Background: UNG1/2 is a major uracil-DNA glycosylase in human cells.
Results: Intracellular processing of U:A and U:G base pairs interferes with the transcription process. For U:A, but not U:G, this effect is enhanced by UNG1/2.
Conclusion: Transcription of uracil-containing DNA declines as a consequence of the base excision.
Significance: Suppression of unwanted transcription may be an important function of UNG1/2.
Keywords: Base Excision Repair (BER), DNA Damage Response, DNA Repair, Flow Cytometry, Gene Transcription, Transfection, Reporter Gene, Uracil in DNA, Uracil-DNA Glycosylase
Abstract
Uracil is an unavoidable aberrant base in DNA, the repair of which takes place by a highly efficient base excision repair mechanism. The removal of uracil from the genome requires a succession of intermediate products, including an abasic site and a single strand break, before the original DNA structure can be reconstituted. These repair intermediates are harmful for DNA replication and also interfere with transcription under cell-free conditions. However, their relevance for cellular transcription has not been proved. Here we investigated the influence of uracil incorporated into a reporter vector on gene expression in human cells. The expression constructs contained a single uracil opposite an adenine (to mimic dUTP misincorporation during DNA synthesis) or a guanine (imitating a product of spontaneous cytosine deamination). We found no evidence for a direct transcription arrest by uracil in either of the two settings because the vectors containing the base modification exhibited unaltered levels of enhanced GFP reporter gene expression at early times after delivery to cells. However, the gene expression showed a progressive decline during subsequent hours. In the case of U:A pairs, this effect was retarded significantly by knockdown of UNG1/2 but not by knockdown of SMUG1 or thymine-DNA glycosylase uracil-DNA glycosylases, proving that it is base excision by UNG1/2 that perturbs transcription of the affected gene. By contrast, the decline of expression of the U:G constructs was not influenced by either UNG1/2, SMUG1, or thymine-DNA glycosylase knockdown, strongly suggesting that there are substantial mechanistic or kinetic differences between the processing of U:A and U:G lesions in cells.
Introduction
Uracil is conceivably one of the most frequently occurring aberrant bases in DNA (1). It originates from two unrelated mechanisms: incorporation of deoxyuracil into nascent DNA strands during replication and hydrolytic deamination of cytosine. De novo incorporation of uracil results in non-mutagenic U:A base pairs, whereas deamination of cytosine generates premutagenic U:G mismatches that lead to G:C → T:A transition mutations upon replication. This is believed to be one of the major sources of mutation in all cell types because several hundred U:G mispairs are generated per human cell per day (1–3). Therefore, the capacity to efficiently remove uracil from the spontaneously arisen U:G mismatches and to faithfully replace it with cytosine is required for the preservation of genomic integrity.
The removal of uracil from genomic DNA takes place primarily by the base excision repair (BER)2 pathway initiated by specific uracil-DNA glycosylases (UDGs), four of which are expressed in human cells (UNG1/UNG2, SMUG1, TDG, and MBD4) (4). The greatest part of the uracil excision activity present in nuclear extracts has been attributed to UNG2 and SMUG1 (5–7). TDG and MBD4 may specialize in excision of deamination and oxidation products of 5-methylcytosine at CpG sites (8–10), whereas UNG1 is the alternatively spliced form of UNG2 present in mitochondria (11). Interestingly, both major UDGs (UNG2 and SMUG1) can excise uracil from both U:A pairs and U:G mismatches in double-stranded DNA and also from single-stranded DNA (6, 12), suggesting the redundant functions of these DNA glycosylases in repair of such substrates. However, because of a better catalytic efficiency and higher protein expression levels (5, 13), UNG2 alone accounts for >90% of the uracil-DNA glycosylase activity in human cell extracts and has a proportional contribution to repair (5, 14). Interestingly, the excision of uracil within the U:A pairs by human UNG is nearly as efficient as the excision of U:G mismatches (15), although there is no obvious reason why this non-mutagenic lesion has to be efficiently removed from DNA. Moreover, UNG1/2 is considered essential for processing of this type of DNA damage because the repair of the U:A pairs by cell extracts is fully suppressed by UNG-specific antibodies while being unaffected by antibodies to SMUG1 or TDG (5, 14).
In addition to causing mutations, uracil can interfere with transcriptional activities by either modulating the binding of transcription factors to the gene regulatory elements (16) or compromising the fidelity of RNA synthesis through the coding regions (17). Moreover, transcription of the uracil-containing DNA templates by protein extracts derived from mammalian cells turned out to be vulnerable to a concurrent intrinsic base excision activity, leading to the generation of single-strand breaks that interfered with transcriptional elongation (18). Considering the high rate of spontaneous generation of uracil in the DNA of living cells, the aim of this work was to examine to which extent uracil or the intermediate products of its repair can interfere with transcription in cells.
EXPERIMENTAL PROCEDURES
Commercial UDG Knockdown Cell Lines
The control glioblastoma LN428 cell line (Trevigen, catalog no. 5503-001-01) and the isogenic UNG (5509-001-01), SMUG1 (5510-001-01), and TDG (5519-001-01) knockdown cell lines were purchased from AMS Biotechnology (Frankfurt am Main, Germany). The presence of at least 70% mRNA knockdown was verified by real-time quantitative PCR (supplemental Table S1) and confirmed by Western blotting or a functional assay.
Stable Knockdown of UNG1/2, SMUG1, and TDG in HeLa Cells
Guided by different algorithms, we chose two candidate sequences for shRNA targeting of the UNG gene (HUGO Gene Nomenclature Committee HGNC:12572) in HeLa cells. The shRNA coding sequences were reconstituted by annealing synthetic oligonucleotides and cloned between the BglII and HindIII restriction sites of the pENTR/pSUPER+ vector (Addgene, Cambridge, MA). HeLa cells were transfected with the shRNA expression vectors together with a pcDNA3 vector (Invitrogen) harboring a neomycin resistance gene (in a molar ratio of 8:1) with the help of Effectene (Qiagen GmbH, Hilden, Germany). Stably transfected clones were selected in DMEM containing 1.1 g/liter G418 (Invitrogen). Single clones were picked after 2–3 weeks, expanded under selection pressure, and screened for UNG1 and UNG2 expression by Western blot analysis with the 2C12 anti-UNG mouse monoclonal antibody (Origene, AMS Biotechnology,). Mouse monoclonal C4 antibody to β-actin or the 119D5-F1 antibody to lamin B1 (Santa Cruz Biotechnology, Inc., Heidelberg, Germany) were used as a loading control. Monoclonal cell lines used in further experiments (including UNGsh-c12 with the lowest UNG protein expression level) were all obtained with the same shRNA expression construct. The sequences of the oligonucleotides used for annealing in this case were 5′-GATCCCCGGGACAGGATCCATATCATTTCAAGAGAATGATATGGATCCTGTCCCTTTTTGGAAA and 5′-AGCTTTTCCAAAAAGGGACAGGATCCATATCATTCTCTTGAAATGATATGGATCCTGTCCCGGG. All synthetic oligonucleotides used in this study were obtained from Eurofins MWG Operon (Ebersberg, Germany).
Stable knockdown of SMUG1 and TDG was performed by the same procedure. Of three shRNA constructs tested in each case, the best performance was achieved when the following oligonucleotides were used for cloning: 5′-GATCCGGCCAAGACAAAGCATGGGACATCTCGAGATGTCCCATGCTTTGTCTTGGTTTTTGGAAA annealed to 5′-AGCTTTTCCAAAAACCAAGACAAAGCATGGGACATCTCGAGATGTCCCATGCTTTGTCTTGGCCG (SMUG1) and 5′-GATCCGGGAACGAAATATGGACGTTCAACTCGAGTTGAACGTCCATATTTCGTTCTTTTTGGAAA annealed to 5′-AGCTTTTCCAAAAAGAACGAAATATGGACGTTCAACTCGAGTTGAACGTCCATATTTCGTTCCCG (TDG). In the case of TDG, single clones were screened by PA5-29140 rabbit polyclonal antibody (Thermo Fisher Scientific, Bonn, Germany). In the case of SMUG1, screening was performed by real-time PCR because none of the three antibodies tested (Origene, catalog nos. TA302931 and TA312730, and Santa Cruz Biotechnology, catalog no. sc-98849), could specifically detect the endogenous protein.
Construction and Verification of Expression Vectors Containing Uracil or a T:G Mismatch
Insertion of a single uracil in the transcribed or the non-transcribed DNA strand within the pEGFP-mODC-ZA expression vector (Fig. 1A) was achieved by nicking one strand of the reporter gene twice with the nicking endonuclease Nt.Bpu10I or Nb.Bpu10I and swapping the excised 18-nt fragment for a synthetic oligonucleotide as described previously (19). Reporter plasmids containing a single thymine mispaired with guanine were obtained by the same methodology that was used for the construction of vectors with a single U:G mismatch. The sequences of the oligonucleotides incorporated into the transcribed and the non-transcribed DNA strands are listed in supplemental Table S2.
FIGURE 1.
Influence of uracil on the expression of the EGFP reporter gene in transiently transfected HeLa cells. A, positions of uracil (underlined) paired with adenine (U:A) or mispaired with guanine (U:G) within the TS and NTS DNA strands of the EGFP gene. The cut positions of the nicking endonucleases used for insertion of the synthetic oligonucleotides are indicated (▿, Nt.Bpu10I; ▴, Nb.Bpu10I). Possible mRNA and protein sequences arising from transcription of templates containing uracil in the TS are on the basis of the specificity of transcriptional mutagenesis reported in Ref. 17. B, verification of uracil incorporation into vector DNA by excision with the E. coli UDG and incision of the resultant AP site by endonuclease IV (E IV). C, time course analyses of the effects of the unique uracils on EGFP expression, measured by flow cytometry as the specific fluorescence in single cells. Reported values are relative to those of the control constructs, obtained by incorporation of the respective unmodified oligonucleotides (T:A and C:G). Shown is a summary of transfection experiments performed with three independent preparations of each type of vector DNA. Data are mean ± S.D.
EGFP Protein Expression Analysis in Transfected Cells
The method for transfection and quantitative EGFP expression analysis in single cells by flow cytometry has been established and validated previously (20, 21). HeLa and the derived cell lines were transfected with a mixture containing equal amounts of the specified EGFP expression constructs (containing uracil or thymine opposite an adenine or a guanine) and the pDsRed-Monomer-N1 vector (Clontech, Saint-Germain-en-Laye, France), which was used as a tracer for transfected cells. Cells were formaldehyde-fixed prior to analyses as described previously. For the time course expression analyses, transfected cells were detached gently at either the 6 or 8 h (as indicated) time point and divided into several parts, one of which was fixed immediately. The rest were replated and fixed at the specified time intervals. Flow cytometry was performed exactly as described previously (20) using FACSCaliburTM and the CellQuestTM Pro software (BD Biosciences).
Analyses of the Uracil Excision Activities in Cell-free Extracts
Exponentially growing cells (4–5 × 107) were harvested in ice-cold phosphate-buffered saline containing 0.5 mm phenylmethanesulfonyl fluoride. After centrifugation (200 × g, 5 min, 4 °C), cell pellets were resuspended in 0.5 ml of lysis buffer (20 mm Tris-HCl (pH 8.0), 1 mm EDTA, and 250 mm NaCl), supplemented with protease inhibitor mixture (Roche Diagnostics) and placed on an ice slurry. Cells were disrupted using a Bachofer GM 70 HD ultrasonic processor (Bachofer GmbH, Reutlingen, Germany) equipped with a microtip (four 40-s sonication cycles at a 20% power setting; duty cycle, 0.4). Insoluble material was removed by ultracentrifugation (105 × g, 45 min, 4 °C), and protein concentration was determined by Bradford assay (22). Single use aliquots were prepared and stored at −80 °C.
Uracil-excising activity in the extracts was measured by cleavage of the resultant apyrimidinic site (AP site). The DNA single strand break was generated by an AP endonuclease (or lyase) activity endogenously present in the extracts. The 15-μl incision reactions contained 100 ng of plasmid DNA and up to 20 μg of cell-free extract protein (as indicated) in 10 mm HEPES (pH 7.5), 200 mm NaCl, 1 mm EDTA, and 33 μg/ml nuclease-free bovine serum albumin (NEB GmbH, Frankfurt am Main, Germany). After incubation at 37 °C for 60 min, the reactions were stopped by adding SDS to 0.1% and heating at 50 °C for 3 min, followed by addition of DNA loading dye and electrophoresis in agarose gels containing ethidium bromide (0.5 mg/liter). Control reactions with uracil-DNA glycosylase and endonuclease IV of Escherichia coli were performed as described previously (19), but the incubation time was increased to 1 h. DNA strand cleavage was determined from the relative intensities of DNA bands by the GelDocTM XR+ molecular imager and Image LabTM software (Bio-Rad) and adjusted for the 2.4-fold difference in the fluorescence yield between the covalently closed and nicked circular DNA (21).
Analysis of the Gene and Transcript Abundances by Real-time Quantitative PCR
Transfected cells were split in three equal parts for RNA, DNA, and protein analyses. Total DNA and RNA were isolated by standard procedures (23). Samples were treated with DNase I supplemented with RiboLockTM RNase inhibitor (Thermo Fisher Scientific) at 37 °C for 5 min, and the integrity of RNA was verified by denaturing agarose gels. RT was accomplished with the RevertAidTM first strand cDNA synthesis kit (Thermo Fisher Scientific). To monitor the RT efficiencies and exclude possible contamination of RNA samples with vector DNA, aliquots were withdrawn prior to the reverse transcription for subsequent comparison with the RT samples.
Real-time quantitative PCR analyses were performed with the LightCycler 1.5 and the LightCycler FastStart DNA MasterPLUS SYBR Green I kit (Roche Diagnostics) following the protocol and criteria described previously (23). Copy numbers of the uracil-containing EGFP constructs were determined on the basis of standard curves obtained with uracil-free DNA from parallel transfections. The obtained values were normalized for transfection efficiencies that were determined from copy numbers of the DsRed-Monomer-N1 vector DNA (internal reference). Differently from the gene copy number analyses, the standard curves for cDNA quantification were built by serial dilutions of DNA recovered from the same transfected sample. Thus, each transcript was directly quantified with respect to its own template DNA.
RESULTS
A Single Uracil in DNA Causes Decreased Gene Expression in Human Cells
To investigate the consequences of a single uracil located in a transcribed region of a gene on transcriptional output, we incorporated synthetic oligonucleotides containing a single deoxyuracil into the EGFP coding sequence of the pEGFP-mODC-ZA expression vector using a methodology described previously (19). The oligonucleotides were inserted into either the transcribed or the non-transcribed strand of the gene, with single uracils placed opposite an adenine (U:A) or a guanine (U:G) (Fig. 1A). Analytical digestion of the vector DNA by a combination of the uracil-DNA glycosylase and endonuclease IV of E. coli confirmed that uracil was incorporated into all vector molecules (Fig. 1B).
Transfection of HeLa cells with the constructed vectors (U:A, U:G, and the respective T:A and C:G control substrates produced by incorporation of the respective unmodified oligonucleotides) showed that uracils in both contexts have a clear negative effect on EGFP expression (Fig. 1C). Protein expression was monitored over a period of between 8 and 48 h after transfection. Provided that several hours are required for protein synthesis and folding of the fluorophore structure, it is expected that the major portion of the EGFP expression observable at the beginning of this period (the 8-h time point) was derived from transcription of the templates containing unrepaired uracil immediately after the gene delivery. Remarkably, the vectors containing uracil (U:A and U:G) were expressed at this time point nearly as well as the vectors containing no base modification, indicating that unrepaired uracil does not interfere with the transcriptional activity. Although uracils in the transcribed DNA strand (TS) are expected to elicit transcriptional mutagenesis in a fraction of mRNA molecules (Fig. 1A), this had no detectable effect on overall EGFP fluorescence because its yields did not differ between the vectors containing uracils in the opposite strands. Barely detectable early after transfections, the inhibitory effect of uracil on gene expression was building up over time, as documented by a progressive loss of EGFP fluorescence, which had the highest rate between 8 and 16 h post-transfection (Fig. 1C). No subsequent recovery of gene expression was detected, indicating that transcription failed to resume for a prolonged period of time. Interestingly, the degree of inhibition of gene expression by the U:G mismatch was somewhat milder than by the U:A base pair, regardless of the DNA strand. The quantitative difference between the U:A and U:G substrates was significant starting from 24 h post-transfection (paired Student's two-tailed t test, p < 0.05).
Very similar dynamics and magnitudes of inhibition of EGFP gene expression were observed for uracils situated in the TS and in the non-transcribed DNA strand (NTS), indicating that a direct arrest of the elongating RNA polymerase II on the damage site was not involved in the mechanism of inhibition of transcription. It is interesting to note that the time course of expression of vectors containing a single uracil (Fig. 1C) greatly resembled the behavior of analogous constructs containing an oxidative base modification, 8-oxo-7,8-dihydroguanine, described previously (23). We recently showed that 8–8-oxo-7,8-dihydroguanine is harmful for transcription in cells only if excised by the specific DNA glycosylase OGG1, indicating that BER interferes with the transcription of genes (21). By analogy, we suggested that a similar mechanism might underlie the inhibition of transcription by uracil.
UNG1/2 Excises Uracil Paired with Adenine and Contributes to the Inhibition of Gene Expression
Because of the evidence that UNG1/2 is the most important human UDG for the removal of uracil paired with adenine (5, 14, 24), we addressed the influence of UNG1/2 on the expression of vectors containing a single U:A base pair. We generated several isogenic cell lines with varying expression levels of the UNG1/2 DNA glycosylase by stable expression of a specific shRNA. A clonal cell line with the highest degree of the protein knockdown (UNGsh-c12) retained 15% of UNG1 and 24% of UNG2 protein expression present in the isogenic cell line stably transfected with the empty vector (Fig. 2, A and B).
FIGURE 2.
Influence of UNG1/2 protein knockdown on the excision of uracil paired with adenine. A, structure and exon specificity (stars) of the shRNA targeting both alternative transcripts of the human UNG gene. B, Western blot analysis of the UNG1 and UNG2 protein expression levels in HeLa clones expressing UNG1/2-specific shRNA (UNGsh) and in the isogenic clone stably transfected with an empty vector (no sh). β-Actin antibody was used for normalization between the samples. C, incision activities in extracts obtained from HeLa cells stably transfected with the UNG1/2 shRNA expression vector (UNGsh-c12) or an empty vector toward the plasmid DNA containing a unique U:A base pair versus the control T:A construct. Excision of uracil, coupled with subsequent incision of the resultant AP site, was detected by conversion of covalently closed DNA into the open circular form. Low molecular weight DNA in gels originated from the cell-free extracts. E IV, endonuclease IV.
To assess to which extent the excision of uracil paired with adenine is influenced by UNG1/2 knockdown, we measured the incision of the vector DNA by protein extracts obtained from the UNGsh-c12 cell line and the control cell line (no sh) with normal UNG1/2 protein levels. Covalently closed circular vector DNA containing a single U:A base pair was efficiently converted into the nicked circular form by incubation with the control extract (Fig. 2C). The efficient strand incision indicates that sufficient AP endonuclease activity was intrinsically present in the extracts to cleave the abasic sites generated by the excision of uracil. The incision of the U:A substrate was proportional to the protein amount, whereas the T:A substrate remained intact under the same reaction conditions, indicating that the assay is suited for detection of UDG activity. The incision activity toward the U:A substrate was reduced by at least a factor of 4 in the extracts obtained from the UNGsh-c12 cell line compared with the control extracts, as judged by the protein amounts required to achieve an equivalent conversion of the substrate.
To inhibit the nonspecific endonuclease activity present in the cell extracts, we had to perform the uracil excision/strand incision reactions described above in the presence of EDTA. Because magnesium cations have been reported to enhance the endonuclease activity of human APE1 (the enzyme essential for AP site cleavage (25)), our reaction conditions could be suboptimal for the incision of the AP sites arising from the excision of uracil. However, the addition of magnesium to the reactions resulted in an increase of the nonspecific endonuclease cleavage of both T:A and U:A substrates (data not shown), therefore necessitating the omission of the metal. To detect whether a fraction of AP sites remained uncleaved under these conditions, we performed parallel reactions supplemented with bacterial endonuclease IV, which does not require magnesium. This did not further enhance the incision of the uracil-containing substrate by the cell extracts (data not shown), demonstrating that the endogenously present AP site endonuclease activity was in excess. Thereby, of the two reaction steps, the excision of uracil was clearly the rate-limiting one.
To measure the effect of UNG1/2 knockdown on the expression of vectors containing uracil paired with adenine, we transfected the vectors containing a single uracil in either the transcribed or the non-transcribed DNA strand into three isogenic HeLa-derived cell lines with varying UNG1/2 protein expression levels and analyzed the EGFP protein expression at 6, 12, and 24 h post-transfection. At the beginning of the time course, expression did not differ between the T:A and U:A constructs (Fig. 3). The same result was registered in all cell lines, regardless of whether the uracil was present in the transcribed or the non-transcribed DNA strand, again indicating that unrepaired uracil causes neither a transcriptional arrest nor EGFP fluorophore misfolding as a result of transcriptional mutagenesis. By 12 h post-transfection, expression of the U:A constructs was reduced by more than half in the control cell line. Importantly, the magnitude of this effect was attenuated significantly by UNG1/2 knockdown. The inhibitory effect of uracil on gene expression was least pronounced in the UNGsh-c12 cell line, which had the lowest UNG1 and UNG2 protein levels of the three cell lines tested and the lowest U:A excision activity (see also Fig. 2). The UNGsh-c6 cell line with intermediate UNG1/2 protein expression levels showed an intermediate level of inhibition of gene expression by uracil. Therefore, the results show that the inhibitory effect of uracil paired with adenine on gene expression are proportional to the cellular UNG1/2 levels, strongly suggesting that BER initiation interferes with transcription of the affected gene. It has to be noted that the negative effect of uracil on gene expression was merely retarded, not fully abolished, by the knockdown because the inhibition achieved the same strength in all three cell lines by the 24-h time point. This result can be explained by the residual UNG1/2 activity in cells.
FIGURE 3.
Expression of the EGFP reporter gene containing a unique uracil paired with adenine (U:A) in HeLa-derived cell lines expressing varying levels of the UNG1 and UNG2 proteins (no sh > UNGsh-c6 > UNGsh-c12). A and B, representative flow cytometry experiments for uracil positioned in the TS (A) and NTS (B) and for the respective T:A control constructs. Shown are overlaid distribution plots of EGFP fluorescence in cell populations gated by the expression of the transfection marker Ds-Monomer. The columns show median EGFP fluorescence in cells. No sh, empty vector. C, relative EGFP expression (U:A/T:A) for three (UNGsh-c6) or four independent experiments, in each of which all cell lines were transfected in parallel. Data are mean ± S.D. **, p < 0.01; ***, p < 0.001; paired two-tailed Student's t test.
Effect of Uracil Opposite a Guanine on Reporter Gene Expression Is Independent from Mismatch Recognition
Previous experiments (Fig. 1) showed that a uracil opposite a guanine inhibits EGFP reporter gene expression in an indirect manner, similar to the uracil paired with adenine, albeit with a smaller magnitude. Because U:G is not a canonical Watson-Crick base pair, we wanted to know whether the inhibition of transcription in this case is caused by BER activity (as in the case of the U:A pairs) or by a mechanism related to mismatch recognition. To answer this question, we compared the expression of vectors containing the U:G and T:G wobble base pairs in HeLa cells. We found that a uracil opposite a guanine had a much stronger negative effect on gene expression than a thymine in the same position (Fig. 4). Furthermore, the expression of the T:G constructs was not decreasing in time (data not shown), in contrast with the result shown for the U:G constructs (Fig. 1), suggesting that the two types of wobble base pairs (U:G and T:G) are processed differently in the host cells and that a higher BER efficiency of the U:G substrate results in a stronger effect on gene transcription.
FIGURE 4.
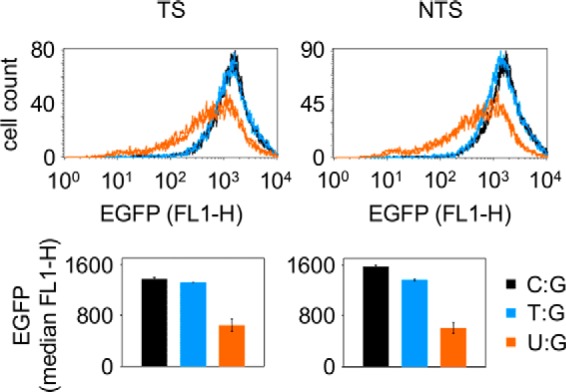
Influence of the U:G and T:G wobble base pairs on the expression of the EGFP reporter gene. Flow cytometry analyses of HeLa cells 24 h post-transfection with vectors containing a unique T:G (blue), U:G (amber), or C:G (black) base pair. Shown are distribution plots of EGFP fluorescence over the transfected cell populations (there are two closely overlaid lines for each construct showing duplicate transfections) and the median values (bars and whiskers represent mean and range for the duplicates). TS and NTS refer to the base opposite the G.
Incision at Uracil Opposite a Guanine by Cell-free Extracts Is Partly Independent from UNG1/2
To compare the excision efficiencies of uracil and thymine (both opposite a guanine), we incubated the respective plasmid constructs with BER-proficient cell extracts (Fig. 5). The yield of the DNA strand scission (and, by inference, also the efficiency of the preceding base excision reaction) was >10-fold higher for uracil than for thymine, as judged by comparison of the protein amounts required to achieve an equivalent degree of the incision. Knockdown of UNG1/2 protein did not influence the excision of thymine, as expected. However, it led to a clear decrease in the excision of uracil. It is noteworthy that the 4-fold reduction of the UNG2 protein level (and a 6- to 7-fold reduction of UNG1) in the UNGsh-c12 cell line compared with the empty vector control (Fig. 2B) resulted in only about a 2-fold decrease of U:G cleavage activity (Fig. 5). This result confirms that UNG1/2 contributes to U:G excision but also indicates a significant input of another uracil-DNA glycosylase (or other glycosylases). Alternatively, the removal of uracil might take place by a recently described mechanism initiated directly by APE1 (26).
FIGURE 5.
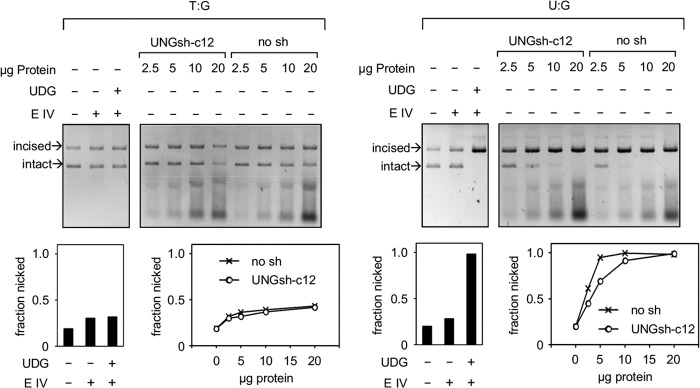
Influence of UNG1/2 knockdown on the excision of uracil opposite a guanine. Incision activities of the cell-free extracts toward the plasmid constructs containing unique U:G and T:G incorrect base pairs. The extracts of cells expressing the UNG1/2 shRNA (UNGsh-c12) and the control extracts (no sh) are the same as in Fig. 2. E IV, endonuclease IV.
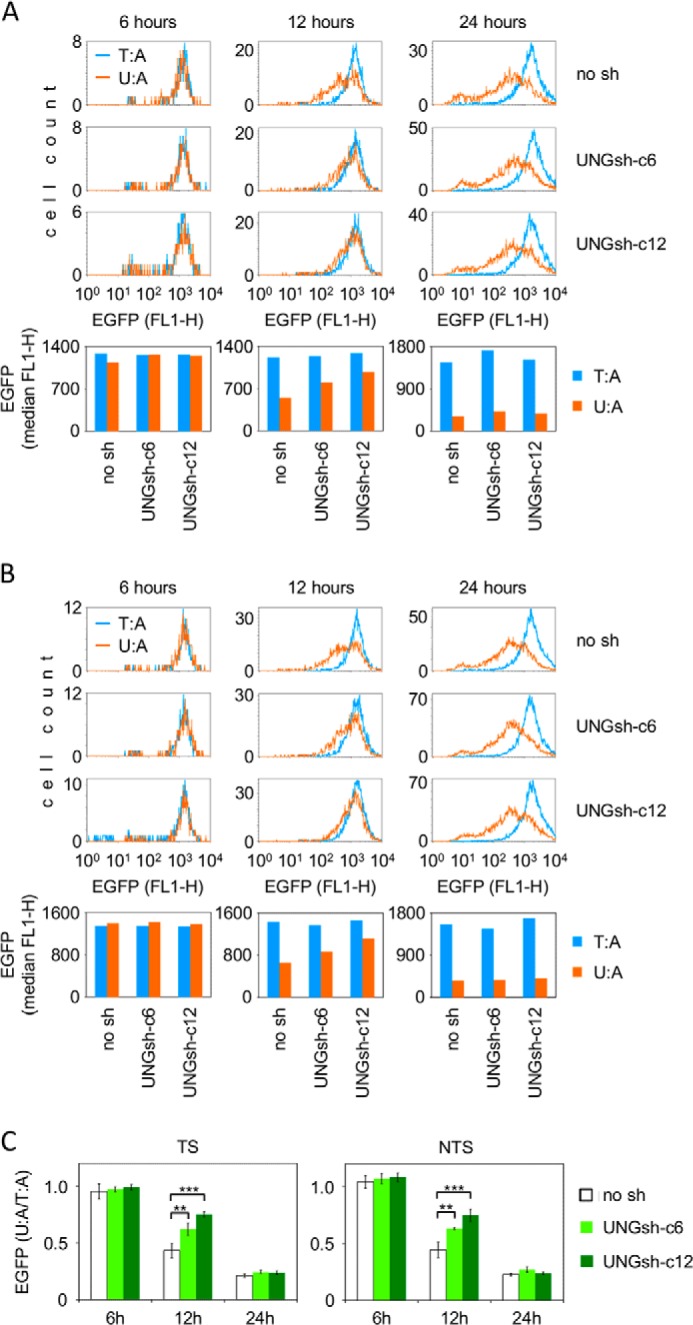
UNG1/2 Has No Effect on the Cellular Expression of the U:G Reporter Construct
Compared with the magnitude of inhibition of transcription by U:A pairs, the effect of a uracil opposite to a guanine was somewhat weaker (Fig. 1), although the excision of this substrate by cell-free extracts takes place with an even higher efficiency (Figs. 2 and 5). This can be explained by a previous finding that AP sites opposite G are repaired at faster rates than those opposite A (5, 14), which would minimize the negative effect on transcription in the case of U:G. Alternatively, the recognition and downstream processing of the U:A and U:G base pairs in cells may follow different pathways. To investigate whether the inhibition of reporter gene expression by uracil mispaired with guanine is mediated by UNG1/2, as shown above for the U:A pairs (Fig. 3), we analyzed the expression of the U:G reporter constructs in the UNG1/2 knockdown cell lines. In contrast with the results obtained for the U:A constructs, there was no difference in EGFP gene expression levels between the knockdown cell lines (UNGsh-c6 and UNGsh-c12) and the control cell line (no sh) (Fig. 6). The initial expression levels (the 6-h time point) for the U:G construct and the control vector containing cytosine paired with guanine were the same in all three cell lines, demonstrating that, also in this configuration, no direct inhibition of transcription takes place in the presence of uracil. At the end of the time course (24 h), the expression of the U:G vector was decreased to the same extent in the three cell lines, showing that processing of uracil mispaired with guanine is harmful to gene expression in all of them. Also at the intermediate time point (12 h), the expression levels were the same between the cell lines (Fig. 6), which is different from the behavior of the U:A substrates discussed above (Fig. 3). This observation was confirmed by additional experiments where other intermediate time points were chosen (data not shown). From the absence of any influence of the cellular UNG1/2 protein levels on the magnitude and dynamics of the inhibition of the gene expression, we infer that, in the cellular context, UNG1/2 most probably does not contribute to excision of uracil mispaired with guanine. Otherwise, a delayed effect would be expected in the UNG1/2 knockdown cell lines, as shown for the U:A construct.
FIGURE 6.
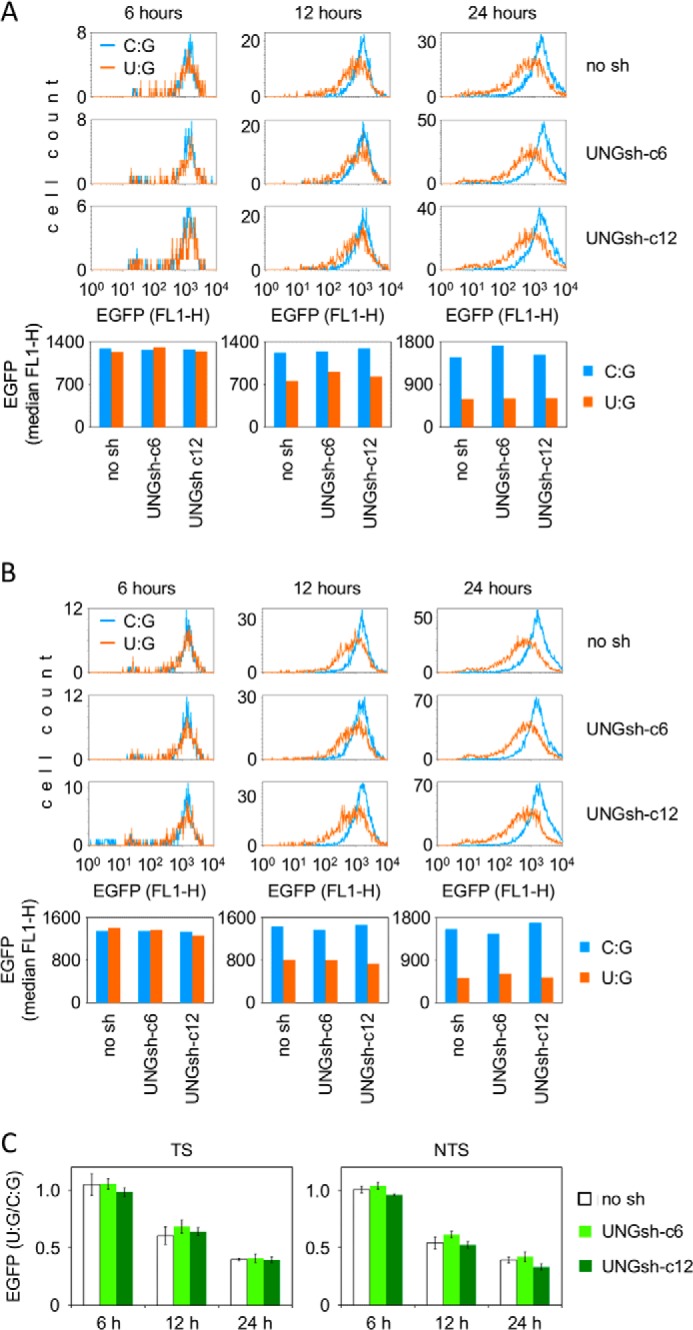
Expression of the EGFP reporter gene containing a unique uracil opposite a guanine (U:G) in HeLa-derived cell lines with varying UNG1 and UNG2 proteins levels. A and B, representative flow cytometry experiments for uracil positioned in the TS (A) and NTS (B) and for the respective C:G control constructs. Shown are overlaid distribution plots of EGFP fluorescence and the respective median EGFP fluorescence values (bottom panels). No sh, empty vector. C, mean relative EGFP expression (U:G/C:G) for three (UNGsh-c6) or four independent experiments. Data are mean ± S.D.
TDG and SMUG1 Do Not Contribute to Cell-free Incision of Uracil-containing Plasmid DNA
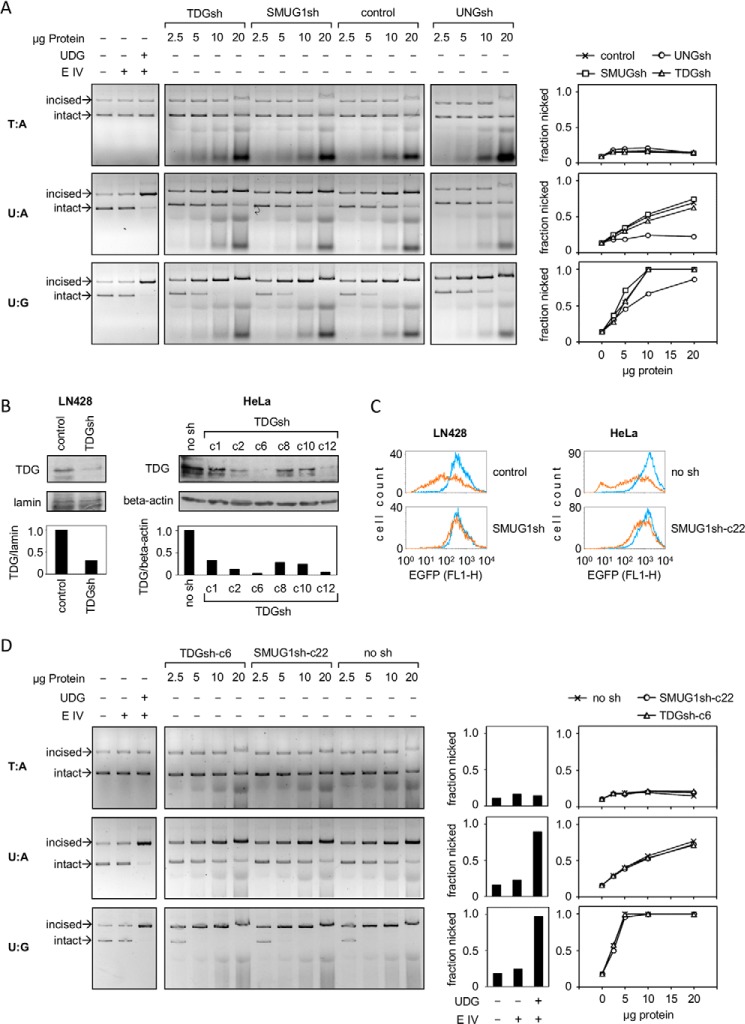
All four human UDG enzymes can excise uracil opposite a guanine in biochemical assays (27) and, hence, could account for the remaining incision activity in the UNG1/2 knockdown cells. Because MBD4 activity is believed to be confined to methylated CpG-rich DNA, we considered SMUG1 and TDG as plausible candidates for the residual U:G incision activity. We tested cleavage activity toward covalently closed plasmid DNA in protein extracts prepared from glioblastoma cell lines where SMUG1 or TDG were knocked down by stable expression of the specific shRNA constructs. Both knockdown cell lines retained the same cleavage activities toward the U:A and U:G substrates as the maternal LN428 cell line (Fig. 7A). At the same time, cleavage of the U:A construct was fully prevented by UNG knockdown, whereas incision at U:G was partly inhibited, very similarly to the results of UNG1/2 knockdown in HeLa cells described previously (Figs. 2 and 5).
FIGURE 7.
Quantification of incision activities at unique U:A and U:G pairs in extracts obtained from SMUG1 and TDG knockdown cells. A, incision of the specified plasmid constructs by LN428 glioblastoma cell-free extracts. Cells were stably transfected with the specified sh constructs. E IV, endonuclease IV. B, Western blot analyses of TDG protein knockdown in LN428 and HeLa cells. C, functional analyses of SMUG1 knockdown (fluorescence distribution plots obtained by flow cytometry) in LN428 and HeLa cells transfected with an expression vector containing multiple 5-hmUs (amber line) and the unmodified vector (blue line). D, incision of the specified plasmid constructs by cell-free extracts obtained from selected HeLa-derived clones stably transfected with the specified sh constructs.
Western blot analyses confirmed that TDG was strongly (∼4-fold) down-regulated at the protein level in the tested cell line (Fig. 7B), which would cause a detectable decrease in the incision activity if TDG were involved. SMUG1 was down-regulated by knockdown to 23% of the control level (measured as the mRNA copy number). However, expression was very low, even in the control cell line. None of the three tested antibodies could specifically detect the endogenous SMUG1 protein in cell extracts. Also, the cleavage activity toward the specific substrate of SMUG1, 5-(hydroxymethyl)uracil (5-hmU) was barely detectable in cell-free extracts (data not shown). However, expression of a transfected construct containing multiple 5-hmUs was impaired in the maternal cell line but not in the SMUG1 knockdown cells (Fig. 7C), indicating that the excision takes place in cells and confirming the potent functional knockdown.
We further performed stable TDG and SMUG1 knockdown in HeLa cells, both of which were successful (Fig. 7, B and C). Analyses of the cleavage activities in HeLa cell-free extracts showed that the U:G substrate is incised with a slightly higher efficiency than the LN428 cell extracts. However, also in this cell model, cleavage activities of both U:A and U:G were not at all affected by knockdown (Fig. 7D). Together, knockdown of SMUG1 and TDG confirmed that UNG1/2 is the only enzyme that accounts for the excision of uracil paired with adenine in cell-free extracts. UNG1/2 also contributes to U:G excision (albeit not to the whole of it), whereas SMUG1 and TDG do not.
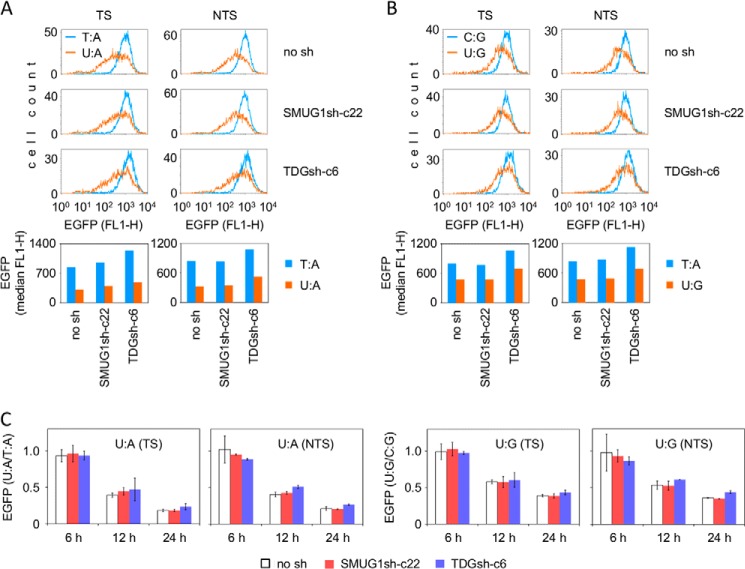
TDG and SMUG1 Are Irrelevant to the Inhibition of Transcription by U:A and U:G Base Pairs
Although no detectable SMUG1 and TDG incision activity toward the U:A and U:G plasmid substrates was present in cell-free extracts, there remained a possibility that these enzymes could be more efficient in a cellular context and initiate some BER in live cells. If present, such cryptic excision activity could contribute to the inhibition of transcription. However, time course expression analyses of the U:A and U:G constructs did not reveal any influence of SMUG1 or TDG knockdown on EGFP levels, indicating that none of the two DNA glycosylases can be relevant for the decline of transcription of the uracil-containing DNA (Fig. 8). We also performed time course expression analyses in the glioblastoma LN428 cell line and the derived SMUG1 and TDG knockdown cells with the same result (data not shown).
FIGURE 8.
Expression of vectors containing a unique uracil in HeLa-derived SMUG1 (SMUG1sh-c22) and TDG (TDGsh-c6) knockdown cell lines. TS and NTS refer to the DNA strand containing uracil. A and B, representative flow cytometry data (12 h post-transfection) for the U:A (A) and U:G (B) constructs. No sh, empty vector. C, relative EGFP expression (U:A/T:A and U:G/C:G) for two independent preparations of each expression construct at the indicated times post-transfection. Data are mean and range.
U:A and U:G Base Pairs Induce Decreased EGFP Transcript Levels
Thanks to the simple fluorophore maturation mechanism, EGFP protein expression levels generally reflect well the gene transcription levels in cell systems where the mRNA lifetime and the translation rate remain constant (20, 28). Therefore, the observed decrease of EGFP protein levels caused by intracellular processing of uracil is very likely to be a result of impaired gene transcription. To verify this, we measured the transcript levels in cells transfected with vectors containing single U:A or U:G base pairs (with uracil located in either the TS or NTS) and of the respective control (T:A and C:G) constructs. The results confirmed that the decreased protein expression levels in cells transfected with the uracil-containing vectors were indeed accompanied by substantially diminished EGFP mRNA levels (Fig. 9), thereby indicating that either the transcriptional activity or the transcript stability are clearly affected.
FIGURE 9.

EGFP transcript levels and the gene copy numbers residing in cells 24 h post-transfection with the U:A and U:G constructs containing the uracil in the TS or the NTS. Real-time quantitative PCR analyses of a representative experiment (quadruplicates). Data are mean ± S.D. Similar results were obtained in three (TS) or four (NTS) independent transfection experiments with separate vector preparations.
In certain cell lines, foreign DNA can undergo extensive deamination of cytidines and subsequent UNG2-dependent degradation (29). Although this process is only known to take place in cells expressing high levels of the APOBEC3 family cytidine deaminases, we measured the amounts of vector DNA persisting in cells 24 h post-transfection to examine whether a single uracil would trigger degradation of the vector DNA in our system. The recovered amounts of vector DNA were slightly reduced for both uracil-containing constructs (by ∼20% for the U:A and <10% for the U:G) relative to the respective control constructs (Fig. 9). This result indicates that the availability of vector DNA to cells is, at least to some extent, affected by the presence of uracil. Therefore, we conclude that the effects of uracil on reporter gene expression are the combined result of a negative influence on the uptake or retention of the vector plus the inhibition of transcription of the available template DNA.
DISCUSSION
Modulation of gene transcription is emerging as an important cellular response to DNA damage (30–32). In this study, we investigated the effects of a single uracil placed in the transcribed region of a mammalian reporter vector on gene expression. Our main finding is that the removal of uracil leads to a declined transcription of the gene, whereas unprocessed uracils are harmless to transcription in human cells. In the case of uracil paired with adenine, the protein knockdown experiments assigned the major roles both in the excision and in the inhibition of transcription to the UNG1/2 DNA glycosylase (Figs. 2 and 3). The uracil excision products (abasic site and single strand breaks) may interfere with transcription in cells either by directly interacting with elongating RNA polymerases, as demonstrated previously in cell-free transcription systems (18, 33, 34), or by generating a signal for persistent repression of gene transcription. Such a scenario is further supported by independent evidence that structurally unrelated oxidative base modifications, 5-hmU (Fig. 7C) and 8-oxo-7,8-dihydroguanine (23), also lead to sustained inhibition of transcription, resulting from cellular processing mediated, respectively, by SMUG1 and OGG1 DNA glycosylases.
The dominant role of UNG1/2 in the excision of U:A base pairs in cells is in agreement with earlier findings under cell-free conditions on the basis of the immunodepletion of different UDGs from fully repair-proficient cell extracts (5, 6, 24). In contrast, the fate of U:G appears to be more complex because SMUG1 and TDG have been suggested to contribute to repair in addition to UNG1/2, at least in cell-free extracts (5, 14). Moreover, alternative to a canonical BER, this lesion can undergo a direct incision by human APE1 (26) and may also be coprocessed by the mismatch repair if a nick or another uracil are present simultaneously in DNA (35). In our substrates containing a unique uracil, it was certainly not the mismatch repair activity that caused the inhibition of transcription (Fig. 4). Also, SMUG1 and TDG DNA glycosylases did not contribute to any significant extent to the U:G incision activity under cell-free conditions (Fig. 7) and to the negative effect on transcription in cells (Fig. 8). Therefore, a direct incision by human APE1 appears as the most likely mechanism for cellular processing of the U:G pairs in addition to BER initiated by UNG1/2.
As a final point, we want to bring attention to the strength of the observed negative effects of single base modifications on the transcriptional output of the affected gene. On the time scale of the performed experiments, a unique U:A pair in the transcribed gene region resulted in an approximately 5-fold (and a U:G pair in an approximately 3-fold) overall decrease of expression of the encoded protein. In both cases, the magnitude of the effect is much higher than reported previously for 8-oxo-7,8-dihydroguanine (21). It is tempting to suggest that suppression of transcriptional activity may be physiologically important under some circumstances, for instance during replication of chromosomal DNA. The interference between the transcription and replication processes is a prominent source of genomic instability (36, 37) that might be prevented by down-regulation of the transcription of newly replicated DNA containing elevated levels of U:A base pairs. UNG1/2 may also contribute to innate cell defense mechanisms against foreign (e.g. viral) uracil-containing DNA by inhibiting transcription and, thus, acting in synergy with the APOBEC-induced DNA editing and degradation pathway described previously (29).
Supplementary Material
Acknowledgments
We thank Julia Allgayer for help with SMUG1 knockdown, Benjamin Hackner and Markus Müller (Carell group, LMU Munich) for oligonucleotides containing 5-hmU, J. Pablo Radicella (CEA Fontenay-aux-roses) for comments regarding the manuscript, and Geir Slupphaug (Norwegian University of Science and Technology) for advice regarding the immunodetection of UNG.
This work was supported by Deutsche Forschungsgemeinschaft Grants KH 263/1 and KH 263/2 (to A. K.).

This article contains supplemental Tables S1 and S2.
- BER
- base excision repair
- UDG
- uracil-DNA glycosylase
- TDG
- thymine-DNA glycosylase
- UNG
- uracil-DNA glycosylase
- EGFP
- enhanced GFP
- AP
- apurinic-apyrimidinic
- TS
- transcribed DNA strand
- NTS
- non-transcribed DNA strand
- 5-hmU
- 5-(hydroxymethyl)uracil.
REFERENCES
- 1. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 2. Frederico L. A., Kunkel T. A., Shaw B. R. (1990) A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 29, 2532–2537 [DOI] [PubMed] [Google Scholar]
- 3. Shen J. C., Rideout W. M., 3rd, Jones P. A. (1994) The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22, 972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krokan H. E., Drabløs F., Slupphaug G. (2002) Uracil in DNA: occurrence, consequences and repair. Oncogene 21, 8935–8948 [DOI] [PubMed] [Google Scholar]
- 5. Kavli B., Sundheim O., Akbari M., Otterlei M., Nilsen H., Skorpen F., Aas P. A., Hagen L., Krokan H. E., Slupphaug G. (2002) hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 277, 39926–39936 [DOI] [PubMed] [Google Scholar]
- 6. Kavli B., Andersen S., Otterlei M., Liabakk N. B., Imai K., Fischer A., Durandy A., Krokan H. E., Slupphaug G. (2005) B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J. Exp. Med. 201, 2011–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nilsen H., Haushalter K. A., Robins P., Barnes D. E., Verdine G. L., Lindahl T. (2001) Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 20, 4278–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Waters T. R., Swann P. F. (1998) Kinetics of the action of thymine DNA glycosylase. J. Biol. Chem. 273, 20007–20014 [DOI] [PubMed] [Google Scholar]
- 9. Cortázar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., Wirz A., Schuermann D., Jacobs A. L., Siegrist F., Steinacher R., Jiricny J., Bird A., Schär P. (2011) Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423 [DOI] [PubMed] [Google Scholar]
- 10. Hendrich B., Hardeland U., Ng H. H., Jiricny J., Bird A. (1999) The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401, 301–304 [DOI] [PubMed] [Google Scholar]
- 11. Nilsen H., Otterlei M., Haug T., Solum K., Nagelhus T. A., Skorpen F., Krokan H. E. (1997) Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 25, 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eftedal I., Guddal P. H., Slupphaug G., Volden G., Krokan H. E. (1993) Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 21, 2095–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doseth B., Visnes T., Wallenius A., Ericsson I., Sarno A., Pettersen H. S., Flatberg A., Catterall T., Slupphaug G., Krokan H. E., Kavli B. (2011) Uracil-DNA glycosylase in base excision repair and adaptive immunity: species differences between man and mouse. J. Biol. Chem. 286, 16669–16680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Visnes T., Akbari M., Hagen L., Slupphaug G., Krokan H. E. (2008) The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair 7, 1869–1881 [DOI] [PubMed] [Google Scholar]
- 15. Slupphaug G., Eftedal I., Kavli B., Bharati S., Helle N. M., Haug T., Levine D. W., Krokan H. E. (1995) Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry 34, 128–138 [DOI] [PubMed] [Google Scholar]
- 16. Verri A., Mazzarello P., Biamonti G., Spadari S., Focher F. (1990) The specific binding of nuclear protein(s) to the cAMP responsive element (CRE) sequence (TGACGTCA) is reduced by the misincorporation of U and increased by the deamination of C. Nucleic Acids Res. 18, 5775–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuraoka I., Endou M., Yamaguchi Y., Wada T., Handa H., Tanaka K. (2003) Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II: implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 278, 7294–7299 [DOI] [PubMed] [Google Scholar]
- 18. Kathe S. D., Shen G. P., Wallace S. S. (2004) Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J. Biol. Chem. 279, 18511–18520 [DOI] [PubMed] [Google Scholar]
- 19. Lühnsdorf B., Kitsera N., Warken D., Lingg T., Epe B., Khobta A. (2012) Generation of reporter plasmids containing defined base modifications in the DNA strand of choice. Anal. Biochem. 425, 47–53 [DOI] [PubMed] [Google Scholar]
- 20. Khobta A., Anderhub S., Kitsera N., Epe B. (2010) Gene silencing induced by oxidative DNA base damage: association with local decrease of histone H4 acetylation in the promoter region. Nucleic Acids Res. 38, 4285–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allgayer J., Kitsera N., von der Lippen C., Epe B., Khobta A. (2013) Modulation of base excision repair of 8-oxoguanine by the nucleotide sequence. Nucleic Acids Res. 41, 8559–8571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bradford M. M. (1976) Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 23. Kitsera N., Stathis D., Lühnsdorf B., Müller H., Carell T., Epe B., Khobta A. (2011) 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 39, 5926–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akbari M., Otterlei M., Peña-Diaz J., Aas P. A., Kavli B., Liabakk N. B., Hagen L., Imai K., Durandy A., Slupphaug G., Krokan H. E. (2004) Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 32, 5486–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barzilay G., Mol C. D., Robson C. N., Walker L. J., Cunningham R. P., Tainer J. A., Hickson I. D. (1995) Identification of critical active-site residues in the multifunctional human DNA-repair enzyme Hap1. Nat. Struct. Biol. 2, 561–568 [DOI] [PubMed] [Google Scholar]
- 26. Prorok P., Alili D., Saint-Pierre C., Gasparutto D., Zharkov D. O., Ishchenko A. A., Tudek B., Saparbaev M. K. (2013) Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc. Natl. Acad. Sci. U.S.A. 110, E3695–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krokan H. E., Bjørås M. (2013) Base excision repair. Cold Spring Harb. Perspect. Biol. 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soboleski M. R., Oaks J., Halford W. P. (2005) Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. FASEB J. 19, 440–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. (2010) APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Warmerdam D. O., Kanaar R. (2010) Dealing with DNA damage: relationships between checkpoint and repair pathways. Mutat. Res. 704, 2–11 [DOI] [PubMed] [Google Scholar]
- 31. Khobta A., Epe B. (2012) Interactions between DNA damage, repair, and transcription. Mutat. Res. 736, 5–14 [DOI] [PubMed] [Google Scholar]
- 32. Lagerwerf S., Vrouwe M. G., Overmeer R. M., Fousteri M. I., Mullenders L. H. (2011) DNA damage response and transcription. DNA Repair 10, 743–750 [DOI] [PubMed] [Google Scholar]
- 33. Roy D., Zhang Z., Lu Z., Hsieh C. L., Lieber M. R. (2010) Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol. 30, 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tornaletti S., Maeda L. S., Hanawalt P. C. (2006) Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem. Res. Toxicol. 19, 1215–1220 [DOI] [PubMed] [Google Scholar]
- 35. Schanz S., Castor D., Fischer F., Jiricny J. (2009) Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc. Natl. Acad. Sci. U.S.A. 106, 5593–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Helmrich A., Ballarino M., Nudler E., Tora L. (2013) Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 20, 412–418 [DOI] [PubMed] [Google Scholar]
- 37. Aguilera A., García-Muse T. (2013) Causes of genome instability. Annu. Rev. Genet. 47, 1–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.