Background: DNA polymerase α-primase synthesizes chimeric RNA/DNA primers for replicative polymerases.
Results: We defined elements that modulate polα and prim activities.
Conclusion: The C-terminal domain of the catalytic subunit of polymerase α and the B-subunit regulate the priming of DNA replication.
Significance: We provide new information on the regulation of RNA/DNA synthesizing complex that is indispensable for replication in eukaryotes.
Keywords: DNA Polymerase, DNA Primase, DNA Replication, Protein-Protein Interaction, RNA, C-terminal Domain, DNA Polymerase α-Primase, DNA Polymerase Activity, DNA Replication Initiation, Primase Activity
Abstract
The initiation of DNA synthesis during replication of the human genome is accomplished primarily by the DNA polymerase α-primase complex, which makes the RNA-DNA primers accessible to processive DNA pols. The structural information needed to understand the mechanism of regulation of this complex biochemical reaction is incomplete. The presence of two enzymes in one complex poses the question of how these two enzymes cooperate during priming of DNA synthesis. Yeast two-hybrid and direct pulldown assays revealed that the N-terminal domain of the large subunit of primase (p58N) directly interacts with the C-terminal domain of the catalytic subunit of polα (p180C). We found that a complex of the C-terminal domain of the catalytic subunit of polα with the second subunit (p180C-p70) stimulated primase activity, whereas the whole catalytically active heterodimer of polα (p180ΔN-p70) inhibited RNA synthesis by primase. Conversely, the polα catalytic domain without the C-terminal part (p180ΔN-core) possessed a much higher propensity to extend the RNA primer than the two-subunit polα (p180ΔN-p70), suggesting that p180C and/or p70 are involved in the negative regulation of DNA pol activity. We conclude that the interaction between p180C, p70, and p58 regulates the proper primase and polymerase function. The composition of the template DNA is another important factor determining the activity of the complex. We have found that polα activity strongly depends on the sequence of the template and that homopyrimidine runs create a strong barrier for DNA synthesis by polα.
Introduction
DNA replication is fundamental to all living organisms. The process of DNA replication of genomic DNA in eukaryotes typically depends on the RNA primers that provide a free 3′-hydroxyl group for the replicative DNA polymerases to bind and start elongation. There is one deviation from this established paradigm. Humans and other mammals possess a PrimPol enzyme, which can prime with either ribo- or deoxynucleotides and also serve as a translesion DNA synthesis polymerase (1). The exact role of the PrimPol is not clear, but it is limited to special situations because the PrimPol knock-out mice are viable.
The major task of accurate nuclear genome replication in eukaryotes is carried out by B-family DNA polymerases (pols)5 (2–6). polϵ is indispensable for the assembly of the replisomes at origins (7), polα-primase (polα-prim) generates primers for the synthesis (8), polδ (9) and polϵ (10) participate in elongation and bulk DNA synthesis (11–13), and polζ is a key player in the synthesis on templates that are difficult to replicate (14, 15). The critical essential step of the replication is the initial primer synthesis by polα-prim, because polδ, polϵ, or polζ cannot start DNA synthesis without a primer with a free 3′-OH end.
polα-prim is the only B-family polymerase complex that possesses RNA and DNA polymerase activities. polα lacks proofreading exonuclease activity, and the synthetic activity of the complex is limited to the synthesis of RNA/DNA hybrids that serve as primers for the synthesis of genomic DNA. During the initiation of DNA synthesis, primase first synthesizes a short RNA primer in a two-phase reaction, the rate-limiting synthesis of dinucleotide, and the subsequent, faster, further elongation to reach around 10 ribonucleotides (16). Then, polα takes over and extends the primer by ∼20 nucleotides of DNA. The ability to determine the size of RNA and DNA parts (called “counting”) is thought to be an intrinsic property of polα and primase (17, 18). This primer is elongated by polδ/polϵ for synthesis of the bulk of chromosomal DNA. The nature of the replication on the lagging strand requires multiple initiations by the polα-prim, which leads to the formation of 165-bp Okazaki fragments that correspond to unit size of nucleosomal repeats (19).
All B-family polymerases are four-subunit proteins (the latest addition to the group was polζ, which shares two accessory subunits with polδ) (20–24) where the largest catalytic subunit has a unique regulatory C-terminal domain binding zinc and iron ions (20, 25–27). These C-terminal domains share sequence similarities and are evolutionarily conserved from yeast to humans (20, 28). Human polα is composed of a 180-kDa catalytic subunit (p180) and a 70-kDa B subunit (p70) (8) (Fig. 1A). The primase part is composed of a 49-kDa catalytic subunit (p49) and a 58-kDa accessory subunit (p58) (8, 29) (Fig. 1A). The two metal binding sites located on the C-terminal domain of p180 (p180C) are occupied by zinc atoms in crystal (26). When p180C was partially purified from bacteria under anaerobic conditions, it contained a detectable level of iron-sulfur clusters (27), which may be a peculiarity of the heterologous expression in bacteria (20). The p58 subunit possesses an evolutionarily conserved, C-terminal four-cysteine motif that coordinates a [4Fe-4S] cluster, which is necessary for the formation and stabilization of the initial di-ribonucleotide of the RNA primer (30–33).
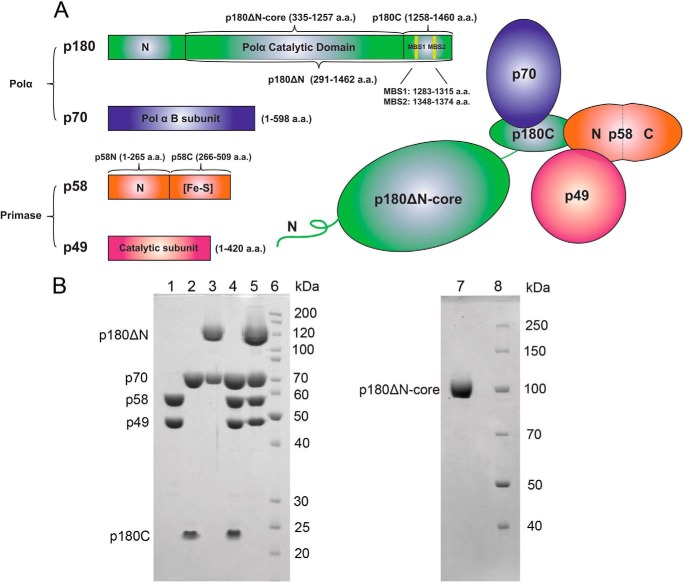
FIGURE 1.
Overall structure of human polα-primase. A, schematic representation of the four-subunit complex, p180 (green), p70 (blue), p58 (orange), and p49 (red). The primary amino acids sequence and the domains of each subunit and various recombinant constructs studied in the current work are shown as stick diagrams on the left. Metal binding motifs in p180 are marked by yellow vertical lines and named two metal binding sites MBS1 and MBS2. B, analysis of the purity of human primase and polα samples. Left panel: lane 1, p49·p58; lane 2, p70·p180C; lane 3, p70·p180ΔN; lane 4, p49·p58·p70·p180C; lane 5, p49·p58·p70·p180ΔN; lane 6, EZ-Run Rec protein ladder (Fisher). Samples were run on 10% SDS-PAGE, and proteins were detected by Coomassie Blue staining. The p180ΔN corresponds to the catalytic subunit with deleted 291 amino acids from the N terminus. Right panel: lane 1, p180ΔN-core, lacking CTD of the catalytic subunit; lane 2, Page Ruler protein ladder (Thermo Scientific). Samples were run on 8% SDS-PAGE and visualized by Coomassie Blue staining.
Mouse and yeast homologs of the polα p180C domain interact with the B subunit and primase, working as a scaffold to tether the catalytic part of p180, B subunit, and primase (34, 35). The large interaction interface (4500 Å2) between the C terminus of yeast pol1 and the B-subunit suggests tight contact between two polypeptides (26). The N-terminal helical domain of the B subunit of polα interacts with SV40 viral helicase, which is required for the activation of the SV40 primosome (36). This structural information is consistent with the idea that p180C·p70 might tether the polα-prim to the other components of replication machinery (34, 37). However, the role of p180C·p70 in the polα-primase function is not well defined.
To get a better understanding of the mechanism of RNA-DNA synthesis required for the beginning of DNA synthesis during replication, we investigated the interactions between the four subunits of the human polα-prim complex and the functional implications of these contacts. We confirmed that the large subunit of primase, p58, interacts with the p180C part of the catalytic subunit of polα via the N-terminal domain. The primase and polymerase assays demonstrated that the p180C·p70 complex regulates both primase and polymerase activities. These activities are also dependent on the ratio of ribo- versus deoxynucleoside triphosphates and the specifics of DNA templates. DNA synthesis by polα proceeds to the end of the standard templates with a scrambled sequence but halts at ∼20 nucleotides of the homopyrimidine (poly-dT) template. This observation limits the applicability of the recent model of primer length counting by yeast polα (38).
Our findings suggest that the proper replication priming is a result of balanced and sequential activities of the components of polα-prim complex and the availability of the proper nucleotides. During the priming, the size of the RNA-DNA hybrid is regulated by the non-catalytic domain, p180C, and the subunit, p70, and depends on the competition between primase, polα, and ribo- versus deoxynucleotides as well as the sequence context of the template.
EXPERIMENTAL PROCEDURES
Materials
We have used the following templates for the primase and DNA polymerase reaction: poly-dT70, 5′-(TTTTTTTTTT)7; 73a, 5′-(TTTTTTT)5AGCGTCTTAATCTAAGCACTCGCTATGTTTTCAAGTTT; 73b, 5′-GTCTGGAATGATGAAGATTACTAGTGAAGATTCTGAGCGTCTTAATCTAAGCACTCGCTATGTTTTCAAGTTT.
For the primer extension assays we have used TYE665 fluorophore-modified oligonucleotides: poly-dA15, 5′-TYE665-AAAAAAAAAAAAAAA; poly-rA15, 5′-TYE665-r(AAAAAAAAAAAAAAA); hetero-DNA, 5′-TYE665-CTTGAAAACATAGCGA; hetero-RNA, 5′-TYE665-r(CUUGAAAACAUAGCGA). All oligonucleotides were from IDT Inc., Coralville, Iowa. The list of bacterial expression plasmid constructs used in the protein-protein interaction study is presented in Table 1.
TABLE 1.
Bacterial expression plasmid constructs used for studies of protein-protein interactions
| Name | Coding sequence (cDNA) | Backbone |
|---|---|---|
| pE1 | cs-p58-His6, cs-SUMOp180C | pETDuet-1 |
| pE2 | cs-p58-His6 | pETDuet-1 |
| pE3 | cs-p49, cs-p58 | pETDuet-1 |
| pE4 | cs-p49, cs-p58N (encoding amino acids 1–265) | pETDuet-1 |
| pE5 | cs-p49, cs-p58C (encoding amino acids 266–509) | pETDuet-1 |
| pC1 | cs-p70-His6, cs-SUMOp180C, cs-p66N | pColaDuet-1 |
| pC2 | cs-p70-His6 | pColaDuet-1 |
| pC3 | cs-SUMOp180C | pColaDuet-1 |
Cloning of Human DNA polα-Primase Genes and the Production of Recombinant Proteins for Pulldown Analysis
The cDNAs for the p49 (420 amino acids (aa)) and p58 (509 aa) subunits of human primase were obtained from Open Biosystems (clone IDs 3686937 and 6148494, respectively). The cDNA for the human B subunit (598 aa) was obtained from Open Biosystems (clone ID 2822514). The pcDNA3/POLA1 encoding for p180 (1462 aa) is a generous gift from Dr. Motoshi Suzuki (Nagoya University Graduate School of Medicine). To demonstrate the direct interaction between p58 and p180C, the coding sequences (cs) for p58-His6 (cs-p58-His6) and cs-SUMO-p180C were cloned together into the MCS1 and MCS2 of pETDuet-1 (vector named as pE1; Table 1), respectively, using a two-step insertion method (39). A single insertion of cs-p58-His6 (pE2) into pETDuet-1 served as a control. Non-tagged cs-p49 and cs-p58 were cloned into the MCS1 and MCS2 of pETDuet-1(pE3), respectively. The cs-p58N (encoding amino acids 1–265) or cs-p58C (aa 266–509) were cloned into the MCS2 of the pETDuet-1 that contains cs-p49 at MCS1. Cloning of the full-length, N-terminally His-tagged human cs-p70 (p70-His6) and SUMO-tagged human cs-p180C (coding for aa 1265–1462; SUMO-p180C) into the pColaDuet-1 vector was described previously (20). To obtain a pColaDuet-1 vector producing only SUMO-p180C (for negative controls), a premature stop codon was introduced into the cs-p70-His6.The pETDuet-1 and pColaDuet-1 constructs were introduced into the Rosetta-2 (DE3) cells by transformation and selected on the LB-Amp+Kan+ medium. Overexpression of the genes of the constructs was done as described previously (20).
Methods
All experiments were repeated at least two times, but most were completed three or more times. The results are illustrated by typical gels.
Yeast Two-hybrid Assay
Coding sequences of human p49, p58, p70, and p180C (encoding for aa 1,258–1,462) (Fig. 1A) were amplified by PCR from vectors described in the previous section and ligated into the pGADT7 and pGBKT7 vectors (Clontech, Mountain View, CA) containing TRP1 and LEU2 selection markers, respectively. The yeast strain AH109 with three reporter genes, HIS3, ADE2, and LacZ, was transformed by combinations of these plasmids. The co-transformations of cs-p49/cs-p58 and cs-p180C/cs-p70 as well as the murine cs-p53/cs-SV40 large T-antigen served as positive controls. The co-transformations of each construct with an empty vector served as negative controls. The cells were allowed to grow three days on Synthetic Complete drop-out plates lacking Leu and Trp (SC−Leu−Trp). Positive colonies were replica-plated on SC−Leu−Trp−His, SC−Leu−Trp-ade, and SC-Leu-Trp-His-Ade plates. Colonies on each plate were counted after 3 days of growth at 30 °C. Activation of both HIS3 and ADE2 reporters was recorded as an interaction if >80% of the colonies with prey and bait genes were His+ and Ade+ (the single HIS3 reporter produces many “false positives”; therefore, only the concomitant Ade+ phenotype suggests that proteins under study really interact).
Nickel-iminodiacetic acid (Ni-IDA) Pulldown Assay
After induction of gene expression in bacterial cultures, cells were harvested and kept in aliquots at −80 °C. Cells were disrupted in ice-chilled water by sonication in lysis buffer containing 20 mm Tris 7.8, 0.15 m NaCl, 10 mm KH2PO4, 3% glycerol, 3 mm β-mercaptoethanol, 0.5 mm PMSF, and 1 μg/ml leupeptin. After centrifugation, 0.2 ml of cleared lysate (corresponding to a 2.5-ml culture volume) was incubated for 1 h with rocking at 4 °C with 10 μl of Ni-IDA resin. The resin was washed 1 time with 0.2 ml of lysis buffer and 2 times with 0.2 ml of the lysis buffer containing 0.5 m NaCl. The bound proteins were eluted by 30 μl of 0.3 m imidazole in the lysis buffer (pH 7.7). Proteins of bacterial lysates and eluted samples from Ni-IDA resin were separated by 12% SDS-PAGE and detected by Coomassie Blue (R250) staining or Western blotting using the Mini-PROTEAN Tetra cell system (Bio-Rad). Anti-His monoclonal antibody and anti-SUMO monoclonal antibodies (6G2A9 and 4G11E9, respectively; GenScript, Piscataway, NJ) were used to detect His6-tagged human p58 and p70 and SUMO-tagged p180C, respectively.
Purification of Human Primase·polα Subunits and Their Complexes
Expression and purification of full-length human primase (p49·p58) was done as in Baranovskiy et al. (39). Expression and purification of p70·p180C was done as in Baranovskiy et al. (20). For expression of the p180 core (aa 335–1257, p180ΔN core) and p70·p180ΔN heterodimer (aa 1–598 and 291–1462, respectively) in insect cells, we cloned them into the pFastBac-1 vector (Invitrogen). In the case of the p180 core, we added cs for the N-terminal His6 tag followed by the tobacco etch virus protease recognition site (ENLYFQ); in the case of p70 we added cs for the N-terminal His6 -tag. Obtaining high titer baculoviruses and protein expression in insect cells was done according to the instructions of the manufacturers. The polα catalytic core (aa 335–1257) with a cleavable N-terminal His6 tag was purified to near homogeneity in four steps, including chromatography on the Ni-IDA column (Bio-Rad), overnight dialysis combined with digestion by tobacco etch virus protease, pass through the Ni-IDA column, and chromatography on Heparin HP HiTrap column (GE Healthcare).
The p70·p180ΔN was purified to near homogeneity in three chromatographic steps, including Ni-IDA (Bio-Rad), heparin HP HiTrap (GE Healthcare), and hydroxyapatite (Bio-Rad) columns. To obtain the tetrameric prim-polα complex (p49·p58·p70·p180ΔN), the pure primase (109 kDa) and polα (200 kDa) samples were mixed at a molar ratio of 1.2:1, incubated for 1 h at 4 °C, and loaded to Superose 12 column (24 ml; GE Healthcare). For all samples used in the activity assay, the peak fractions from the last column were combined, concentrated to 2 mg/ml, and diluted 5-fold in the buffer containing 30 mm HEPES-KOH (pH 7.9), 50 mm NaCl, 1 mm DTT, and 1% glycerol and flash-frozen in aliquots. The results of analysis of the purity of our proteins are presented in Fig. 1B.
Primase Assay (de Novo Synthesis of RNA Primers on Single-stranded Templates)
The activity of full-length human primase with or without p70·p180ΔN or p70·p180C, was tested on poly-dT70 (IDT Inc., Coralville, Iowa). Reactions (20 μl) were assembled on ice in the following order: 100 nm template was added to reaction buffer (30 mm HEPES-KOH (pH 7.9), 1 mm DTT, 7 mm magnesium acetate) followed by the addition, depending on the experiment, of 100 μm ribo-ATP or 1 mm deoxy-ATP (dATP) with 0.15 μm [α-32P]rATP (>3000 Ci/mmol) or 0.15 μm [α-32P]dATP (>3000 Ci/mmol; MP Biomedicals, Santa Ana, CA). Then 0.1 μm primase with or without 0.025, 0.05, or 0.1 μm p70·p180C or 0.2 μm p70·p180ΔN was added, and reaction tubes were incubated at 37 °C for 20 min in a thermo cycler (Thermolyne Amplitron I). A molecular weight marker was created by 5′ phosphorylation of the 70-mer poly(dT) single-stranded by T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) following the manufacturer's instructions with [γ-32P]rATP (4500 Ci/mmol, MP Biomedicals). Reaction products were mixed with formamide loading buffer (95% formamide, 5 mm EDTA, 0.02% bromphenol blue, and 0.02% xylene cyanol), heated at 65 °C for 10 min, and resolved by 20% urea-PAGE (UreaGel System (19:1 acrylamide/bisacrylamide); National Diagnostics, Atlanta, GA) for 5 h at 2000 V. The gel was dried at 65 °C for 45 min using a Bio-Rad 583 gel dryer. The reaction products were visualized and quantified by phosphorimaging (Typhoon 9410, GE Healthcare).
Primer Extension Assay
Activities of the p180ΔN-core, p70·p180ΔN, or p70·p180ΔN·p49·p58 were compared in reactions (20 μl) that contained the 0.75 μm poly-dT70 template with 0.50 μm 5′-TYE665 fluorophore-labeled poly-dA15 DNA or poly-rA15 RNA oligos or 73a (73b) templates pre-annealed with hetero-DNA or RNA primers. The annealing of the 73a or the 73b template with RNA or DNA primers was done by a decrease of temperature from 80 to 25 °C in 0.5 °C/min gradient in the thermo cycler (50 μl volume, 50 mm NaCl). The activity of primase p49·p58 heterodimer was analyzed using close to an equal 1:1.5 ratio of enzyme to template. The activities of primase p49·p58·p70p·180ΔN heterotetramer was analyzed using a low 1:15 ratio of enzyme to primer/template. The low ratio of primase to template allowed us to approach single-hit conditions, whereas at the equal ratio the primase was able to synthesize longer products due to multiple binding events with substrate. This allowed us better access the inhibitory effect of dATP. Reactions were assembled on ice in the following order. The primer/template was added to the buffer containing 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 10 mm MgCl2, 0.2 mg/ml BSA, and 2 mm DTT followed by the addition of dATP (200 μm or otherwise indicated under “Results”) or 200 μm dNTPs for the poly-dT70 and 73a (or 73b) template, respectively. Then, the p180ΔN-core, p70·p180ΔN, or p70·p180ΔN·p49·p58 was added into the reaction to concentrations indicated in the figure legends. Reactions were held for the indicated time points at 37 °C in the thermo cycler. T4 DNA polymerase (Promega Corporation; stock concentration 25 nm) was used as a control for the primer extensions on hetero-DNA and RNA. Reaction products were mixed with formamide loading buffer (95% formamide, 0.025% Orange G, 5 mm EDTA, and 0.025% SDS) and heated at 70 °C for 5 min. The reaction products were separated as described previously. Visualization of the products used the Typhoon 9410 imager (emission of fluorescence at 645 nm).
RESULTS
The Role of polα in Primase Function
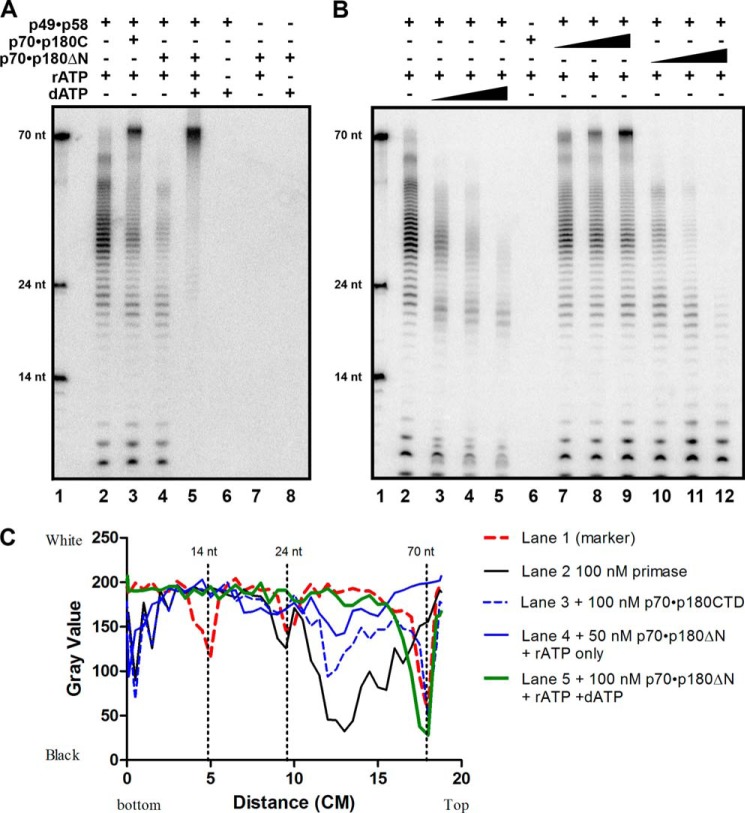
To study the functional relevance of the subunit contacts in polα-prim, we purified several variants of recombinant polα with a combination of different parts of the primase functional domains (Fig. 1A): B-subunit (p70) with the catalytically proficient catalytic DNA pol subunit, p180ΔN, or C-terminal of p180 (p180C). Similar to yeast polα (17), where the N terminus interacts with Ctf4 for the integration of polα into the replication fork (40), the N terminus of p180 is poorly folded and has no conserved motifs necessary for DNA polymerizing activity, so it has been deleted to increase protein solubility and simplify its purification. All protein samples used for activity analysis were purified to the high level of homogeneity (Fig. 1B). We analyzed de novo RNA synthesis and the extension of these primers on the poly-dT template with radioactive [α-32P]ATP by the p49·p58 heterodimer of primase alone or in the complex with heterodimer of p70·p180C or p70·p180ΔN. The heterodimeric primase synthesized RNA primers of an average of 8–10 nucleotides unit-length and multiples of this unit. They were extended to around 20 nucleotides and, more frequently, to around 30 nucleotides, and less efficiently, they were extended further to 50 and 60 nucleotides (Fig. 2A, lane 2). The addition of dATP dramatically inhibited the activity of primase (Fig. 2B, lanes 3–5). In the presence of p70·p180C the majority of products became longer, around 70 nucleotides in length (Fig. 2A, lane 3, and titration of p70·p180C concentration in Fig. 2B, lanes 7–9). The explanation for the predominance of the long products could be the elevated activity/processivity of the primase and non-random initiation/extension by primase in the presence of p70·p180C biased toward the 3′-end of the template. The presence of catalytically active heterodimeric polα, conversely, inhibited the extension of unit-length primers when there was no dATP in the reaction (Fig. 2A, lane 4, and titration of p70·p180ΔN in Fig. 2B, lanes 10–12). This observation is consistent with earlier studies (41). The addition of dATP with p70·p180ΔN, as expected, led to a rapid synthesis of the full-length products (Fig. 2A, lane 5). The quantification of the product distribution and band intensity in the case of primase alone plus p70·p180C or p70·p180ΔN with or without dATP (Fig. 2C) provides support to the visual analysis of the gel shown in Fig. 2A.
FIGURE 2.
Regulation of de novo primase activity by polα. polα-primase activity assay was done on a poly-dT 70-mer template (0.1 μm) as described under “Experimental Procedures.” A, effect of polα on primase activity. Lane 1, nucleotide size marker; lane 2, RNA primers synthesized by 0.1 μm p49·p58 alone; lane 3, RNA primers synthesized by 0.1 μm p49·p58 with the addition of 0.1 μm p70·p180C; lane 4, RNA primers synthesized by 0.1 μm p49·p58 with the addition of 0.05 μm p70·p180ΔN; lane 5, hybrid RNA-DNA fragments synthesized by 0.1 μm p49·p58·p70·p180ΔN complex with both the rATP and the dATP in reaction; lanes 6–8, negative controls. nt, nucleotides. B, primase activity assays with the titration of the polα variants. Lane 1, nucleotide size marker; lane 2, RNA primers synthesized by p49·p58 alone; lanes 3–5, RNA synthesized by p49·p58 in the presence of increasing concentrations of dATP (25, 50, 100 μm); lane 6, no activity by p70·p180C with rATP (negative control); lanes 7–9, the effect of increasing concentrations of p70·p180C (0.025–0.1 μm) on RNA synthesized by p49·p58; lane 10-12, RNA synthesized by p49·p58 with 0.05–0.2 μm p70·p180ΔN. C, quantification of gel shown in panel A. The mean gray value of each lane was selected from the bottom to the top of the gel image and analyzed by Image J. The distribution of the band (x axis: distance from bottom to the top of the gel) versus the intensity of the band (y axis: gray value closer to 0 means black, 250 means white).
The Inhibitory Effect of dATP on Primase Activity
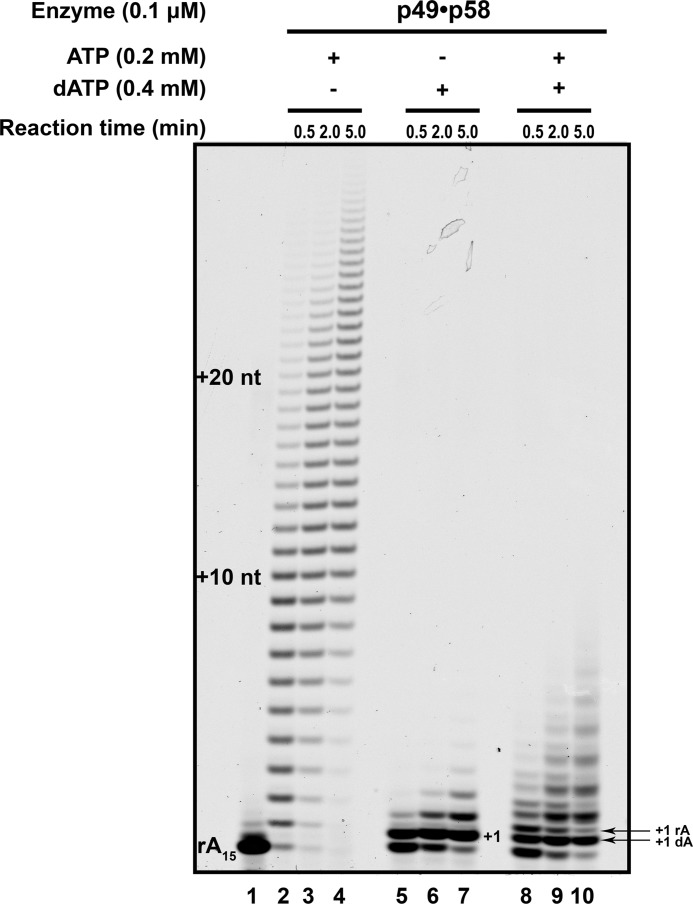
To further study how the primase activity is inhibited by dATP (Fig. 2B, lanes 3–5), we analyzed the ability of the heterodimeric primase to extend ribonucleotide primers in the presence of ribo- versus deoxynucleotides or a mixture of both. We have used the high ratio of enzyme to template (“Experimental Procedures”) to push the primase to its limits to detect the incorporation of dNTPs. Primase robustly extended the poly-rA15 primers on the poly-dT70 template with rATP. The extent of primer utilization and the length of products increased with longer reaction times, indicating the multiple cycles of binding and synthesis (Fig. 3, lanes 2–4). dATP was a poor substrate for the primase reaction; it was incorporated in the +1 position quite efficiently, but further incorporations were not favored, with no products longer than +4 (Fig. 3, lanes 5–7). The primase extended about half of the RNA primers with dAMP in 30 s, whereas the addition of a second dAMP was almost 10 times slower. The primer extension was also limited when both rATP and dATP were present (Fig. 3, lanes 8–10). The primase incorporated both dATP and rATP with comparable efficiency (Fig. 3, lanes 8–10, note that oligonucleotides containing only RNAs migrate slower than RNA primers extended with dNTPs). These results indicate that primase does not greatly discriminate against dATP, but the products with incorporated dAMP are poor substrates for the following extension.
FIGURE 3.
Primase robustly extends RNA primers only in the presence of ribonucleotides. The poly-rA15 primer (enzyme to primer/template ratio = 1:1.5) was extended by heterodimeric primase (p49·p58) in the presence of rATP, dATP, or both. Lane 1, reaction without nucleotides; lanes 2–4, reaction with 0.2 mm ATP for 0.5, 2.0, and 5.0 min, respectively; lanes 5–7, reaction with 0.4 mm dATP for 0.5, 2.0, and 5.0 min, respectively; lanes 8–10, reaction with 0.2 mm ATP plus 0.4 mm dATP for 0.5, 2.0, and 5.0 min, respectively. nt, nucleotides.
The C-terminal Domain (CTD) of the Catalytic Subunit of Human polα Interacts with the N-terminal Part of the Large Subunit of Primase
Mammalian polα interacts with the large subunit of primase (29). The structure of artificially made, chimeric primase-CTD of polα (amino acids 1445–1462) protein suggested that the CTD of polα interacts with the N terminus of the large subunit of primase (38). To examine the intersubunit interactions between primase and polα in vivo we have used a yeast two-hybrid assay (“Experimental Procedures”). As expected, a strong interaction was detected between the two primase subunits as well as the C-terminal domain of polα (p180C) with the second subunit of polα (p70). No interaction was detected between p180C and p49. The presence of two plasmids containing cs-p58 fused to activation domains and cs-p180C (encoding for aa 1258–1462) fused to binding domains led to a positive signal for both HIS3 and ADE2 reporters. However, there was no activation of reporters when p58 was fused to the binding domain and p180C was fused to the activation domain. We concluded that the large subunit of human primase interacts with the CTD domain of polα.
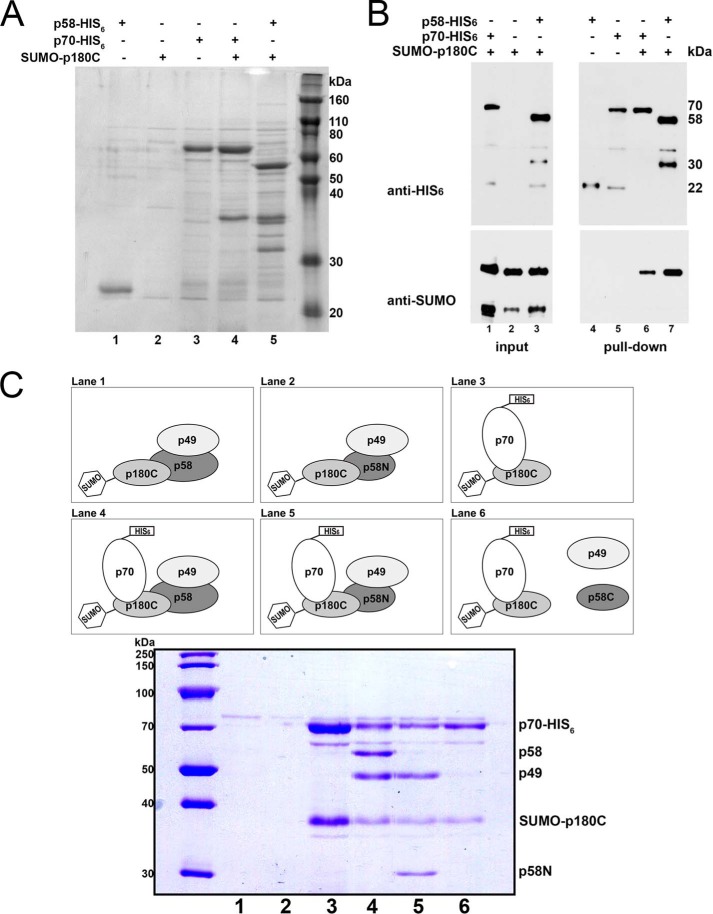
To get more precise information about the interaction, we explored the binding of the same proteins produced in bacteria in Rosetta-2 (DE3) cells (Fig. 4). Co-expression of the genes encoding the SUMO-tagged p180C and His6-tagged p58 allowed us to obtain a stable binary complex by affinity purification on the Ni-IDA resin (Fig. 4A, lane 5). The stoichiometry of the binding between p58-His6 and SUMO-p180C was similar to the positive control (Fig. 4A, lane 4). The identities of p58-His6 and p70- His6 purified by Ni-IDA resin as well as the SUMO-p180C were verified by the Western blot analysis (Fig. 4B).
FIGURE 4.
N terminus of the large subunit of primase tethers it to polα. Analysis of the pulldown of proteins separated on SDS-PAGE and stained by Coomassie Blue (R250) (see “Experimental Procedures”). A, direct interaction between p58 and p180C. Lane 1, p58-His6 is self-cleaved to a 24-kDa product in the absence of its binding partner; lane 2, SUMO-p180C does not bind to Ni-IDA resins; lane 3, p70-His6 pulldown by Ni-IDA resin; lane 4, SUMO-p180C binds to p70-His6; lane 5, p180C sumo binds to p58-His6. B, Western blot detection of the interacting proteins seen in A. C, finding a region of p58 that is responsible for its interaction with polα. Lanes 1 and 2, p49, p58, p58N, or SUMO-p180C do not bind to Ni-IDA resin in the absence of p70-His6 (negative controls); lane 3, SUMO-p180C binds to p70-His6 (positive control). Lane 4, p58·p49 interacts with polα (SUMO-p180C·p70-His6; positive control); lane 5, p49·p58N interacts with SUMO-p180C·p70-His6; lane 6 shows the absence of interaction of p58C or p49 with SUMO-p180C·p70-His6.
In the crystal structure, p58N is tightly bound to the p49 subunit of primase (38), although it lacks the C-terminal domain with the [Fe-S] cluster (amino acids 287–509). Weiner and co-workers (31, 32) noted that the purification of the p58N alone is problematic because it is largely insoluble in the absence of p49. In our experiments, only a 24-kDa proteolytic product was seen (Fig. 4, A, lane 1, and B, lane 4) in the pulldown of p58-His6 alone, which likely represents the C-terminal part of the p58 subunit. To investigate the role of the primase domain containing the [Fe-S] cluster in interaction with polα, plasmids encoding for two subunits of primase (p49·p58) and two subunits of polα (SUMO-p180C·p70-His6) were constructed, and the four-subunit complex was produced. An N-terminal SUMO tag was added to the p180C to address the problem of poor solubility of the protein in Escherichia coli. The levels of primase are typically higher than polα when produced in E. coli cells. The His6 tag, therefore, was placed in the N-terminal of the p70 subunit to obtain a quaternary complex with more even stoichiometry after pulldown. Similar to the p49·p58·SUMO-p180C·p70-His6 (positive control; Fig. 4C, lane 4), the p49·p58N and SUMO-p180C·p70-His6 are represented by stable quaternary complexes after affinity purification on the Ni-IDA resin (Fig. 4C, lane 5). After the production of p49·p58C and SUMO-p180C·p70-His6, only a binary complex SUMO-p180C·p70-His6 was present on the gel after pulldown (Fig. 4C, lane 6). We concluded that the polα interaction with primase is through the p180C-p58N. Neither p49 nor the part containing the [Fe-S] domain of p58 plays a role in the interaction. Our data are consistent with the recent finding that the last 18 residues of the C-terminal region of the yeast polα catalytic subunit are important for binding to the N terminus of the primase large subunit (34, 38).
DNA Sequence Dependence of the Extent of Synthesis by polα
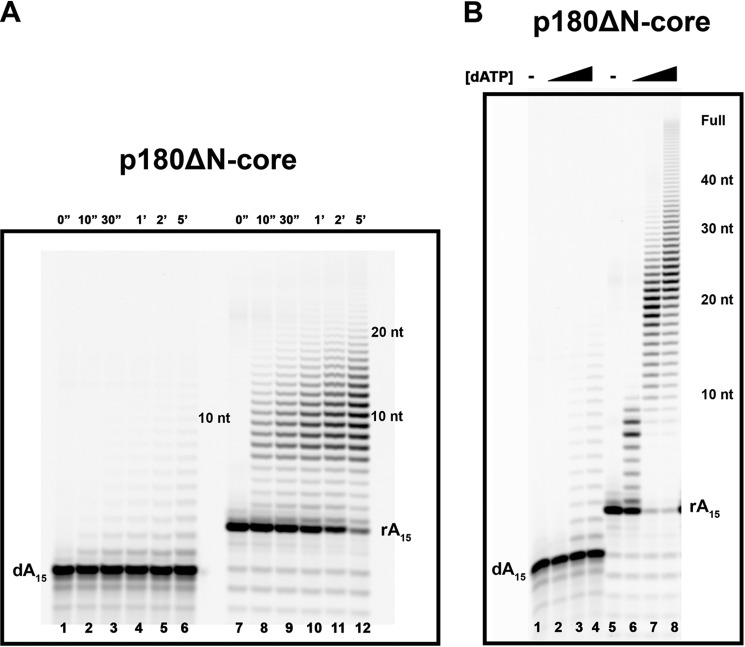
To examine the parameters of the primer extension on different templates, we utilized a version of an enzyme with a core polymerase domain of human polα (p180ΔN-core; aa 335–1257). This fully catalytically proficient protein lacks both the N-terminal part of p180 and the C-terminal domain of p180 with two metal binding sites necessary for the interaction with p70 (Fig. 1A). In the same time of reaction on the poly-dT70 template, the efficiency of the extension of the 15-mer DNA primer by p180ΔN-core is much lower in comparison to the 15-mer RNA primer (Fig. 5A; note the different electrophoretic mobility of DNA versus RNA). The efficiency of this reaction with both types of primers is increased when the concentration of dATP goes up (Fig. 5B). However, most products remain short, about 20 nucleotides of newly synthesized DNA, despite overall stimulation. It appears that the addition of 20 nucleotides of DNA attached to 15-mer DNA or RNA is around the upper limit of the synthetic potential of the p180ΔN-core on this template (Fig. 5B, lanes 7 and 8). The result is in agreement with the previous findings for yeast polα with the same poly-dT template (17).
FIGURE 5.
Preferential RNA primer extension by human polα polymerase domain (p180ΔN-core) on the homopolymeric (dT)70 template. A, extension of the poly-dA15 and poly-rA15 primers (enzyme:primer/template ratios = 1:50 and 1:80, respectively) in the presence of 0.2 mm dATP. The products of reactions at the indicated time points were analyzed as described under “Experimental Procedures.” B, the dependence of the extension of the poly-dA15 and poly-rA15 primers (enzyme:primer/template ratios = 1:50 and 1:80, respectively) on the dATP concentration. The black triangle indicates that the reactions were carried out with the increase of dATP concentrations (0.01, 0.1, and 1 mm, respectively). nt, nucleotides. The reaction time was 20 min.
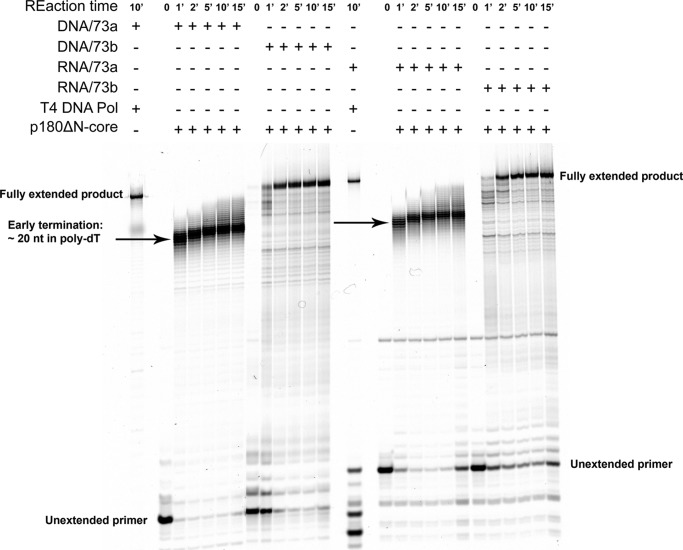
To explore whether this “20-mer rule” is a general or template-specific property of polα, we examined DNA synthesis by the same core enzyme in the extension of RNA and DNA 16-mer primers with identical heterogeneous sequences. We designed two different templates with a heterogeneous sequence and a single, fixed site of primer annealing (“Materials,” under “Experimental Procedures”). The first template, 73a, included a 35 homopolymeric-dT run at the 5′-end. The second template, 73b, contained all heterogeneous sequences with the 3′ 38-mer sequence identical to the template 73a. Surprisingly, the efficiency of the extension of DNA and RNA primers by the p180ΔN-core was identical in the matched primer/template pair (Fig. 6). Interestingly, the synthesis by the p180ΔN-core on template 73a was prone to termination at positions around 20 nucleotides deep into the 35 homo-dT run. No such pausing was detected on the heteropolymeric 73b template, and polα reached the end of the template, similar to T4 DNA polymerase. We concluded that polα has a limited capacity for DNA synthesis only on the poly-dT template.
FIGURE 6.
Same efficiency of the extension of DNA and RNA primers on hetero-homopolymeric hybrid and heteropolymeric DNA templates by the p180ΔN-core. For control of the full extension of the primers, we used reactions with T4 DNA polymerase, which robustly and completely extended DNA as well as RNA primers (left lane in the left and right halves of the gel). Arrows show the zones of termination of DNA synthesis. All reactions contain 0.2 mm dNTPs, and the enzyme to primer/template ratio was 1:50, except for the T4 DNA pol (∼1:2500).
The C-terminal Domain of polα Regulates DNA Polymerase Activity
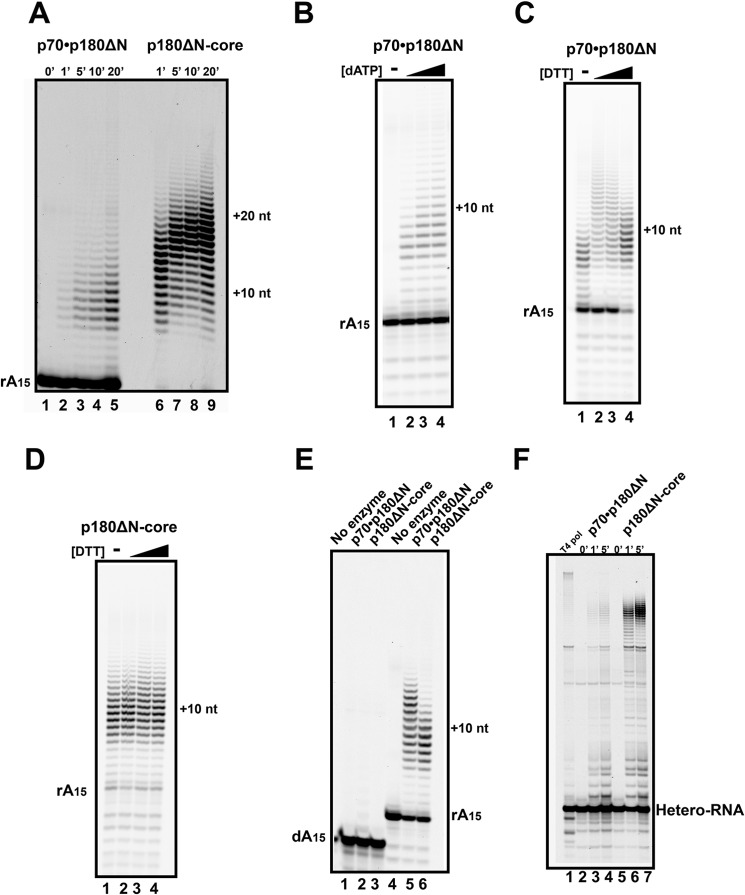
In comparison to the p180ΔN core, the primer extension activity of the dimeric polα complex (p180ΔN·p70) on the poly-dT70 template with RNA primer is much lower (note a 2-fold higher enzyme concentration of p180ΔN·p70; Fig. 7A). It is interesting that the average length of the synthesized fragments is also decreased to ∼10 nucleotides. The increase of concentrations of dATP in reaction with the RNA primer on the poly-dT70 template did not result in a stimulation of the primer usage. The slight increase in the product size (Fig. 7B) is negligible in comparison to the 10-fold stimulation in the p180ΔN-core by dATP (Fig. 5B, lanes 5–8). The presence of CTD with metal binding sites rendered the dimeric enzyme sensitive to DTT (Fig. 7C), whereas DDT did not modulate the activity of the p180ΔN-core (Fig. 7D). With an increased reaction time and greater than a 5-fold excess of the enzyme, we obtained a comparable activity of the p70·p180ΔN and p180ΔN-core on RNA15/poly-dT70. The synthesis by the dimeric polα was still terminated a few nucleotides earlier than the synthesis by the p180ΔN-core (Fig. 7E). The lower activity of the p70·p180ΔN dimer compared with the p180ΔN-core was also observed on the hybrid homo-heteropolymeric 73a template with the RNA primer (Fig. 7F).
FIGURE 7.
Inhibitory effect of CTD and p70 on the primer extension by polα. A, the time course of the extension of the poly-rA15 primers on the poly-dT70 template by p70·p180ΔN (enzyme to primer/template ratio = 1:50; lane 2-5) or by the p180ΔN-core (enzyme to primer/template ratio = 1:100; lanes 6–9). nt, nucleotides. B, extension of the poly-rA15 primers by p70·p180ΔN on the poly-dT70 template (enzyme to primer/template ratio = 1:50, reaction time was 5 min). The black triangle indicates that the reactions were carried out with the increase of dATP concentrations of 0.01, 0.1, and 1 mm, respectively. C and D, effect of DTT on the ability of polα to extend the poly-rA15 primer. C, polα p70·p180ΔN (enzyme to primer/template ratio = 1:50, reaction time 10 in). D, p180ΔN-core (enzyme to primer/template ratio = 1:100, reaction time 5 min). The black triangle indicates that the reactions were carried out with the increase of DTT concentrations of 0, 0.5, 1.0, 2.0 mm DTT. E, smaller fragments in primer extension by excess of dimeric polα (p70·p180ΔN) in comparison to the polα core. Extension of the poly-dA15 and poly-rA15 primers in long reactions by the excess of p70·p180ΔN (enzyme to primer/template ratio = 1:50, reaction time 15 min) or by the p180ΔN-core (enzyme to primer/template ratio = 1:250, reaction time 2 min) with 0.2 mm dATP. F, lower activity of dimeric polα in comparison to the polα core on hybrid homo-heteropolymeric DNA template. Extension of the RNA primers on the DNA template 73a by p70·p180ΔN (0.025 μm) or by p180ΔN-core (enzyme to primer/template ratio = 1:50) for the indicated time points is shown. As a control, T4 DNA polymerase (∼1:2500, reaction time 2 min) was used to obtain a completely extended, full-length product.
Primase and polα Activities of Quaternary polα-prim Complex
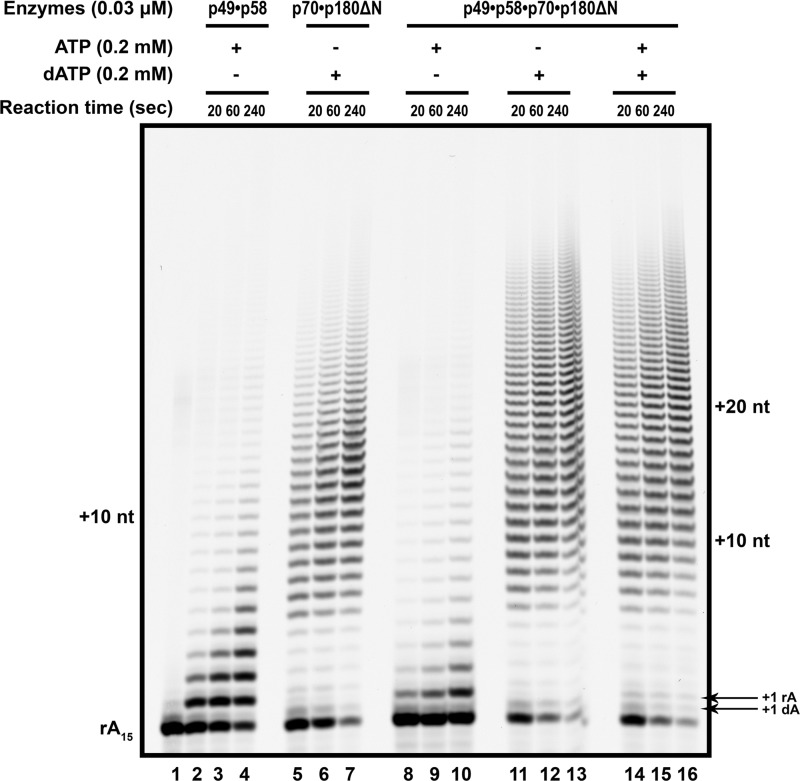
During the initiation of DNA replication, the utilization of ribonucleotides and deoxynucleotides by primase·polα should happen in a sequential order. Primase first synthesizes RNA, then polα synthesizes the DNA part of the primer. To study the primase activity in the polα-prim complex, we analyzed the activities of primase and polα within the four-subunit complex in the primer extension assay with ribo- versus deoxynucleotides or a mixture of both. In these experiments we used a low 1:15 ratio of enzyme to template to bring reactions closer to single-hit conditions. Primase p49·p58 and polα p70·p180ΔN were used as internal controls (Fig. 8). The primase is a non-processive enzyme and was able to incorporate only a few nucleotides (up to 10) (lanes 2–4). Oligonucleotides with a higher number of incorporations seen after 4 min of reaction most likely represent rebinding of the enzyme (lane 4). The heterodimer of polα processively added substantially more nucleotides with the median around position +15. In complex with polα, primase activity was less than the primase alone (Fig. 8, compare lanes 8–10 and 2–4). This was likely due to the handover of primers from primase to polα. polα robustly extended the primer in complex with primase with only dATP (Fig. 8, lanes 11–13). In comparison to p70·p180ΔN alone (Fig. 8, lanes 5–7), there is a slight increase of primer usage and product length. In the reactions with both rATP and dATP, only a few rAMPs were attached by primase to the rA15 (position +1), and polα rapidly outcompeted primase for the primer 3′-end and robustly extended it (Fig. 8, lanes 14–16).
FIGURE 8.
Primase and DNA polymerase activities of the tetrameric polα-primase complex during the extension of the RNA primer on the polydT template. The poly-rA15 primer annealed to polydT and extended by primase (lanes 2–4: 0.2 mm rATP), polα (lanes 5–7: 0.2 mm dATP), or the polα-prim complex (lanes 8–16) is shown. Lane 1, reaction without nucleotides; lanes 8–10, reactions containing 0.2 mm rATP; lanes 11–13, reactions containing 0.2 mm dATP; lanes 14–16, reactions containing 0.2 mm mixture of rATP and dATP. All reactions have an enzyme to primer/template ratio of 1:15 for balanced RNA and DNA pols activities. In each case, the time course of the reaction included points of 20, 60, and 240 s. nt, nucleotides.
DISCUSSION
The eukaryotic primases involved in the bulk replication of large genomes are embedded into a four-subunit protein complex with the DNA polα (Fig. 1A). The understanding of the mechanism of regulation of the synthesis of the RNA-DNA hybrid primer during replication is fragmentary, in part due to the lack of knowledge of the protein-protein interaction within the four-subunit complex. One of the key parameters of the polα-prim reaction is the regulation of the length of the RNA and DNA parts of the primers made available for major replicative DNA polymerases. The limitation of the current models of counting is that they are based on structures of either the primase or the polα parts of the complex.
Several x-ray crystal structures of the parts of the eukaryotic primase are available. The most complete is a structure of the human dimeric primase lacking the CTD of the large subunit (38). The analysis suggests that the same set of residues in the small catalytic subunit is responsible for the initiation and extension step of primer synthesis. The role of the highly conserved CTD of the large subunit, necessary for the initial dinucleotide synthesis, could only be indirectly inferred from this structure. The x-ray crystal structures of the p58C and its analog from yeast are available (33, 42, 43). The high structural similarity to the DNA photolyase/cryptochrome family of flavoproteins that binds flavin adenine dinucleotide (FAD) (43) suggests that this domain may be involved in the binding and stabilizing of the initial dinucleotide synthesized by primase to the template for further elongation (38). The folding of the region that binds single-stranded DNA indicates that the N terminus of the p58C can adapt to become α-helical or β-sheet structure, which suggests possible conformational change during priming (42). Based on our data that p58C does not interact with the other parts of the polα-prim complex (Fig. 4C), it is possible that the p58C domain is flexible by itself and with respect to other parts of the complex. This domain may act as an extra finger domain that binds to the first 5′ nucleotide for the critical rate-limiting step of dinucleotide synthesis.
polα is composed of a large catalytic subunit and a smaller B-subunit. Only the catalytic domain of the large subunit is necessary for the DNA synthesis, whereas its CTD and B-subunit are required for the replication and cell viability (44, 45). The contact between the B-subunit and the CTD with primase is thought to tether primase to the replisomes and origins (26, 35, 46, 47). The exact role of the polα CTD and B-subunit in primer synthesis is unknown. In the SV40 replication system, polα synthesizes the whole viral genome (5.2 kb) in the presence of the viral helicase (large-T antigen) through the helicase interaction with p49, p70, and p180 (37, 48, 49). The most critical contacts seem to be with the N terminus of p70 (37). We have found that the presence of p70·p180C sharply increases the lengths of the reaction products, most likely affecting the primase processivity (Fig. 2). The binding to p180C can stabilize the p58N domain, which is proposed to be involved in the interaction with the DNA-RNA hybrid duplex (50). Furthermore, p180C by itself may participate in stabilizing the primase complex with the RNA-primed template because it is located near p58N. Yeast p70·p180C heterodimer has an oligonucleotide/oligosaccharide-binding fold and zinc finger domains and possesses a micromolar affinity to dsDNA (26).
The deoxynucleotides act as natural competitive inhibitors of primase during the primer extension reaction (Fig. 2B, lanes 3–5). We observed that primase inserts first dNTP with the efficiency close to that of rNTPs but slowly extends from them (Fig. 3). We concluded that primase lacks significant selectivity between ribo- and deoxynucleotides at insertion step, and the inhibitory effect of deoxyribonucleotides is mediated by an inefficient extension. Primase has two NTP binding sites: so-called initiation and elongation sites. In the course of dinucleotide synthesis the “initiation” site binds NTP, which becomes the 5′-terminal nucleotide of the primer. Biochemical and structural data accumulated so far indicate that primase uses the same set of functional residues for dinucleotide synthesis and its extension, which means that the 3′-terminal nucleotide of primer is located in the initiation site (38, 51, 52). Our data indicate that the initiation site is more selective for rNTPs than the elongation site. Primase is known to synthesize the RNA primer 8 to 10 nucleotides long, which is then extended by polα to 30 nucleotides (2, 16, 53). In vitro biochemical evidence from our study and others suggests that the heterodimeric primase is a low processivity enzyme, but it is capable of synthesis far beyond 10 nucleotides by iterative cycles of synthesis (30, 32). We confirmed that the interaction of primase in a tetrameric complex with polα is mediated by binding of pri2N to p180C (Fig. 4C). This stabilizes the complex and enhances primase activity (Fig. 2). The presence of polα inhibited the primase activity in the reactions containing rNTPs but no dNTPs. This suggests that polymerase and primase activities of the complex are competitive in vitro. In the crystal structure, polα establishes an elaborated network of contacts with 9 bp of DNA-RNA duplex (17) and should effectively capture an RNA-primed template when the primase synthesizes the unit-length primer. This reduces the effective amount of primers available for the primase.
Ribonucleotides at high concentrations inhibit B-family pols in a step preceding base pairing (54). polα strongly prefers dNTPs over NTPs (55); therefore, in the absence of deoxynucleotides it does not participate significantly in primer extension. We also observed a limited RNA and robust DNA pols activity of the tetrameric complex in reaction with a mixed pool of nucleotides (Fig. 8). These results demonstrate that the primase activity is further suppressed in the polα-prim complex. In the presence of primase in the polα complex, the product length increased (Fig. 8, compare lanes 5–7 to 11–13). polα-primase complex has two active sites for the primed DNA template, which increases the probability of substrate binding. After initial binding in the primase active site and after extension, the substrate might be translocated to the polα active site without dissociation from the complex (29).
During primer synthesis, the RNA synthesized by primase is extended by polα. The RNA primer (poly-rA15) and single-stranded DNA template (poly-dT70) form an unusual A-form RNA/DNA duplex, which is extended by yeast polα more efficiently than the DNA primer (poly-dA15), which forms a B-form duplex with the template (17). The extension by polα comes to a stop when synthesis reaches 20 nucleotides long. This effect was elegantly explained and thought to be the intrinsic property of yeast polα undergoing conformational change when it reaches a certain distance from the A-form of DNA (17). Our results on the extension of the 5′-end labeled fluorescent DNA (poly-dA) and RNA (poly-rA) primers by human polα agree with this observation (Fig. 5). The increase of the dATP concentration increased the efficiency of these reactions, but a termination zone around 20 nucleotides still existed. Surprisingly, the extension assay using RNA and DNA primers with a random sequence as well as a template with a combination of terminal 5′-poly-dT and heterogeneous sequences provided a completely different overall picture (Fig. 6). First, there was no difference for RNA versus DNA primers for the p180ΔN-core to extend. Second, no termination zone was seen on these templates until the synthesis proceeded into the polyT stretch. Obviously, a sharp contrast between RNA and DNA primers and the “20-nucleotide rule” could only be seen for the templates with poly-dT homopolymeric runs. The DNA strands of such runs are known to form an atypical structure (i.e. bending, H-form variants) in the neutral pH condition and in the presence of divalent metal ions (i.e. Mg2+) (56–58). We conclude that polα typically utilizes RNA and DNA primers with equal efficiency and the homopyrimidine (dT) run in duplex with homopurine (dA) forms an unusual structure, which prevents the moving of polα above a certain distance. This specific property of polα may apply to other types of polypyrimidine tracts (59). Our data suggest that counting by polα itself is an exception rather than the rule, and there are additional external signals in vivo that regulate the length of primers synthesized by polα.
The comparison of the catalytic activity of the polα p180ΔN-core and p70·p180ΔN indicates that the catalytic activity of polα is negatively regulated by p70·p180C. The active conformation of the yeast polα core appears to be in tighter contact with the DNA-RNA duplex (17) in comparison to RB69 polymerase (gp43), a prototype of the eukaryotic B-family pols α, δ, ϵ, and ζ, which lacks the distinctive C-terminal domain and B-subunit (60, 61). The C-terminal domain and the B-subunit of polα are not required for its catalytic activity. It is currently unknown how these additional elements fit into the tertiary complex. We propose that the p70·p180C in the polα complex interferes with the formation of the ternary complex including the primer/template duplex and deoxynucleotides. It is possible that the p70·p180C is positioned in the complex in a way that lowers accessibility of the pol active site.
Additional evidence that supports that the C-terminal domain and B-subunit plays a role in regulation of polα activity is the suppression of the inhibitory effect of p70·p180C on pol activity by the optimal concentration of DTT (Fig. 7, C and D; note that the p180ΔN-core works completely independently of the DTT concentration). It is noticeable that certain concentrations of DTT also result in a change of the distribution of sizes of DNA fragments synthesized by p70·p180ΔN. DTT is a well known agent modulating DNA polymerases activities in vitro (49). It is thought to reduce the intramolecular disulfide linkages that may affect p180C folding (62). The p180C contains two highly conserved four-Cys motifs, which bind either two zinc atoms or one zinc and one [Fe-S] cluster (17, 20, 27). At the present time there is not enough structural information on the folding of the polα complex and the effects of oxidation on this folding to explain the effect of DDT. It is possible that the effect of Cys oxidation that inhibits polα and the natural regulatory effect of the C-terminal domain/B-subunit to polα activity represent separate mechanisms.
Acknowledgments
We thank Vinod B. Agarkar and Corinn Grabow for participation in the initial steps of this study and Dr. Motoshi Suzuki for the plasmid with human polα. We thank Kristi Berger and Hollie Siebler for expert editing of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM101167 (NIGMS; to T. H. T.) and CA129925 (NCI; to Y. I. P.).
- pol
- DNA polymerase
- polα-prim
- polα-primase
- polδ
- DNA polymerase δ
- polϵ
- DNA polymerase ϵ
- polζ
- DNA polymerase ζ
- p49
- catalytic subunit of human primase
- p58
- large subunit of human primase
- p58N
- N-terminal domain of human primase large subunit
- p180C
- C-terminal domain of human polα catalytic subunit
- p180ΔN
- catalytic subunit of human polα
- p180ΔN-core
- catalytic domain of human polα
- Fe-S
- iron-sulfur cluster
- aa
- amino acids
- cs
- coding sequence
- Ni-IDA
- nickel-iminodiacetic acid
- dATP
- deoxy-ATP
- rATP
- ribo-ATP
- SUMO
- small ubiquitin-like modifier
- CTD
- C-terminal domain.
REFERENCES
- 1. García-Gómez S., Reyes A., Martínez-Jiménez M. I., Chocrón E. S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I. J., Blanco L. (2013) PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 52, 541–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garg P., Burgers P. M. (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 3. Pavlov Y. I., Shcherbakova P. V., Rogozin I. B. (2006) Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 255, 41–132 [DOI] [PubMed] [Google Scholar]
- 4. Kunkel T. A. (2009) Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol. 74, 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. MacNeill S. (2012) Composition and dynamics of the eukaryotic replisome: a brief overview. Subcell. Biochem. 62, 1–17 [DOI] [PubMed] [Google Scholar]
- 6. Waisertreiger I. S., Liston V. G., Menezes M. R., Kim H. M., Lobachev K. S., Stepchenkova E. I., Tahirov T. H., Rogozin I. B., Pavlov Y. I. (2012) Modulation of mutagenesis in eukaryotes by DNA replication fork dynamics and quality of nucleotide pools. Environ. Mol. Mutagen 53, 699–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanaka S., Araki H. (2013) Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 5, a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pellegrini L. (2012) The Pol α-primase complex. Subcell. Biochem. 62, 157–169 [DOI] [PubMed] [Google Scholar]
- 9. Tahirov T. H. (2012) Structure and function of eukaryotic DNA polymerase δ. Subcell. Biochem. 62, 217–236 [DOI] [PubMed] [Google Scholar]
- 10. Hogg M., Johansson E. (2012) DNA polymerase ϵ. Subcell. Biochem. 62, 237–257 [DOI] [PubMed] [Google Scholar]
- 11. Kunkel T. A., Burgers P. M. (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pavlov Y. I., Shcherbakova P. V. (2010) DNA polymerases at the eukaryotic fork-20 years later. Mutat Res 685, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johansson E., Dixon N. (2013) Replicative DNA polymerases. Cold Spring Harb. Perspect. Biol. 5, a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lawrence C. W., Hinkle D. C. (1996) DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 28, 21–31 [PubMed] [Google Scholar]
- 15. Northam M. R., Robinson H. A., Kochenova O. V., Shcherbakova P. V. (2010) Participation of DNA polymerase ζ in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics 184, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuchta R. D., Stengel G. (2010) Mechanism and evolution of DNA primases. Biochim Biophys Acta 1804, 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perera R. L., Torella R., Klinge S., Kilkenny M. L., Maman J. D., Pellegrini L. (2013) Mechanism for priming DNA synthesis by yeast DNA polymerase α. Elife 2, e00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zerbe L. K., Kuchta R. D. (2002) The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41, 4891–4900 [DOI] [PubMed] [Google Scholar]
- 19. Smith D. J., Whitehouse I. (2012) Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baranovskiy A. G., Lada A. G., Siebler H. M., Zhang Y., Pavlov Y. I., Tahirov T. H. (2012) DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J. Biol. Chem. 287, 17281–17287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson R. E., Prakash L., Prakash S. (2012) Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. U.S.A. 109, 12455–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Makarova A. V., Stodola J. L., Burgers P. M. (2012) A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 40, 11618–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gómez-Llorente Y., Malik R., Jain R., Choudhury J. R., Johnson R. E., Prakash L., Prakash S., Ubarretxena-Belandia I., Aggarwal A. K. (2013) The architecture of yeast DNA polymerase ζ. Cell Rep. 5, 79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee Y. S., Gregory M. T., Yang W. (2014) Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc. Natl. Acad. Sci. U.S.A. 111, 2954–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dua R., Levy D. L., Campbell J. L. (1998) Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase ϵ in DNA replication and the S/M checkpoint pathway. J. Biol. Chem. 273, 30046–30055 [DOI] [PubMed] [Google Scholar]
- 26. Klinge S., Núñez-Ramírez R., Llorca O., Pellegrini L. (2009) 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. EMBO J. 28, 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Netz D. J., Stith C. M., Stümpfig M., Köpf G., Vogel D., Genau H. M., Stodola J. L., Lill R., Burgers P. M., Pierik A. J. (2012) Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tahirov T. H., Makarova K. S., Rogozin I. B., Pavlov Y. I., Koonin E. V. (2009) Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol ϵ and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol. Direct 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Copeland W. C., Wang T. S. (1993) Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J. Biol. Chem. 268, 26179–26189 [PubMed] [Google Scholar]
- 30. Klinge S., Hirst J., Maman J. D., Krude T., Pellegrini L. (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carr A. M., Paek A. L., Weinert T. (2011) DNA replication: failures and inverted fusions. Semin. Cell Dev. Biol. 22, 866–874 [DOI] [PubMed] [Google Scholar]
- 32. Weiner B. E., Huang H., Dattilo B. M., Nilges M. J., Fanning E., Chazin W. J. (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282, 33444–33451 [DOI] [PubMed] [Google Scholar]
- 33. Vaithiyalingam S., Warren E. M., Eichman B. F., Chazin W. J. (2010) Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc. Natl. Acad. Sci. U.S.A. 107, 13684–13689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kilkenny M. L., De Piccoli G., Perera R. L., Labib K., Pellegrini L. (2012) A conserved motif in the C-terminal tail of DNA polymerase α tethers primase to the eukaryotic replisome. J. Biol. Chem. 287, 23740–23747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mizuno T., Yamagishi K., Miyazawa H., Hanaoka F. (1999) Molecular architecture of the mouse DNA polymerase α-primase complex. Mol. Cell. Biol. 19, 7886–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang H., Weiner B. E., Zhang H., Fuller B. E., Gao Y., Wile B. M., Zhao K., Arnett D. R., Chazin W. J., Fanning E. (2010) Structure of a DNA polymerase α-primase domain that docks on the SV40 helicase and activates the viral primosome. J. Biol. Chem. 285, 17112–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou B., Arnett D. R., Yu X., Brewster A., Sowd G. A., Xie C. L., Vila S., Gai D., Fanning E., Chen X. S. (2012) Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase α-primase. J. Biol. Chem. 287, 26854–26866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kilkenny M. L., Longo M. A., Perera R. L., Pellegrini L. (2013) Structures of human primase reveal design of nucleotide elongation site and mode of Pol α tethering. Proc. Natl. Acad. Sci. U.S.A. 110, 15961–15966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baranovskiy A. G., Gu J., Babayeva N. D., Agarkar V. B., Suwa Y., Tahirov T. H. (2014) Crystallization and preliminary X-ray diffraction analysis of human DNA primase. Acta Crystallogr. F Struct. Biol. Commun. 70, 206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simon A. C., Zhou J. C., Perera R. L., van Deursen F., Evrin C., Ivanova M. E., Kilkenny M. L., Renault L., Kjaer S., Matak-Vinkovi D., Labib K., Costa A., Pellegrini L. (2014) A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510, 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sheaff R. J., Kuchta R. D., Ilsley D. (1994) Calf thymus DNA polymerase α-primase: “communication” and primer-template movement between the two active sites. Biochemistry 33, 2247–2254 [DOI] [PubMed] [Google Scholar]
- 42. Agarkar V. B., Babayeva N. D., Pavlov Y. I., Tahirov T. H. (2011) Crystal structure of the C-terminal domain of human DNA primase large subunit: Implications for the mechanism of the primase-polymerase α switch. Cell Cycle 10, 926–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sauguet L., Klinge S., Perera R. L., Maman J. D., Pellegrini L. (2010) Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS ONE 5, e10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mizuno T., Ito N., Yokoi M., Kobayashi A., Tamai K., Miyazawa H., Hanaoka F. (1998) The second-largest subunit of the mouse DNA polymerase α-primase complex facilitates both production and nuclear translocation of the catalytic subunit of DNA polymerase α. Mol. Cell. Biol. 18, 3552–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feng W., D'Urso G. (2001) Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase ϵ are viable but require the DNA damage checkpoint control. Mol. Cell. Biol. 21, 4495–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mäkiniemi M., Pospiech H., Kilpeläinen S., Jokela M., Vihinen M., Syväoja J. E. (1999) A novel family of DNA-polymerase-associated B subunits. Trends Biochem. Sci 24, 14–16 [DOI] [PubMed] [Google Scholar]
- 47. Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., Labib K. (2009) A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang S. G., Weisshart K., Gilbert I., Fanning E. (1998) Stoichiometry and mechanism of assembly of SV40 T antigen complexes with the viral origin of DNA replication and DNA polymerase α-primase. Biochemistry 37, 15345–15352 [DOI] [PubMed] [Google Scholar]
- 49. Bohn E. W., Wilson S. H. (1974) Studies on the activity of the a particle-associated DNA polymerase. Cancer Res. 34, 1977–1981 [PubMed] [Google Scholar]
- 50. Lao-Sirieix S. H., Nookala R. K., Roversi P., Bell S. D., Pellegrini L. (2005) Structure of the heterodimeric core primase. Nat. Struct. Mol. Biol. 12, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 51. Copeland W. C., Tan X. (1995) Active site mapping of the catalytic mouse primase subunit by alanine scanning mutagenesis. J. Biol. Chem. 270, 3905–3913 [DOI] [PubMed] [Google Scholar]
- 52. Vaithiyalingam S., Arnett D. R., Aggarwal A., Eichman B. F., Fanning E., Chazin W. J. (2014) Insights into eukaryotic primer synthesis from structures of the p48 subunit of human DNA primase. J. Mol. Biol. 426, 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Frick D. N., Richardson C. C. (2001) DNA primases. Annu. Rev. Biochem. 70, 39–80 [DOI] [PubMed] [Google Scholar]
- 54. Yao N. Y., Schroeder J. W., Yurieva O., Simmons L. A., O'Donnell M. E. (2013) Cost of rNTP/dNTP pool imbalance at the replication fork. Proc. Natl. Acad. Sci. U.S.A. 110, 12942–12947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mirkin S. M., Frank-Kamenetskii M. D. (1994) H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 23, 541–576 [DOI] [PubMed] [Google Scholar]
- 57. Chomilier J., Sun J. S., Collier D. A., Garestier T., Hélène C., Lavery R. (1992) A computational and experimental study of the bending induced at a double-triple helix junction. Biophys. Chem. 45, 143–152 [DOI] [PubMed] [Google Scholar]
- 58. Oguey C., Foloppe N., Hartmann B. (2010) Understanding the sequence-dependence of DNA groove dimensions: implications for DNA Interactions. PLoS ONE 5, e15931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hile S. E., Eckert K. A. (2004) Positive correlation between DNA polymerase α-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J. Mol. Biol. 335, 745–759 [DOI] [PubMed] [Google Scholar]
- 60. Franklin M. C., Wang J., Steitz T. A. (2001) Structure of the replicating complex of a pol α family DNA polymerase. Cell 105, 657–667 [DOI] [PubMed] [Google Scholar]
- 61. Bebenek A., Carver G. T., Dressman H. K., Kadyrov F. A., Haseman J. K., Petrov V., Konigsberg W. H., Karam J. D., Drake J. W. (2002) Dissecting the fidelity of bacteriophage RB69 DNA polymerase: site-specific modulation of fidelity by polymerase accessory proteins. Genetics 162, 1003–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aguirre J. D., Chifotides H. T., Angeles-Boza A. M., Chouai A., Turro C., Dunbar K. R. (2009) Redox-regulated Inhibition of T7 RNA polymerase via establishment of disulfide linkages by substituted Dppz dirhodium(II,II) complexes. Inorg. Chem. 48, 4435–4444 [DOI] [PubMed] [Google Scholar]