasis. Recent studies have indicated that the miRNAs comprising the miR-23b/27b/24 cluster might influence tumor metastasis, although the precise nature of this regulation remains unclear. Here, expression of the miR-23b/27b/24 cluster is found to correlate with metastatic potential in mouse and human breast cancer cell lines and is elevated in metastatic lung lesions in human breast cancer patients. Ectopic expression of the miRNAs in the weakly metastatic mouse 4TO7 mammary tumor cell line had no effect on proliferation or morphology of tumor cells in vitro but was found to increase lung metastasis in a mouse model of breast cancer metastasis. Furthermore, gene expression profiling analysis of miRNA overexpressing 4TO7 cells revealed the direct targeting of prosaposin (PSAP), which encodes a secreted protein found to be inversely correlated with metastatic progression in human breast cancer patients. Importantly, ectopic expression of PSAP was able to suppress the metastatic phenotype in highly metastatic 4T1 and MDA-MB-231 SCP28 cells, as well as in cells ectopically expressing miR-23b/27b/24. These findings support a metastasis-promoting function of the miR-23b/27b/24 cluster of miRNAs, which functions in part through the direct inhibition of PSAP.
Introduction
MicroRNAs (miRNAs)2 have been recognized as regulatory RNAs with significant roles in multiple pathological processes. Individual miRNAs have been shown to regulate hundreds of downstream genes, and studies have implicated improper miRNA expression as a causal factor in tumorigenesis and metastasis (1–3). Alterations in miRNA levels are a frequent occurrence in cancer, and miRNA expression patterns can be utilized to stratify cancer subtypes, even in poorly differentiated tumors of ambiguous origins (4, 5). In addition, miRNAs have been identified as significant prognostic and predictive markers during tumor progression. For example, miR-10b (6) and the miR-200 family of miRNAs (7) are correlated with breast cancer metastasis, whereas miR-15b is associated with recurrence in melanoma (8).
Multiple methods have been utilized to examine miRNA expression changes during tumor progression, including sequencing, microarrays, and qRT-PCR approaches (9). Array-based miRNA expression profiling has uncovered broad miRNA changes in cancer cell lines, which can be correlated with metastatic capacity. Tavazoie et al. (10) examined the MDA-MB-231 human breast cancer cell line, along with sublines possessing differing metastatic potential to lung or bone, revealing miR-126 and miR-335 as suppressors of metastasis. In addition, a screen of 51 human breast cancer cell lines revealed tight clustering of miRNA expression changes that corresponded with the breast cancer subtype (11). Importantly, miRNAs have also been associated with causal roles during metastasis, both as mediators (miR-21, miR-373, miR-520c, miR-17–92, miR-10b, miR-9, miR-200) and inhibitors (miR-24, miR-34, miR-15–16) of metastatic progression (3). Many of these miRNAs play functional roles in various defined hallmarks of cancer, including migration and invasion (miR-10b, miR-373, miR-520c, miR-200, miR-34), proliferation and cell death (miR-17–92, miR-21, miR-24), angiogenesis (miR-17–92, miR-210), and colonization (miR-200) (6, 7, 12–14).
Previous studies have implicated the miR-23/27/24 miRNAs in the regulation of tumor progression. These miRNAs are expressed in two separately regulated clusters, comprising 23a/27a/24–2 and miR-23b/27b/24 (15). Expression of the miR-23/27/24 clusters has been linked to bone morphogenetic protein and TGFβ signaling, although the exact regulatory mechanism of each cluster remains unclear (16–18). Sun et al. (16) found that the miRNAs within the miR-23b/27b/24 cluster were independently regulated upon treatment with bone morphogenetic protein-2, with miR-23b decreasing in expression, miR-27b showing no significant changes, and miR-24 levels increasing. In addition, there are conflicting reports regarding the functional role of the miR-23/27/24 clusters during tumor development and metastatic progression. miR-23b has been shown to decrease migration and invasion in vitro (19) and to decrease colon cancer lung metastasis (20) and breast cancer lymph node metastasis in vivo (21). Conversely, ectopic expression of the entire miR-23a/27a/24 cluster increased migration and invasion in breast cancer cells and promoted hepatic metastasis (22). Similarly, miR-24 has been shown to inhibit apoptosis and promote the formation of micrometastases within the lungs of subcutaneously injected mice (23). Finally, although miR-27b has been shown to decrease primary colorectal cancer tumor growth and inhibit angiogenesis (24), miR-27a has been shown to positively correlate with breast cancer progression (25). Thus, the miR-23ab/27ab/24 miRNAs have been shown to possess dichotomous roles during tumor progression. Although it is possible that these contradictory findings are a result of the different model systems employed in the previous reports, further research is required to identify the precise role of the miRNAs during breast cancer invasion and tumor progression.
In this study, we examined the expression of the miR-23/27/24 clusters across multiple isogenic cell line series containing sublines with diverse metastatic propensities. Expression of miR-23b/27b/24 was found to be elevated in the metastatic variants within these progression series, as well as within lung metastasis samples relative to matched primary human breast cancer tissues. Furthermore, ectopic expression of all three miRNAs within the lowly metastatic 4TO7 mouse breast cancer cell line enhanced lung metastasis. Microarray expression analysis for targets of the miRNAs further uncovered prosaposin (PSAP) as a direct target of miR-24 and miR-27b.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
HeLa, 67NR, 168FARN, 4TO7, 66cl4, 4T1, MDA-MB-231 parental and sublines, as well as H29 cells, a packaging cell line used for retrovirus production, were maintained in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) containing 10% fetal bovine serum, supplemented with 1 mm l-glutamine and penicillin/streptomycin (Invitrogen). MCF10A, MCF10AT, MCFCA1h, and MCFCA1a cells were maintained in 1:1 DMEM:Hams F12 medium (Invitrogen) containing 5% horse serum, supplemented with 20 ng/ml EGF, 0.5 mg/ml hydrocortizone, 10 mg ml−1insulin, 1% penicillin/streptomycin and 50 ng/ml cholera toxin. miR-23b/27b/24 was amplified from mouse genomic DNA (miR-23b/27b/24, TGTCTGTAATGCCACACAGG (forward primer) and ACGTACACAATAGCTGCTAG (reverse primer)) using the Phusion High-fidelity DNA Polymerase kit (New England Biolabs) and cloned into the pMSCV-puro vector for stable expression in 4TO7 cells. Human and mouse PSAP were cloned from the respective cDNA (hPSAP, CGCAAGCTTGCCACCGGTTAGCGCCTGCGCTC (forward primer) and CGGCTCGAGCCAATGCTGTGGTTTCTGCC (reverse primer); mPSAP, CGCAAGCTTGCCACCTGCGCACTGGCGTCTG (forward primer) and CGGCTCGAGCCCTAGACCCACAAGTAGGTGAC (reverse primer)) and cloned into the pMSCV-puro vector.
Analysis of Clinical Samples and KM-plotter Data Set
Patient samples from the IRCCS Institute of Romagna for the Study and Treatment of Cancer (IRST) were collected and purified as described previously (7). Briefly, women with resected breast cancer were selected from patients followed from 1995 to 2010 in IRST (Meldola, Italy). Tumor specimens were deidentified and were considered exempt samples in accordance with the institutional review board of the Local Ethic Committee (Forlì, Italy). Surgical tumor specimens were fixed in formalin and embedded in paraffin. Ten primary breast tumors and 10 lung metastatic tissues derived from primary breast lesions were collected. For six patients, matched primary-metastatic tissues were available. Total RNA was collected from 20-mm-thick sections from formalin-fixed paraffin-embedded tissue blocks using the formalin-fixed paraffin-embedded RNA/DNA purification kit (Norgen) according to the manufacturer's instructions and subjected to RT-PCR analysis. For KM-Plotter investigation, distant metastasis-free survival analysis was performed utilizing the latest available breast cancer data set, comprising 1610 patient samples and the 200871_s_at PSAP probe (26).
RNA Isolation and Microarray
RNA for qRT-PCR and microarray was isolated from cultured cells using a miRVana total RNA isolation kit (Ambion) according to the manufacturer's instructions. For RNA isolation from tumor lesions, the lesions were macroscopically dissected and pulverized using a liquid nitrogen cooled mortar and pestle, followed by RNA isolation with the miRVana kit. For microarrays, samples were analyzed with the Agilent Whole Mouse Genome 4 × 44k arrays. RNA samples were labeled with Cy5 using the Agilent low RNA input linear amplification kit and were hybridized with the Cy3-labeled human reference RNA (Stratagene). Duplicate arrays were performed for each sample and analyzed with an Agilent G2565BA scanner and Agilent Feature Extraction software (version 9.5). The Cy5/Cy3 ratios were calculated by median signal and normalized by the array median. Probes with >2.5-fold changes were identified as potential direct targets of miR-23/27/24.
Quantitative Real-time PCR
Mature miR-23ab, miR-27ab, and miR-24 were reverse transcribed using the TaqMan Reverse Transcription Kit (Applied Biosystems) followed by real-time PCR using TaqMan miRNA assays (Applied Biosystems). mRNA was analyzed by synthesizing cDNA using the Superscript III First-strand kit (Invitrogen), and qPCR was performed using the Power SYBR green PCR master mix (Applied Biosystems). All analysis was performed using an ABI 7900HT PCR machine according to the manufacturer's instructions. A standard curve was created from serial dilutions from cDNA for each gene of interest. Values were normalized by the expression of GAPDH for mRNA or RNU6B for miRNA. Primers used for quantitative PCR were: mouse Rai4, 5′-CGATACAAACGAGTGGAACAAGA-3′ (forward) and 5′-TCGCTGTCATGCTTCGTGG-3′ (reverse); mouse Hs3st2, 5′-GTGGACGTGTCTTGGAACG-3′ (forward) and 5′-CACTGACGAAGTGGATCTGGG-3′ (reverse); mouse Myh2, 5′-AAGTGACTGTGAAAACAGAAGCA-3′ (forward) and 5′-GCAGCCATTTGTAAGGGTTGAC-3′ (reverse); mouse Otx2, 5′-TATCTAAAGCAACCGCCTTACG-3′ (forward) and 5′-AAGTCCATACCCGAAGTGGTC-3′ (reverse); mouse Pftk1, 5′-AAGAGCAAAATCCGTCCCTAGC-3′ (forward) and 5′-TCATCTCAACGAAGATACAGCCA-3′ (reverse); mouse Psap, 5′-CCTGTCCAAGACCCGAAGAC-3′ (forward) and 5′-AAGGAAGGGATTTCGCTGTGG-3′ (reverse); mouse Rnf144, 5′-AGCTGTGCCTCGGGGAATA-3′ (forward) and 5′-GCAGTTTCTAGCCCTTCTTTGAT-3′ (reverse); mouse Sacs, 5′-TGGAGACCTGACTTCTAAAACCT-3′ (forward) and 5′-TGGTGGAGTTGTTTGACCAAAT-3′ (reverse); and mouse Gapdh, 5′-AGGTCGGTGTGAACGGATTTG-3′ (forward) and 5′-TGTAGACCATGTAGTTGAGGTCA-3′ (reverse).
Transwell Migration Assay
105 4TO7 control or miRNA-overexpressing cells were seeded into the upper chamber of an 8-μm pore transwell migration insert (BD Biosciences) containing serum-free media. Serum-containing DMEM was used as an attractant in the bottom chamber, and cells were allowed to migrate for 12 h. At the conclusion of the experiment, the inserts were removed, and trypisized cells were counted from the bottom chamber, providing a quantitative value for migrated cells across the membrane. For invasion assays, 1 mg/ml Matrigel (BD Biosciences) was polymerized in the top chamber prior to the addition of cells, which were allowed to invade for 20 h before quantification.
Tumor Xenografts and Bioluminescence Analysis
All procedures involving mice and experimental protocols were approved by Institutional Animal Care and Use Committee of Princeton University. For experimental lung metastasis studies, 105 tumor cells were injected into the lateral tail vein of female BALB/c mice. For orthotopic injections, 106 cells were injected into the mammary gland as described previously (27). Development of primary tumors was analyzed every 4 days, with final measurements performed at end point. Lungs collected from tail vein-injected mice were washed briefly in PBS, followed by overnight fixation in Bouin's fixative (Sigma) and were stored in 70% ethanol. Lungs from orthotopic injections were minced and digested with 300 units/ml type 1A collagenase (Sigma, C2674) and 100 units/ml hyaluronase (Sigma, H3506) for 60 min at 37 °C. Dissociated cells were passed through a 70-μm strainer and plated into 15-cm plates. 60 μm 6-thioguanine was used to select for 4TO7 tumor cells, which were grown for 2 weeks, followed by staining with crystal violet for 30 min for quantification.
Luciferase Reporter Assay
Wild-type and mutant 3′-UTRs were PCR-amplified from human genomic DNA. The 3′-UTRs were cloned into the pMIR-REPORT vector (Ambion) downstream of firefly luciferase. Mutations in miRNA target sites were generated using the QuikChange multisite-directed mutagenesis kit (Stratagene). 5 × 104 HeLa cells were plated in 24-well plates 24 h prior to transfection. 200 ng of reporter plasmid was co-transfected with the Renilla luciferase control plasmid and 10 pm pre-miRNA precursor or precursor control (Ambion) using Lipofectamine 2000 (Invitrogen). Cells were lysed 24 h after transfection and assayed for luciferase activity using the Glomax 96 Luminometer (Promega). Primer sequences for cloning or mutagenesis were: human RAI14, 5′-GGATTCTTTGGCAGGACACTT-3′ (forward) and 5′-AATAAATACGCATCCTGACACAA-3′ (reverse); human HS3ST2, 5′-GTCTCCAAGGTACAAGGGCTCT-3′ (forward) and 5′-TTAAGGACACGAGAGAGCCATATT-3′ (reverse); human MYH2, 5′-CCCGATGCTGTGGAATGAC-3′ (forward) and 5′-CTTGCAAAGGAACTTGGGCT-3′ (reverse); human OTX2, 5′-CCTGTAGAAGCTATTTTTGTGGGT-3′ (forward) and 5′-GCATCTAGGACAATCAGTCGC-3′ (reverse); human PFTK1, 5′-CCTGAATCGGTTCTCTTCTGC-3′ (forward) and 5′-TGCTATGTGTCATGTTGAAGC-3′ (reverse); human PSAP, 5′-TTTCCCAGCTGCAGAAGTCACCTAC-3′ (forward) and 5′-GCTGTCTCCTGGCAACCGGAG-3′ (reverse); human RNF144, 5′-TCAAGCTCCCTAATCAAAGATGG-3′ (forward) and 5′-GGGAGACAGACTACATAAGACACAA-3′ (reverse); human SACS, 5′-AGAGACAGGAAGTTCCCATTG-3′ (forward) and 5′-GTCATGGGTCTTAGGTGTTTATT-3′ (reverse); human PSAP-miR-27, 5′-CCCTGGTTCATCAGAGTTTTGGCTTTCCC-3′ (forward) and 5′-GGGAAAGCCAAAACTCTGATGAACCAGGG-3′ (reverse); human PSAP-miR-24-1, 5′-AGGCGGGTGGAGTGGCTGAGGGCAGGA-3′ (forward) and 5′-TCCTCCCTCAGCCACTCCACCCGCCT-3′ (reverse); and human PSAP-miR-24-2, 5′-GAGCATTTTGTCACCGCTTTCTTGGTGTGTGG-3′ (forward) and 5′-ACCAAGAAAGCGGTGACAAAATGCTC-3′ (reverse).
Western Blot Analyses
SDS lysis buffer (0.05 mm Tris-HCl, 50 mm 2-mercaptoethanol, 2% SDS, 0.1% bromphenol blue, 10% glycerol) was used to collect protein from cells. Samples were sonicated, and heat-denatured protein was equally loaded, separated on a 10% SDS-PAGE gel, transferred onto a pure nitrocellulose membrane (Bio-Rad), and blocked with 5% milk. Anti-β-actin (1:5000 dilution, Abcam) and anti-PSAP (1:1000, BD Biosciences) were incubated with the membrane overnight at 4 °C. Membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (1:2000 dilution, GE Healthcare) or anti-rabbit secondary antibody (1:2000 dilution, GE Healthcare) for 1 h, and chemiluminescence signals were detected by ECL substrate (GE Healthcare).
RESULTS
miR-23/27/24 Expression Correlates with Breast Cancer Lung Metastasis
Previous reports have implicated the miR-23ab/27ab/24 miRNAs as key mediators of metastasis, although there exists uncertainty as to the precise role of the miRNAs during breast cancer metastasis. These clusters are thought to originate from a highly conserved duplication event; miR-23a and -23b and miR-27a and -27b contain minute differences within the mature miRNA sequences, although the seed sequences of the related miRNAs are conserved across clusters, resulting in two highly conserved clusters (Fig. 1A). To examine expression of the miRNAs within metastasis, we utilized matched primary tumor and lung metastasis samples isolated from microdissected tumor samples from breast cancer patients. qRT-PCR analysis of these samples revealed a consistent up-regulation of all miRNAs in the metastatic tissues (Fig. 1B).
FIGURE 1.
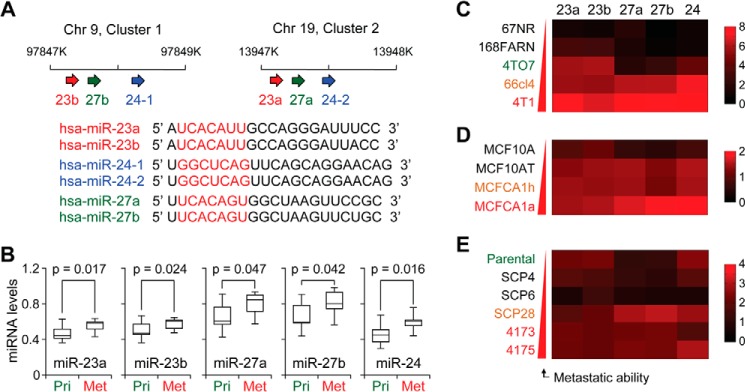
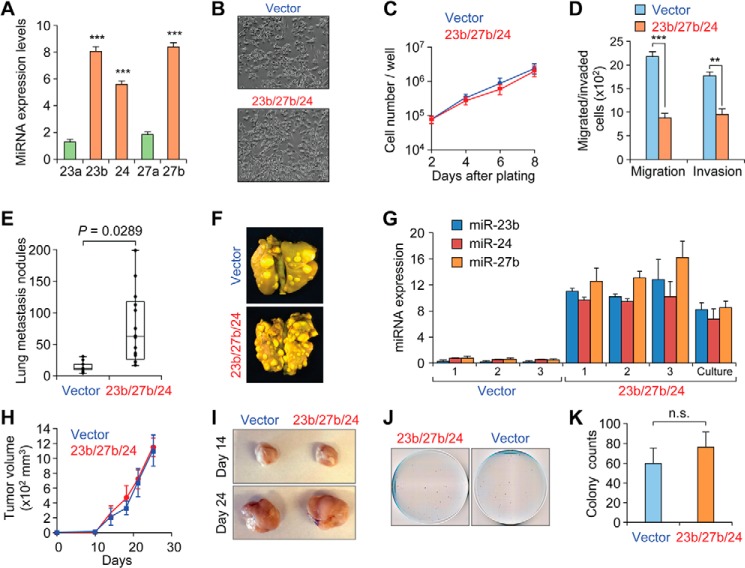
The miR-23a/27a/24 and miR-23b/27b/24 clusters correlate with breast cancer metastasis. A, upper panel, schematic representing the chromosomal locations of human miR-23ab/27ab/24. Lower panel, sequence alignment of the miR-23ab/27ab/24 clusters. Seed sequences are highlighted in red, and miRNA members with identical seed sequences are color-coded. B, box plots showing normalized expression levels of indicated miRNAs in human primary (Pri) tumor and metastasis (Met) samples (n = 10). p values were based on Student's t test. C–E, normalized qRT-PCR expression of miR-23ab, miR-27ab, and miR-24 in 4T1 (C), MCF10A (D), or MDA-MB-231 (E) cell line series, as shown by heat maps. Data represent average, U6-normalized expression.
To better examine the function these miRNAs during tumor progression, three breast cancer isogenic cell line series were selected that model the progression of breast cancer with increasing metastatic ability: the mouse 4T1 cell line series and the human MCF10A series and MDA-MB-231 series. The 4T1 series, consisting of 67NR, 168FARN, 4TO7, 66cl4, and 4T1 cells, are a syngenic model derived from a spontaneously arising BALB/cfC3H tumor (28). This series displays varied metastatic potential upon orthotopic injection into the mammary gland of mice, with 4T1 and 66cl4 cells producing spontaneous lung metastases, 4TO7 forming micrometastases in the lungs, and 67NR and 168FARN forming primary tumors but lacking metastases (28–30). The MCF10A series includes the immortalized, but otherwise normal and non-tumorigenic MCF10A mammary epithelial cell line, as well as in vivo selected derivatives of H-RAS-transformed parental cells with varying metastatic potentials (31). These include the relatively benign MCF10AT cells, which do not typically progress into carcinomas, as well as the low-grade, poorly metastatic carcinoma line MCF10CA1h and the metastatic MCF10CA1a cells. The MDA-MB-231 cell line consists of a weakly metastatic parental line, as well as clonally selected, single cell-derived progenies (SCPs) that feature varying metastatic propensity and organotropism (32, 33). Specifically, although SCP4 and SCP6 cells are essentially non-metastatic, the SCP28, 4173, and 4175 lines are capable of forming overly lung metastases (33).
miR-23a/27a/24 and miR-23b/27b/24 expression levels were examined in these cells by qRT-PCR. Expression of miR-23a, miR-27a, and miR-24 showed a correlation with metastatic potential within the 4T1 (Fig. 1C), MCF10A (Fig. 1D), and MDA-MB-231 progression series (Fig. 1E). miRNA expression of all three members of the miR-23b/27b/24 cluster showed an overall similar but slightly stronger association with metastatic potential (Fig. 1, C–E). Given the similarity of these two clusters, we chose to focus on functional analysis of the role of the miR-23b/27b/24 cluster.
The mouse miRNAs were stably overexpressed in the weakly metastatic 4TO7 cells using the pMSCV retroviral vector, as verified by qPCR (Fig. 2A). Expression of all three miRNAs in the miR-23b/27b/24 cluster were elevated by ∼6–8 fold relative to vector transduced cells, at a level similar to the endogenous miRNA expression found in 4T1 cells (Fig. 1C). Importantly, the overexpression was specific, with no significant changes observed in miR-23a and miR-27a within these cells. Overexpression of miR-23b/27b/24 had no effect on morphology or proliferation within the 4TO7 cells (Fig. 2, B and C). Interestingly, miRNA overexpressing cells reduced transwell migration and decreased Matrigel invasion (Fig. 2D), consistent with previous reports of decreased migration upon ectopic expression of miR-23b in prostate cancer cell lines (19).
FIGURE 2.
Analysis of ectopic miR-23b/27b/24 expression in 4TO7 cells. A, qRT-PCR miRNA expression levels of indicated miRNAs in 4TO7-miR-23b/27b/24 cells. Data represent average ± S.D. normalized to vector-transduced cells. ***, p < 0.001. B, phase contrast images of 4TO7 pMSCV-puro vector control or miR-23b/27b/24-overexpressing cells. C, in vitro growth curves showing similar growth rates for either vector or miRNA-overexpressing cells during the exponential growth phase. Data represent average ± S.D. with triplicate for each line at each time point. D, migration and invasion of 4TO7 vector or miRNA-overexpressing cells toward serum-containing media. Data represent average number of cells counted from bottom well from triplicate experiments ± S.D. **, p < 0.01; ***, p < 0.001. E, quantification of overt metastatic lesions on lungs from BALB/c mice injected with the indicated cell lines (105 cells per injection) and allowed to grow for 14 days. p value from Student's t test (n = 10). F, representative images of Bouin's fixed lungs from mice in E. G, normalized qRT-PCR miRNA expression levels of indicated miRNAs in overt lung lesions isolated from mice injected with indicated cells or non-injected miRNA-overexpressing cells in culture. Data represent average ± S.D. H, in vivo primary mammary tumor volume from mice injected with 106 4TO7 vector control or miR-23b/27b/24 overexpressing cells (n = 10). Data represent average ± S.D. I, representative images of resected primary tumors from experiment in H at the indicated time point. J, representative crystal violet-stained cell culture plates showing tumor colonies derived from dissociated lungs from mice in H. K, quantification of dissociated lung colonies from J. Data represent average ± S.D. No significance (n.s.) by Student's t test.
The miR-23b/27b/24 Cluster Promotes Lung Metastasis
To investigate the influence of the miR-23b/27b/24 cluster miRNAs on metastatic colonization in the lungs, mice were inoculated with vector control or miRNA-overexpressing 4TO7 cells via the lateral tail vein and subsequently monitored for the incidence of lung metastasis. Lungs were collected 2 weeks post-injection, and overt lesions were counted. This quantification revealed that miR-23b/27b/24 expression significantly increased the number of lung lesions relative to control (p = 0.0289, Fig. 2, E and F). Interestingly, qRT-PCR analysis revealed that miR-23b/27b/24 levels were modestly elevated in isolated lung lesions beyond that of the in vitro population prior to injection, suggesting that the metastasis process may have selected for clones with elevated expression (Fig. 2G).
To examine the influence of miR-23b/27b/24 on the early steps of metastatic progression, 4TO7 cells were inoculated orthotopically into the mammary fat pad of BALB/c mice. Primary tumor growth was comparable between both parental and miR-23b/27b/24-overexpressing 4TO7 cells (Fig. 2, H and I), both of which spontaneously disseminated to the lungs but failed to develop into overt lung metastases. Instead, the micrometastatic burden was estimated by culturing tumor colonies from dissociated lungs; quantification of crystal violet-stained colonies revealed a trend toward increased metastasis from mice injected with the miR-23b/27b/24-overexpressing cells, although this did not reach significance (Fig. 2, J and K). Together, these in vivo results indicate that the miR-23b/27b/24 cluster promotes lung metastasis with most of the influence exerted on the step of colonization.
Genomic Identification of miR-23b/27b/24 Target Genes
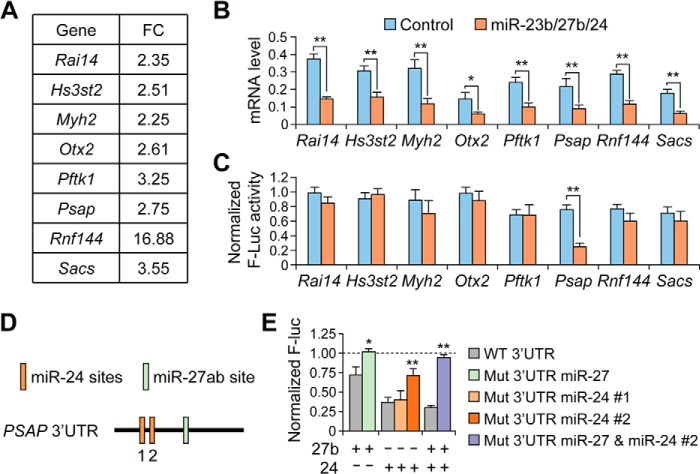
To elucidate the molecular mechanism of miR-23b/27b/24 in enhancing lung metastasis, we employed a combined approach, utilizing microarray analysis and the TargetScan computational target prediction algorithm (34). The microarray analysis detected 24,705 unique genes, revealing 402 genes with >2.5-fold decreased expression in the miRNA-overexpressing cells. To identify potential direct targets of miR-23b/27b/24, the subset of down-regulated genes was analyzed for putative miRNA-binding sites. Candidate target genes were then examined for predicted miRNA binding sites using TargetScan, and eight predicted targets were chosen for further analysis (Fig. 3A). qRT-PCR results validated repression of all eight genes after ectopic miRNA expression (Fig. 3B).
FIGURE 3.
Identification of putative miR-23b/27b/24 cluster targets. A, average gene expression (expressed as fold change) for candidate target genes from microarray analysis of 4TO7-miR-23b/27b/24 cells relative to vector control. B, qRT-PCR mRNA expression levels for putative target genes in 4TO7 vector cells or miR-23b/27b/24 overexpressing cells. **, p < 0.05; **, p < 0.01 by Student's t test. Data represent average ± S.D. C, normalized firefly luciferase expression from reporter plasmids containing indicated 3′-UTR 24 h after co-transfection with control or miR-23b/27b/24 miRNA oligonucleotides. **, p < 0.01. Data represent average ± S.D. D, schematic representing the predicted miR-27ab and miR-24 binding sites within the human PSAP 3′-UTR. MiR-24 contains two predicted sites, referred to here as 1 and 2, whereas miR-27ab contains one predicted binding site. E, normalized firefly luciferase (Luc) expression from PSAP reporter constructs similar to following site-directed mutagenesis of predicted miRNA binding sites and co-transfection with miR-27b or miR-24 as indicated. *, p < 0.05; **, p < 0.01 by Student's t test. Data represent average ± S.D.
To confirm direct targeting by the miRNAs, the 3′-UTRs from the putative target genes were cloned into the pMIR-REPORT luciferase plasmid. Reporter plasmids were co-transfected into HeLa cells along with the miRNAs of interest and a Renilla luciferace plasmid for normalization. Analysis of luciferase expression revealed that only the prosaposin (Psap) reporter expression was suppressed after ectopic miRNA expression (Fig. 3C). PSAP is a secreted protein that has been described previously as both a metastasis promoter and suppressor (35–37). PSAP mRNA contained three predicted binding sites for the miR-23/27/24 clusters within the 3′-UTR, including two for miR-24 and one for miR-27 (Fig. 3D). We confirmed direct targeting through mutagenesis of the predicted miRNA binding sites: mutation of the miR-27 binding site and one of the two miR-24 partially rescued luciferase signal, whereas combined mutagenesis of both miR-24 and miR-27 sites fully rescued luciferase signal (Fig. 3E).
PSAP Expression Is Correlated with Lung Metastasis
Examination of PSAP expression in our cell line models revealed that the mRNA and protein expression level of PSAP decreased in the metastatic variants of the 4T1 and MCF10A isogenic progression series (Fig. 4, A–D). In particular, a significant reduction of PSAP protein and mRNA expression was found in the 4T1 and MCFCA1a cell lines, which represent the most metastatic ones and the only ones capable of forming macrometastases in each series. These findings implicate PSAP as a putative metastasis suppressor, we next sought to examine the effect of PSAP overexpression on metastatic progression. To this end, mouse and human PSAP was ectopically expressed in 4T1 and MDA-MB-231-SCP28 cells, respectively. Ectopic expression was confirmed by qRT-PCR and Western blot analyses (Fig. 5A), and cells were injected via the lateral tail-vein into mice. Analysis of lung lesions revealed that PSAP suppressed metastasis in both 4T1 and SCP28 cells (Fig. 5B). Importantly, when Psap was ectopically expressed in 4TO7-miR-23b/27b/24 cells, there was a significant decrease in lung lesions, indicating that re-expression of Psap was capable of partially neutralizing the metastasis-promoting effects of the phenotype resulted from the overexpression of the miR-23b/27b/24 cluster (Fig. 5C).
FIGURE 4.
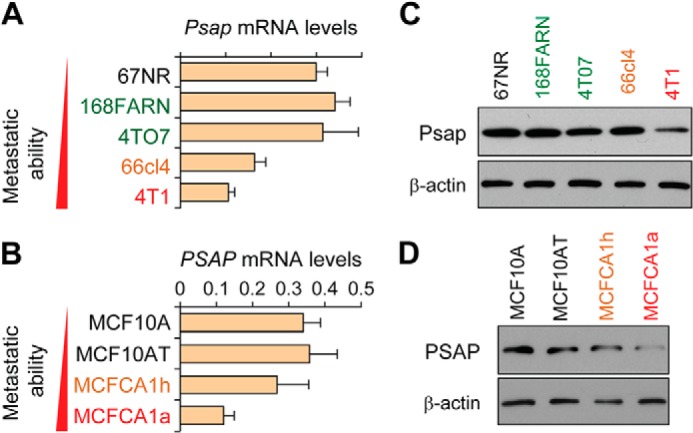
PSAP decreases in cells with elevated metastatic potential. A and B, qRT-PCR expression levels of Psap in indicated cell lines from 4T1 (A) and MCF10A (B) series. Data represent average ± S.D. C and D, Western blot analysis of PSAP or β-actin from indicated cells lines as in A and B.
FIGURE 5.
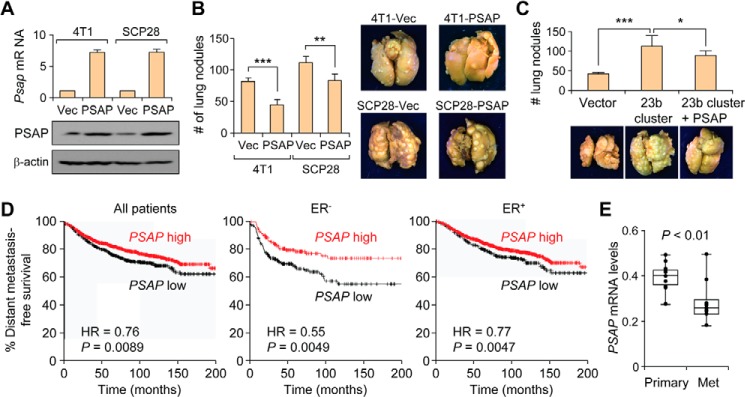
PSAP is a metastasis suppressor and a poor prognosis factor in breast cancer. A, qRT-PCR (upper panel) and Western blot analysis of the expression levels of PSAP in 4T1 or SCP28 cells after transfection of the expression plasmid or vector (Vec). Data represent average ± S.D. B, lung lesion quantification and representative images of lungs from mice injected with vector or PSAP-overexpressing cells from A (p values by Student's t test). Data represent average ± S.D. C, lung lesion quantification and representative images of lungs from mice injected with indicated SCP28 cells. D, Kaplan-Meier curves showing the distant metastasis free survival of patients with high or low expression of PSAP in breast tumors. p values were computed by a log rank test. E, box plots showing PSAP expression levels in 10 primary and metastasis (Met) samples as quantified by qRT-PCR analysis. p values computed by Student's t test. Data represent mean ± S.E. normalized to GAPDH expression. ER, estrogen receptor; HR, hazard ratio.
To further examine the clinical significance of PSAP as a metastasis factor, we analyzed the expression levels of PSAP in publically available clinical datasets, as well as the microdissected primary breast cancer or lung metastasis patient samples from before. Analysis of breast cancer patients using the KM-plotter data set (26) revealed a significant correlation between PSAP expression and increased distant metastasis-free survival, particularly in estrogen receptor-negative breast cancer patients (Fig. 5D). In addition, analysis of patient samples revealed that PSAP expression decreased in lung metastasis samples compared with the primary mammary tumors (Fig. 5E). Together, these results indicate that PSAP is a direct target of the miR-23/27/24 clusters and is reduced during metastasis, both in cell lines and clinical samples.
DISCUSSION
miRNA regulation has been implicated in various critical aspects of tumor progression, both as clinical biomarkers and functional regulators of metastasis. Previous in vitro and in vivo studies have implicated the miR-23/27/24 family of miRNAs as both positive and negative regulators of the metastatic cascade. For example, expression of miR-23b and miR-27b has been shown to inhibit migration and anchorage-independent growth in prostate cancer cells (19), whereas miR-23a/27a/24 increases migration and metastasis in breast cancer lines (22). In this study, we show that the expression of the miR-23b/27b/24 cluster is elevated in lung metastasis in human breast cancer and that ectopic expression of the miRNAs promotes lung metastasis in the 4TO7 mouse mammary cell line. Therefore, the miR-23b/27b/24 is likely to have a metastasis-promoting role in lung metastasis of breast cancer.
It is curious that ectopic expression of the miRNAs was capable of increasing the metastatic potential upon intravenous inoculation while possessing much weaker effects on spontaneous metastasis. It is possible that this effect can be attributed in part to limitations within the assay; both 4TO7 vector and miRNA overexpression cells develop visible lesions upon lateral tail vein injection but were unable to develop into overt metastasis in orthotopic experiments. In addition, it is possible that the observed defect in migration and invasion could offset any increase of metastatic colonization abilities conferred by the miRNAs. Future studies will be necessary to further elucidate the influence of the miRNAs on the stepwise progression of breast cancer lung metastasis.
Multiple targets have been identified for members of the miR-23b/27b/24 cluster, including SPRY, ANXA2, ARHGEF6, CFL2, LIMK2, PIK3R3, PLAU, VEGFC, PTPN9, and PTPRF (21–24). We have additionally uncovered the direct targeting of PSAP by miR-24 and miR-27b. PSAP, a highly conserved precursor for saposins A, B, C, and D, has been identified previously as a functional regulator of metastatic progression (36, 37). Our results show that PSAP mRNA and protein expression is correlated with reduced metastatic potential and that ectopic PSAP expression is capable of inhibiting metastasis. Previous studies found that PSAP was down-regulated in metastatic variants of the PC3 prostate cancer and MDA-MB-231 breast cancer cells (36). Finally, PSAP has been shown to increase thrombospondin 1 expression in PC3 cells, and PSAP or a short PSAP-mimetic peptide (DWLPK) inhibited prostate cancer lung metastasis (38). Future studies will be required to further analyze the functional mechanism of PSAP in suppressing metastatic colonization in the lungs.
Enhanced migration and invasion were canonically believed to increase the metastatic potential of tumor cells, although recent reports have identified highly metastatic cells with decreased migratory capacity (39). Indeed, this is observed within the 4T1 series, in which the most highly metastatic 4T1 cells are actually less migratory (7, 29). Thus, it is not possible to predict metastatic potential through migratory capacity alone. In our current study, overexpression of miR-23b/27b/24 inhibits invasion and migration, while dramatically increasing metastatic colonization in the lungs. This observation is similar to what we previously reported about the miR-200 family miRNAs (7). miRNAs have been shown to influence the expression of hundreds of downstream genes, so it is perhaps not surprising that ectopic expression of three miRNAs might simultaneously influencing separate aspects of the metastatic cascade. The dichotomous functions of these miRNAs in early and late steps of the metastatic cascade may complicate their utilization as therapeutic targets. One possible approach to sidestep this complication is to use the direct target of miR-23b/27b/24, PSAP, or its peptide mimetics, as therapeutic agents against metastasis. Future studies should assess whether PSAP or its functional peptide mimetics may have a therapeutic influence on metastatic progression. The combination of clinical correlation and direct functional inhibition in preclinical mouse models strongly suggests that PSAP is functioning as a metastasis suppressor. Future studies will be required to asses the utility of PSAP as a therapeutic strategy, both as a diagnostic marker for disease progression and as a direct pharmaceutical target.
Acknowledgments
We thank M. Korpal and Y. Wei for expert technical advice and support and members of the Kang laboratory for insightful discussions and technical suggestions. Additionally, we thank F. Miller for the 4T1 cell line series.
Footnotes
- miRNA
- microRNA
- qRT-PCR
- quantitative RT-PCR
- SCP
- single cell-derived progeny.
REFERENCES
- 1. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harquail J., Benzina S., Robichaud G. A. (2012) MicroRNAs and breast cancer malignancy: an overview of miRNA-regulated cancer processes leading to metastasis. Cancer Biomark 11, 269–280 [DOI] [PubMed] [Google Scholar]
- 3. Iorio M. V., Croce C. M. (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 4, 143–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 5. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma L., Teruya-Feldstein J., Weinberg R. A. (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682–688 [DOI] [PubMed] [Google Scholar]
- 7. Korpal M., Ell B. J., Buffa F. M., Ibrahim T., Blanco M. A., Celià-Terrassa T., Mercatali L., Khan Z., Goodarzi H., Hua Y., Wei Y., Hu G., Garcia B. A., Ragoussis J., Amadori D., Harris A. L., Kang Y. (2011) Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 17, 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Satzger I., Mattern A., Kuettler U., Weinspach D., Voelker B., Kapp A., Gutzmer R. (2010) MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int. J. Cancer 126, 2553–2562 [DOI] [PubMed] [Google Scholar]
- 9. Farazi T. A., Hoell J. I., Morozov P., Tuschl T. (2013) MicroRNAs in human cancer. Adv. Exp. Med. Biol. 774, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tavazoie S. F., Alarcón C., Oskarsson T., Padua D., Wang Q., Bos P. D., Gerald W. L., Massagué J. (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riaz M., van Jaarsveld M. T., Hollestelle A., Prager-van der Smissen W. J., Heine A. A., Boersma A. W., Liu J., Helmijr J., Ozturk B., Smid M., Wiemer E. A., Foekens J. A., Martens J. W. (2013) miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 15, R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dews M., Homayouni A., Yu D., Murphy D., Sevignani C., Wentzel E., Furth E. E., Lee W. M., Enders G. H., Mendell J. T., Thomas-Tikhonenko A. (2006) Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 38, 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Q., Gumireddy K., Schrier M., le Sage C., Nagel R., Nair S., Egan D. A., Li A., Huang G., Klein-Szanto A. J., Gimotty P. A., Katsaros D., Coukos G., Zhang L., Puré E., Agami R. (2008) The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 10, 202–210 [DOI] [PubMed] [Google Scholar]
- 15. Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun F., Wang J., Pan Q., Yu Y., Zhang Y., Wan Y., Wang J., Li X., Hong A. (2009) Characterization of function and regulation of miR-24-1 and miR-31. Biochem. Biophys. Res. Commun. 380, 660–665 [DOI] [PubMed] [Google Scholar]
- 17. Sun Q., Zhang Y., Yang G., Chen X., Zhang Y., Cao G., Wang J., Sun Y., Zhang P., Fan M., Shao N., Yang X. (2008) Transforming growth factor-β-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 36, 2690–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rogler C. E., Levoci L., Ader T., Massimi A., Tchaikovskaya T., Norel R., Rogler L. E. (2009) MicroRNA-23b cluster microRNAs regulate transforming growth factor-β/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology 50, 575–584 [DOI] [PubMed] [Google Scholar]
- 19. Ishteiwy R. A., Ward T. M., Dykxhoorn D. M., Burnstein K. L. (2012) The microRNA -23b/-27b cluster suppresses the metastatic phenotype of castration-resistant prostate cancer cells. PLoS One 7, e52106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang H., Hao Y., Yang J., Zhou Y., Li J., Yin S., Sun C., Ma M., Huang Y., Xi J. J. (2011) Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat. Commun. 2, 554. [DOI] [PubMed] [Google Scholar]
- 21. Pellegrino L., Stebbing J., Braga V. M., Frampton A. E., Jacob J., Buluwela L., Jiao L. R., Periyasamy M., Madsen C. D., Caley M. P., Ottaviani S., Roca-Alonso L., El-Bahrawy M., Coombes R. C., Krell J., Castellano L. (2013) miR-23b regulates cytoskeletal remodeling, motility and metastasis by directly targeting multiple transcripts. Nucleic Acids Res. 41, 5400–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X., Liu X., Xu W., Zhou P., Gao P., Jiang S., Lobie P. E., Zhu T. (2013) c-MYC-regulated miR-23a/24–2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J. Biol. Chem. 288, 18121–18133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du W. W., Fang L., Li M., Yang X., Liang Y., Peng C., Qian W., O'Malley Y. Q., Askeland R. W., Sugg S. L., Qian J., Lin J., Jiang Z., Yee A. J., Sefton M., Deng Z., Shan S. W., Wang C. H., Yang B. B. (2013) MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J. Cell Sci. 126, 1440–1453 [DOI] [PubMed] [Google Scholar]
- 24. Ye J., Wu X., Wu D., Wu P., Ni C., Zhang Z., Chen Z., Qiu F., Xu J., Huang J. (2013) miRNA-27b targets vascular endothelial growth factor C to inhibit tumor progression and angiogenesis in colorectal cancer. PLoS One 8, e60687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang W., Zhu J., Su S., Wu W., Liu Q., Su F., Yu F. (2012) MiR-27 as a prognostic marker for breast cancer progression and patient survival. PLoS One 7, e51702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gyorffy B., Lánczky A., Szállási Z. (2012) Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 19, 197–208 [DOI] [PubMed] [Google Scholar]
- 27. Chakrabarti R., Hwang J., Andres Blanco M., Wei Y., Lukačiašin M., Romano R. A., Smalley K., Liu S., Yang Q., Ibrahim T., Mercatali L., Amadori D., Haffty B. G., Sinha S., Kang Y. (2012) Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat. Cell Biol. 14, 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aslakson C. J., Miller F. R. (1992) Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 52, 1399–1405 [PubMed] [Google Scholar]
- 29. Lou Y., Preobrazhenska O., auf dem Keller U., Sutcliffe M., Barclay L., McDonald P. C., Roskelley C., Overall C. M., Dedhar S. (2008) Epithelial-mesenchymal transition (EMT) is not sufficient for spontaneous murine breast cancer metastasis. Dev. Dyn. 237, 2755–2768 [DOI] [PubMed] [Google Scholar]
- 30. Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R. A. (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 [DOI] [PubMed] [Google Scholar]
- 31. Tang B., Vu M., Booker T., Santner S. J., Miller F. R., Anver M. R., Wakefield L. M. (2003) TGF-beta switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J. Clin. Invest. 112, 1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minn A. J., Kang Y., Serganova I., Gupta G. P., Giri D. D., Doubrovin M., Ponomarev V., Gerald W. L., Blasberg R., Massagué J. (2005) Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Invest. 115, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Campana W. M., O'Brien J. S., Hiraiwa M., Patton S. (1999) Secretion of prosaposin, a multifunctional protein, by breast cancer cells. Biochim. Biophys. Acta 1427, 392–400 [DOI] [PubMed] [Google Scholar]
- 36. Kang S. Y., Halvorsen O. J., Gravdal K., Bhattacharya N., Lee J. M., Liu N. W., Johnston B. T., Johnston A. B., Haukaas S. A., Aamodt K., Yoo S., Akslen L. A., Watnick R. S. (2009) Prosaposin inhibits tumor metastasis via paracrine and endocrine stimulation of stromal p53 and Tsp-1. Proc. Natl. Acad. Sci. U.S.A. 106, 12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu S., Delorme N., Liu Z., Liu T., Velasco-Gonzalez C., Garai J., Pullikuth A., Koochekpour S. (2010) Prosaposin down-modulation decreases metastatic prostate cancer cell adhesion, migration, and invasion. Mol. Cancer 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Catena R., Bhattacharya N., El Rayes T., Wang S., Choi H., Gao D., Ryu S., Joshi N., Bielenberg D., Lee S. B., Haukaas S. A., Gravdal K., Halvorsen O. J., Akslen L. A., Watnick R. S., Mittal V. (2013) Bone marrow-derived Gr1+ cells can generate a metastasis-resistant microenvironment via induced secretion of thrombospondin-1. Cancer Discov. 3, 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xue B., Krishnamurthy K., Allred D. C., Muthuswamy S. K. (2013) Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat. Cell Biol. 15, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]