Background: The integrated stress response (ISR) is necessary to help the tumor adapt to its microenvironment.
Results: ATF4 activates a PEPCK-M-dependent pro-survival pathway under amino acid deprivation or ER stress (ISR) by binding to its proximal promoter.
Conclusion: PEPCK-M participates in supportive adaptations of cancer cells under stress.
Significance: A previously unappreciated role for PEPCK-M in cancer cells opens a therapeutic window and enhances our understanding of cancer metabolism.
Keywords: Cell Metabolism, ER Stress, Transcription Regulation, Tumor Metabolism, Unfolded Protein Response (UPR), AAR, AARE, ATF4, PEPCK-M, UPR
Abstract
Mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M), encoded by the nuclear PCK2 gene, links TCA cycle intermediates and glycolytic pools through the conversion of mitochondrial oxaloacetate into phosphoenolpyruvate. In the liver PEPCK-M adjoins its profusely studied cytosolic isoform (PEPCK-C) potentiating gluconeogenesis and TCA flux. However, PEPCK-M is present in a variety of non-gluconeogenic tissues, including tumors of several origins. Despite its potential relevance to cancer metabolism, the mechanisms responsible for PCK2 gene regulation have not been elucidated. The present study demonstrates PEPCK-M overexpression in tumorigenic cells as well as the mechanism for the modulation of PCK2 abundance under several stress conditions. Amino acid limitation and ER stress inducers, conditions that activate the amino acid response (AAR) and the unfolded protein response (UPR), stimulate PCK2 gene transcription. Both the AAR and UPR lead to increased synthesis of ATF4, which mediates PCK2 transcriptional up-regulation through its binding to a putative ATF/CRE composite site within the PCK2 promoter functioning as an amino acid response element. In addition, activation of the GCN2-eIF2α-ATF4 and PERK-eIF2α-ATF4 signaling pathways are responsible for increased PEPCK-M levels. Finally, PEPCK-M knockdown using either siRNA or shRNA were sufficient to reduce MCF7 mammary carcinoma cell growth and increase cell death under glutamine deprivation or ER stress conditions. Our data demonstrate that this enzyme has a critical role in the survival program initiated upon stress and shed light on an unexpected and important role of mitochondrial PEPCK in cancer metabolism.
Introduction
Phosphoenolpyruvate carboxykinase (PEPCK)4 (GTP; EC 4.1.1.32) activity, which catalyzes the conversion of oxaloacetate to phosphoenolpyruvate (PEP), is distributed both in the cytosol and mitochondria as a result of two enzymatically indistinct isozymes, PEPCK-C and PEPCK-M (1, 2), encoded by different nuclear genes (Pck1 and Pck2, respectively) (3). PEPCK-C, the first committed step of gluconeogenesis and glyceroneogenesis, is restricted to differentiated liver, small intestine, kidney cortex, and adipose tissue, whereas PEPCK-M appears not to be constrained to those tissues as it is expressed in a variety of cell types, including T- and B-cells, pancreatic β-cells, or neurons. It is important to note that the PEPCK reaction is the only pathway that is able to communicate mitochondrial intermediates with the glycolytic intermediary pool above PEP. Although the role of the mitochondrial isozyme remains largely unknown, recent reports have shown a complex interaction with its cytosolic counterpart toward hepatic gluconeogenesis through cataplerosis of mitochondrial precursors (4). In pancreas, PEPCK-M was highly expressed in β-cells where it enhanced TCA cycle dynamics via its recycling of GTP generated at the succinyl-CoA synthase reaction (5). Thus, PEPCK-M activity pumped the TCA cycle and sourced PEP toward pyruvate formation to feed acetyl-CoA for the citrate synthase reaction (pyruvate cycling), altogether regulating glucose-stimulated insulin secretion. Interestingly, and in contrast to its cytosolic counterpart, hormones or nutrients that are known to regulate gluconeogenesis do not transcriptionally regulate the gene coding for PEPCK-M, PCK2.
Eukaryotes have evolved complex mechanisms to allow cells to confront and adapt to variable conditions such as nutrient limitations. One such process, known as integrated stress response (ISR), collectively groups several signaling pathways that converged on the phosphorylation of eIF2α and comprise responses triggered by amino acid starvation (amino acid response (AAR)) or endoplasmic reticulum stress (unfolded protein response (UPR)) leading to the activation of target genes. The interplay of downstream activating pathways with the severity and duration of stress determines the fate of the cell, thus ensuring cell viability or activation of cell death. Importantly, tumors dynamically activate ISR to allow cancer cells to cope with metabolic limitations.
In the present work we describe the selective expression of PCK2 in several human tumors and all cancer cell lines studied, suggesting that PEPCK-M activity might support specific purposes in the context of tumor metabolic adaptations. Expression is sensitive to several cues that signal through the canonical ISR response, including chemical inducers of UPR and amino acid limitations. The mechanism of PCK2 gene regulation in cancer cells under ISR requires recruiting ATF4 to a consensus amino acid response element (AARE) sequence located in the PEPCK-M proximal promoter. Finally, we provide evidence on the importance of this gene as a pro-survival mechanism under conditions of stress, probably by channeling TCA intermediates into the triose-phosphate pool. Hence, PEPCK-M up-regulation is a novel metabolic adaptation in cancer.
EXPERIMENTAL PROCEDURES
Cell Culture
Human breast (MCF7), cervix (HeLa), and colon (HCT116) carcinoma cell lines and mouse wild type NIH-3T3 and Kras-V12 carcinogenic NIH-3T3 (NIH-3T3Kras) (a gift from Dr. Capella, The Bellvitge Institute for Biomedical Research (IDIBELL), Spain) and PKR-like endoplasmic reticulum-resident kinase (PERK)-deficient mouse embryonic fibroblasts (a gift from Dr. Douglas R. Cavener, Pennsylvania State University) were maintained in DMEM supplemented with 10% FBS, 10000 units/ml penicillin, 10 mg/ml streptomycin, and 2 mm l-glutamine (Biological Industries) and incubated in a humidified atmosphere of 5% CO2 at 37 °C. Thapsigargin, tunicamycin, actinomycin D, and etoposide were purchased from Sigma.
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted using UltraspecTM RNA isolation system (Biotecx, TX). cDNA synthesis from 2 μg of RNA was performed using a High Capacity cDNA Reverse Transcription kit (Invitrogen). cDNA was analyzed using real-time quantitative RT-PCR assays in a 7900HT real-time RT-PCR system (Applied Biosystems, Carlsbad, CA). Data analysis is based on the ΔΔCt method.
Western Blot
Cells were homogenized in radioimmunoprecipitation assay buffer supplemented with protease and phosphatase inhibitors and centrifuged at 15,000 × g for 15 min at 4 °C. Western blots were performed with 20–50 μg of tissue extract. Proteins were separated in 8–12% SDS-PAGE and transferred to an Immobilon membrane (Millipore, Bedford, CA). Horseradish peroxidase activity linked to secondary antibody was detected with ECL substrate (Pierce) in a Fujifilm LAS 3000 Intelligent Dark Box IV imaging system. Antibodies used to detect PEPCK-M were rabbit anti-PEPCK-M (Abcam; ab70359) and goat anti-PEPCK-M (Fig. 1A) (Everest Biotech; EB06944). Antibodies against ATF4, GCN2 (general control nonderepressible 2), C/EBPα C/EBPβ, and α-actinin were from Santa Cruz. α-Tubulin and γ-tubulin antibodies were purchased from Sigma. Phospho-specific antibody against eIF2α (Ser-51) was from Calbiochem. Anti-SOD1 was from Novocastra. Anti-SOD2 was from Stressgen. Anti-co-chaperone p23 was from Affinity Bioreagents. Anti-green fluorescent protein (GFP) was from Abcam. Antibodies detecting total eIF2α, VDAC (voltage-dependent anion channel protein), caspase-7, and PARP (poly(ADP-ribose) polymerase) were from Cell Signaling. Antibodies against cytosolic PEPCK were a generous gift of Dr. Daryl K. Granner (Vanderbilt University, Nashville, TN).
FIGURE 1.
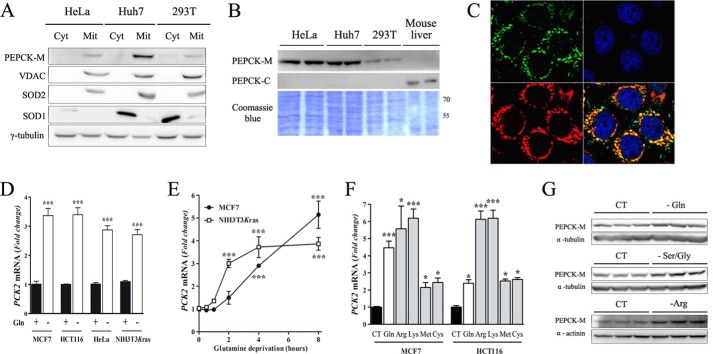
PEPCK-M is expressed in cancer cells, and its expression is regulated by amino acid limitation. A, PEPCK-M protein expression detection in different cell lines analyzed by Western blot after subcellular fractionation. Cytosolic and mitochondrial superoxide dismutases (SOD1 and -2, respectively) and the mitochondrial voltage-dependent anion channel protein (VDAC) transporter were used as markers. Cyt, cytosol; Mit, mitochondria. B, mitochondrial and cytosolic PEPCK (PEPCK-M and PEPCK-C, respectively) were measured in the same cell lines shown in A; mouse liver was used as positive control for PEPCK-C, and Coomassie Blue was used as the loading control. C, indirect immune detection using confocal microscopy of human PEPCK-M endogenously expressed in MCF7 cells. PEPCK-M staining is shown in green. Mitochondria are visualized using MitoTracker Red CMXRos (red), and nuclei are stained in blue using TO-PRO®3. PEPCK-M and mitochondria colocalize as seen by the yellow in the superimposed image. C–E, quantitative PCR with human PCK2-specific primers. D, four different cell lines (see “Experimental Procedures” for more information) were incubated with media lacking glutamine for 16 h. E, MCF7 and NIH3T3Kras cells were glutamine-deprived for the indicated times. F, MCF7 and HCT116 cells were cultured in media lacking glutamine, arginine, lysine, methionine, or cysteine for 8 h. CT, control. In all cases, the -fold induction of PCK2 with respect to basal conditions is represented. Data are presented as the mean ± S.E., n = 3–4. *, p < 0.05; ***, p < 0.001 relative to basal conditions; one-way ANOVA, with a Newman-Keuls post-hoc test. G, PEPCK-M protein induction was verified by Western blot in MCF7 cells incubated for 24 h in media lacking glutamine, serine and glycine, or arginine as indicated. α-Tubulin or α-actinin were used as the loading control.
Histology and Immunofluorescence
PEPCK-M was immunostained as previously described (4). For mitochondrial labeling, cultured cells were incubated in culture media containing a 0.1 μm final concentration of MitoTracker Red CMXRos (Invitrogen) dissolved in dimethyl sulfoxide for 20 min in a 37 °C, 5% CO2 gas incubator. The cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized by incubating with 0.2% Triton X-100 in PBS for 10 min. Cells were then blocked by incubating with 3% BSA in PBS for 30 min and treated with the PEPCK-M primary antibody (ab70359; Abcam) for 16 h. After 4 washes with 0.1% Triton in PBS, cells were incubated with anti-rabbit Alexa Fluor® 488 secondary antibody ((Invitrogen) for 1 h and during this incubation TO-PRO3 was added to stain nuclei. Samples were examined using a confocal microscope (TCS SL, Leica), and LCS Lite software (Leica) was used to collect digital images.
Subcellular Fractionation
Cells were incubated with digitonin (50 μg/ml) for 4 min on ice and centrifuged at 1200 × g for 10 min to remove large cell and tissue fragments and cell nuclei. Pellet 1 was discarded, and mitochondria were pelleted by centrifuging the supernatant at 12,000 × g for 10 min. Cytosol was mixed with 4× SDS sample buffer (0.250 m Tris, pH 6.8, 8% SDS, 40% glycerol, 4% β-mercaptoethanol, 0.04% bromphenol blue). Pellet-containing mitochondria were resuspended in the same SDS buffer (25 mm Tris-HCl, 1 mm EDTA, 1 mm EGTA, 1% SDS, pH 7.4) and mixed with 4× SDS sample buffer to reach total final volume equal to cytosol. Western blots were performed with equal volumes of cytosolic and mitochondrial fractions to maintain a correct correlation relative to fresh material. Proteins were separated in 12% SDS-PAGE.
Plasmid Constructs
The PCK2 promoter region was amplified from human genomic DNA and cloned into the luciferase reporter vector PGL3-basic (Promega, Mannheim, Germany). The three sequences of the recombinant plasmids, PCK2–1825/+42, PCK2–808/+42, and PCK2–214/+42, were amplified and subcloned independently using an initial amplification product as template (5′-CTCAGCAGCTCCAGGCTTATATTC and 5′-CATGCCAGTTAAGCCGCAGGCCAG) and introducing SacI and HindIII restriction sites in the PCR primers (SacI-1-fw, 5′-ACTCAGCAGAAAGAGAGCTCACTTCC-3′; SacI-2-fw, 5′-GCAGGCGGATCACGAGCTCAGGAGT-3′; SacI-3-fw, 5′-TGTGCGGCTGGGAGCTCGGTGGCGGATGGG-3′; HindIII-rev, 5′-GAGCGCGGCGAAGCTTGCCACAGCAGAGGC-3′). Point mutations in the AARE sequences were introduced by PCR. The primer sequences were as follows (the AAREs are underlined, and the mutated nucleotides are shown in bold in the forward sequences): AARE1-fw, 5′-AGTCCTCAAAGGCATAGTATGTGCGGCTG-3′; AARE1-rev, 5′-CAGCCGCACATACTATGCCTTTGAGGACT-3′; AARE2-fw, 5′-GGGCAGATGAGGCTACTTATTGGATTGTG-3′; AARE2-rev, 5′-CACAATCCAATAAGTAGCCTCATCTGCCC-3′.
Transfection and Luciferase Assays
Cell transfection was performed using polyethyleneimine (linear polyethyleneimine, Mr 25,000, Sigma). The different promoter plasmids (2 μg) and 0.25 μg of pSV40-β-galactosidase control vector (Promega) were co-transfected into 6-well plates containing 80% confluent cells and then distributed into 24-well plates. Cells were maintained overnight in complete medium and treated as indicated. Luciferase activity was measured in a TD 20/20 luminometer (Turner Designs) using the Luciferase Assay System (Promega) as previously described (6). In overexpression experiments, the transcription factor expression plasmids (100 ng of each) were co-transfected with the promoter constructs, and luciferase activity was measured 24 h after transfection. The total amount of transfected DNA was kept constant by the addition of empty pcDNA3.1 plasmid. The luciferase values were normalized to protein content and to β-galactosidase activity using the luminescent β-galactosidase detection kit II (Clontech). C/EBP (CCAAT/enhancer-binding protein) vectors were a gift of Richard W. Hanson (Case Western Reserve University, Cleveland, Ohio). ATF4 and ATF3 cDNA vectors were gifts of Diego Haro (Universitat de Barcelona, Barcelona, Spain).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as described previously (6). Briefly, chromatin from treated MCF7 cells was sonicated to an average fragment size of 400–500 bp in a Branson Sonifier (8 cycles of 10 s sonication with a 30-s rest time at 4 °C). Rabbit or mouse IgG (Sigma) was used as a control for nonspecific interaction of DNA. Input was prepared with 10% of the chromatin material used for an immunoprecipitation. Later, a 1:10 dilution of input material was performed before PCR amplification. PCR products were resolved in agarose gel. Densitometry was performed using Multi Gauge software. Primers sequences were: PCK2-AARE1, 5′-CTCGTTCCTAGCTTGTTTGCCACC-3′ and 5′- GCCAAACCCATCTCTCCCATTGGA-3′; PCK2-AARE2, 5′-GTGGGTGATATCCAGTGGTCTCAG-3′ and 5′-GGCCTGGTCATTCTTTCTCTCCA-3′; ASNS, 5′-TGGTTGGTCCTCGCAGGCAT-3′ and 5′-CGCTTATACCGACCTGGCTCCT-3′.
RNA Interference (RNAi)
Small interfering RNA (siRNA) oligonucleotides targeting human PCK2 (PCK2#1, 5′-CAAGGAUGUGGCACGAGUATT-3′; PCK2#2, 5′-GGGCCAUCAACCCUGAGAATT-3′; PCK2#3, 5′-UGGCUACAAUCCAGAGUAATT-3′), ATF4 (5′-GCCUAGGUCUCUUAGAUGATT-3′), GCN2 (5′-CAGCAGAAAUCAUGUACGATT-3′), and control non-silencing siRNA (scramble, SCR: (5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Shanghai GenePharma. MCF7 and HeLa cells were transfected using Oligofectamine transfection reagent (Invitrogen) as specified by the manufacturer. Cells were used in experiments 48 h after siRNA transfection. PCK2 was silenced using a pool of three individual siRNA. For PCK2 knockdown with shRNA, GIPZ Lentiviral shRNAs (Open Biosystems, clone IDs: V3LMM_427490 and V3LHS_328126, denominated B and C, respectively) were used. GIPZ non-silencing lentiviral shRNA (clone ID RHS-4348) was used as negative control. After transduction, cells were selected with puromycin and sorted by fluorescein-activated cell sorter (FACS) based on turboGFP fluorescence.
Glycolytic Rate Measurement
Glycolytic rate was determined by measuring the conversion of d-[5-3H]glucose to 3H2O, released by the enolase step of glycolysis (7). Briefly, cells were incubated with 5 μCi of d-[5-3H]glucose (American Radiolabeled Chemicals, St. Louis, MO) at 37 °C for 60 min. After incubation, triplicate 50-μl aliquots were transferred to uncapped PCR tubes containing 50 μl of 0.2 n HCl, and each tube was transferred to a scintillation vial containing 0.5 ml of H2O so that the water in the vial and the contents of PCR tube were not allowed to mix. The vials were sealed, and the diffusion took place for 24 h. The amount of diffused and undiffused 3H2O was determined by scintillation counting and compared with controls of d-[5-3H]glucose and 3H2O alone.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Proliferation Assay
For the MTT assay, (0.5 mg/ml final concentration) was added to each well, and then the plates were incubated at 37 °C for 3 h. The formazan product was dissolved in isopropyl alcohol HCL, and the absorbance at 570 nm was measured using a microplate reader.
Analysis of Viability by Phosphatidylserine Exposure and Propidium Iodide Uptake
Cell viability in siRNA-treated MCF7 cells was determined by analyzing phosphatidylserine exposure and membrane integrity by double staining with annexin V-FITC and propidium iodide before flow cytometric analysis (FACSCalibur, BD Biosciences) using the annexin V-FITC Apoptosis Detection kit (eBioscience, San Diego, CA). Data analysis was performed with CellQuest software (BD Biosciences). For GFP expressing MCF7-shRNA clones, annexin V-allophycocyanin, and 7-aminoactinomycin D (eBioscience) were used and analyzed on FACS Canto (BD Biosciences).
Statistical Analysis
Results are expressed as the means ± S.E. Statistical analysis was performed by one-way ANOVA (Newman-Keuls post-hoc test) and two-tailed Student's t test using GraphPad Prism® software. p < 0.05 was considered significant.
RESULTS
PCK2, Not PCK1, Is Present in Tumors and Tumorigenic Cell Lines
Data mining through curated array data at BioGPS pointed at tumor-specific expression of PCK2 that correlated with aggressive growth and was evident in either tumor-bearing mice, human tumors, or chemically induced tumors. To confirm, we assayed PCK2 mRNA levels in tumor cell lines of human and compared them to those of the cytosolic isoform (PCK1; PEPCK-C). Table 1 shows raw Ct expression of both genes, confirming that PCK2 is highly expressed in all tumorigenic cell lines tested to date, whereas PCK1 is below the detection level. However, in immortalized fibroblasts or differentiated C2C12 or NIH3T3-L1 a higher amount of PCK1 mRNA was observed. Interestingly, Kras-activated NIH-3T3 (NIH-3T3Kras) cells do not express significant levels of Pck1 mRNA, whereas greatly overexpress Pck2 mRNA in contrast to non-tumorigenic parent cell line (NIH-3T3). Protein concentration was also assayed in three cell lines by subcellular fractionation (Fig. 1A), confirming that mitochondrial PEPCK (PEPCK-M; PCK2) was the only PEPCK activity expressed in these cell lines and was properly localized to the mitochondria. As expected, no PEPCK-C protein was present in these cell lines (Fig. 1B). Immunocytochemistry localized PEPCK-M exclusively in the mitochondria of MCF7 cells (Fig. 1C).
TABLE 1.
mRNA abundance of PEPCK isozyme encoding genes in several human and mouse cell lines
| Species | Cell line | Raw Ct values |
|
|---|---|---|---|
| PCK2/Pck2 | PCK1/Pck1 | ||
| Human | MCF7 | 27.4 | >35 |
| HeLa | 28.7 | >35 | |
| Jurkat | 28.9 | >35 | |
| Saos | 25.6 | >35 | |
| T98G | 26.5 | >35 | |
| U2OS | 25.4 | >35 | |
| U87 | 26.5 | >35 | |
| SH-SY5Y | 34.3 | >35 | |
| Mouse | MEF | 24.2 | >35 |
| C2C12 | 29.1 | 32.7 | |
| 3T3 L1 | 24.1 | 32.4 | |
| MC-3T3 | 24.8 | 33.9 | |
| NIH-3T3KrasV12 | 22.0 | >35 | |
Nutrient Restriction Up-regulates PCK2 Expression
Because PEPCK-M regulates TCA cycle dynamics and cataplerosis, we hypothesized that it might also regulate carbon flux during nutrient stress, as the TCA is key to mobilize intermediates toward biosynthetic processes. Glutamine represents the quantitatively most important TCA cycle fuel in cancer cells; consequently, we investigated whether PCK2 mRNA is regulated by withdrawal of these amino acids. qRT-PCR analysis confirmed that PCK2 mRNA content increased at least 2-fold after glutamine deprivation in all cell lines tested (Fig. 1D). PCK2 mRNA measured in MCF7 and NIH-3T3Kras cell lines at various treatment times (0–8 h) demonstrates a rapid (within 2–4 h) and robust induction in response to the lack of glutamine (Fig. 1E). To determine whether this induction could be reproduced by the withdrawal of other amino acids, MCF7 and HCT116 cells were incubated for 8 h in media lacking a particular amino acid. qRT-PCR analysis confirmed that PCK2 expression was induced by the depletion of either single amino acid tested, even at similar or higher levels of those seen upon glutamine deprivation (Fig. 1F). Increased mRNA levels correlated with protein concentration at 24 h after depletion of glutamine or serine and glycine in MCF7 cells (Fig. 1G).
ER Stress Increases PEPCK-M Content
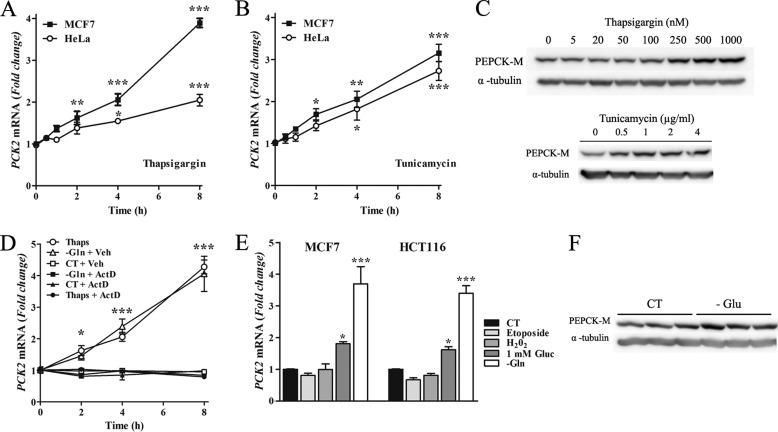
AARs and ER stress share a common downstream effector mechanism; inhibition of translation by phosphorylation of eIF2α. Therefore, we examined the hypothesis that ER stress would also regulate PCK2 expression. To test this hypothesis, chemical inducers of ER stress response, thapsigargin (a sarcoendoplasmic Ca2-ATPase inhibitor), or tunicamycin (a glycosyltransferase inhibitor) were submitted to MCF7 or HeLa cells for 0 to 8 h; DMSO was added in control wells (vehicle). As seen in Fig. 2, PCK2 levels were induced up to 3-fold upon thapsigargin or tunicamycin treatment (Fig. 2, A and B). We next examined the effects of ER stressors (increasing doses of thapsigargin or tunicamycin for 24 h) on PEPCK-M protein content. A robust dose-dependent accumulation of PEPCK-M was observed in MCF7 cells treated with tunicamycin (Fig. 2C) and HCT116 treated with both drugs (data not shown). PCK2 up-regulation was the consequence of direct transcriptional activation downstream of ER stress signaling as actinomycin-D, a RNA polymerase II inhibitor, prevented the accumulation of PCK2 mRNA by either glutamine deprivation or thapsigargin (Fig. 2D).
FIGURE 2.
PEPCK-M is activated by ER stress. MCF7 and HeLa cells were incubated with media containing 500 nm thapsigargin (A) or 2 μm tunicamycin (B) for the indicated times. C, PEPCK-M protein induction was verified by Western blot in MCF7 cells exposed for 24 h to different concentrations of thapsigargin (upper panel) or tunicamycin (lower panel). α-Tubulin was used as the loading control. D, MCF7 cells were incubated for the indicated time points in control media (CT + Veh), in media lacking glutamine, or media containing thapsigargin in the presence or absence of 5 μg/ml actinomycin D (ActD). DMSO was used as vehicle (Veh). E, MCF7 and HCT116 cells were cultured in media containing 10 μm etoposide, 0.4 mm H202, 1 mm glucose, or media lacking glutamine for 8 h. In all cases, the -fold induction of PCK2 with respect to control conditions is represented. Data are presented as the mean ± S.E., n = 3. *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to basal conditions; one-way ANOVA, with a Newman-Keuls post-hoc test. F, PEPCK-M protein levels were analyzed by Western blot in MCF7 cells incubated for 16 h in media containing 1 mm glucose.
To rule out an indirect effect on PCK2 transcription by stress conditions other than ER stress, we examined the modulation of PCK2 mRNA levels by H2O2, an effector of oxidative stress pathways, and the topoisomerase II inhibitor etoposide, which triggers DNA damage and stimulates apoptotic response. Results shown in Fig. 2F demonstrate that these stressors do not modulate PCK2 levels, ruling out an indirect activation of PCK2 transcription by chemical inducers of ER stress. On the other hand, to assess PCK2 transcription regulation in physiologic conditions of ER stress, its mRNA content and protein levels were quantified in MCF7 cells on a low glucose medium. As observed in Fig. 2, E and F, glucose limitation was also capable of up-regulating mitochondrial PEPCK in MCF7.
An AARE Consensus Site at the PCK2 Promoter Recruits ATF4 to Activate Transcription under AAR or ER Stress
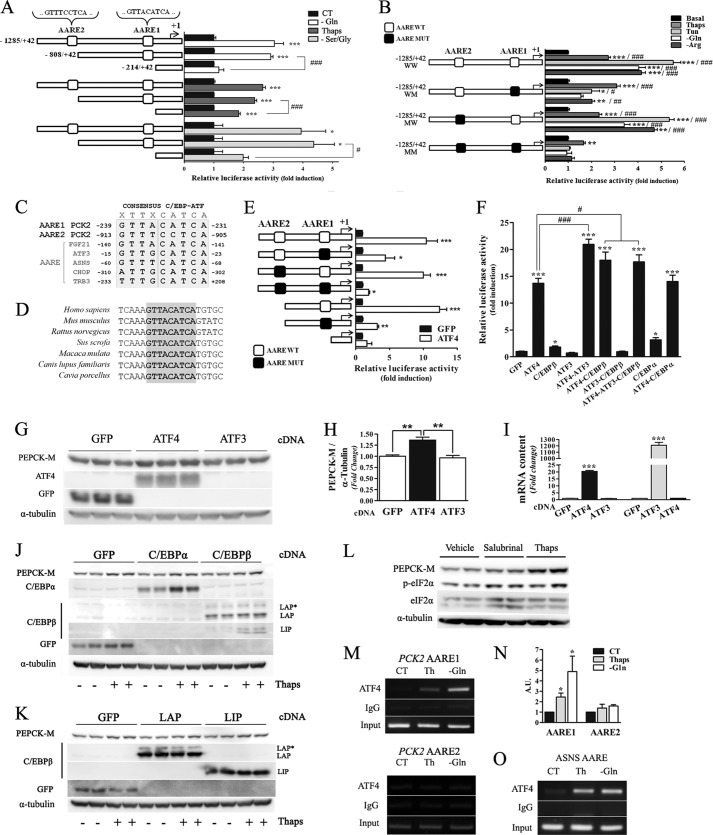
Transcriptional up-regulation was responsible for increased PEPCK-M protein levels observed; therefore, we evaluated whether a downstream transcriptional effector of ER or amino acid stress response directly mediated this induction. Promoter sequences ranging in size from −214 to −1285 upstream of the transcriptional start site of the human PCK2 gene were engineered as luciferase reporter constructs as shown in Fig. 3A. Luciferase activity from MCF7 cells transiently transfected with these constructs (PCK2/−1285, PCK2/−808, and PCK2/-214) was studied under glutamine or serine plus glycine deprivation or thapsigargin stimulus. Although PCK2/−1285 and −808 sustained identical induction by stress stimuli, PCK2/−214 had a blunted response (Fig. 3A). These data suggest that the sequence from −808 to −214 of the human PCK2 gene promoter may contain a relevant element responsible for the increase in luciferase activity after treatments. Detailed in silico analysis of the promoter sequence revealed the presence of different putative consensus binding sites for the ATF (activating transcription factor) family of transcription factors, referred to as the C/EBP-ATF composite site and also known as AARE. AARE sequences bind monomers and homo- and heterodimers of ATF and C/EBP proteins, both subfamilies of the larger bZIP (basic region/leucine zipper) transcription factor family.
FIGURE 3.
Transcriptional control of PCK2 under amino acid limitation and ER stress is mediated by ATF4 through AARE consensus sequences. A, MCF7 cells were transiently transfected with the PCK2 full-length promoter-luciferase reporter construct (−1327 from the translational start) and truncated promoter fragments lacking one or both putative amino acid response elements (AARE1 and AARE2). When indicated, cells were glutamine- or serine- and glycine-deprived (−Ser/Gly) or treated with thapsigargin (Thaps; 500 nm) for 16 h. B, MCF7 cells were transiently transfected with the wild type PCK2 full-length promoter-luciferase reporter constructs or constructs AARE1 and AARE2. When indicated, cells were glutamine- or arginine-deprived (−Arg), treated with thapsigargin (500 nm), or with tunicamycin (Tun; 2 μm) for 16 h. C, comparison of the PCK2 AARE sequences with functional C/EBP-ATF binding sites present in other genes. D, alignment of the highly conserved PCK2 AARE1 sequence in different mammals. E, luciferase activity of the wild type, truncated, and mutated PCK2 promoter constructs co-transfected with an expression plasmid for human ATF4 during 24 h. GFP expression vector was used as control. F, luciferase activity of the wild type PCK2 promoter construct co-transfected with expression plasmids for GFP, ATF4, ATF3, C/EBPα, C/EBPβ, or combinations during 24 h. In A, B, E, and F luciferase and β-galactosidase activities were measured in cell extracts. -Fold induction of luciferase activity of treated cells with respect to basal condition or control vector is represented. G, PEPCK-M protein levels were measured by Western blot after ATF4 and ATF3 cDNA transfection, and GFP was used as cDNA control and α-tubulin as loading control. H, quantification of the PEPCK-M levels (normalized to α-tubulin) in the experiment detailed in G. The average of three experiments is shown. I, ATF4 and ATF3 mRNA levels after transfection were determined by quantitative PCR PEPCK-M levels determined by Western blot after C/EBPα and C/EBPβ (J) and C/EBPβ-LAP and C/EBPβ-LIP (K) overexpression with and without thapsigargin treatment (500 nm; 24 h). MCF7 cells were treated 5 μm salubrinal or 500 nm thapsigargin for 24 h (L) and ChIP analysis of ATF4 recruitment to the PCK2 promoter in response to glutamine deprivation and thapsigargin (500 nm) treatment during 8 h before chromatin preparation for ChIP in MCF7 cells (M). Amplification of input DNA (representing 1% of immunoprecipitated material) is shown for comparison. N, the level of amplified DNA was quantified by densitometric analysis of three different gels and corrected for the amount of input DNA. Values are expressed as the -fold change with respect to the control media. O, as the positive control, ChIP analysis of ATF4 recruitment to the asparagine synthetase (ASNS) AARE sequence in the experiment detailed above is shown. Data are presented as the mean ± S.E., n = 3. *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to basal conditions; #, p < 0.05; ##, p < 0.01; ###, p < 0.001 relative to minimal promoter (A and E), double-mutated (B) constructs, or GFP overexpression (F). One-way ANOVA, with a Newman-Keuls post-hoc test. CT, control.
The relative contribution of these sites to promoter activity was then assessed using reporter constructs with specific mutations at each of the AARE sites (AARE1 and AARE2) found in the proximal promoter of the PCK2 gene (see “Experimental Procedures”). Gene reporter assays showed that mutations in the AARE1 response element severely abrogated response, both to ER stress induction by thapsigargin and amino acid limitation (Fig. 3B). In contrast, mutations in the AARE2 binding element had minor effects on transcriptional activity. The AARE1 site (GTTACATCA), located at position −917 bp upstream of the nucleotide designated as +1, in the 5′-flanking region of the human PCK2 promoter matches a functional C/EBP-ATF composite site present in the FGF21 gene promoter that binds ATF4 and C/EBPβ (8) (Fig. 3C). PCK2 AARE1 sequence is highly conserved in mammals (Fig. 3D). Therefore, it was likely that an ATF-C/EBP family member also binds to and transactivates, through the PCK2 AARE-1. To determine whether these proteins affected PCK2 gene transcription, we studied the transcriptional response of the reporter constructs upon overexpression of ATF4, ATF3, c/EBPα, and C/EBPβ and GFP (as control) in MCF7 cells. Fig. 3E shows that ATF4 overexpression robustly stimulated the transcriptional activity of the wild type construct containing both AARE1 and AARE2 sites, whereas constructs lacking the AARE1 site remained unstimulated, suggesting that AARE1 may be the site responsible for ATF4-mediated activation. Even though the overexpression of ATF3 did not produce a significant increase in promoter activity by itself, its co-expression with ATF4 potentiated transcriptional activity (Fig. 3F). Consistent with these results overexpression of ATF4, but not ATF3, was sufficient to produce a robust increase in PEPCK-M levels (Fig. 3, G–I). Overexpression of either C/EBPα or C/EBPβ also potentiated the ATF4 effect on luciferase activity, whereas both had minor effects when overexpressed alone (Fig. 3F). Neither c/EBPα, C/EBPβ (Fig. 3J), LAP (activator), nor LIP (inhibitor) isoforms of C/EBPβ independently (Fig. 3K) affected PEPCK-M protein levels, under basal or stressed conditions. Finally, salubrinal, an inhibitor of eIF2α dephosphorylation that induces ATF4 production, caused a small but significant increase in basal PEPCK-M protein levels, suggesting that eIF2α activation is sufficient to initiate the cascade leading to PCK2 up-regulation (Fig. 3L).
To confirm that ATF4 binds to the human PCK2 promoter in vivo, ChIP assays were performed. Chromatin fragments isolated from MCF7 cells treated with thapsigargin or incubated in the absence of glutamine for 8 h were immunoprecipitated with an anti-ATF4 antibody. The immunoprecipitate was tested for PCK2 promoter sequences by PCR amplification, demonstrating ATF4 recruitment to the PCK2 promoter region containing the AARE1 site in vivo (Fig. 3M; quantification provided in Fig. 3N) and supporting the conclusion that ATF4 is the protein responsible for PCK2 promoter transactivation upon AAR and UPR induction. As a positive control, ATF4 recruitment in the AARE site of the asparagine synthase gene was also confirmed (Fig. 3O).
Both GCN2 and PERK Are Involved in the AAR and UPR-mediated Induction of PCK2 through ATF4
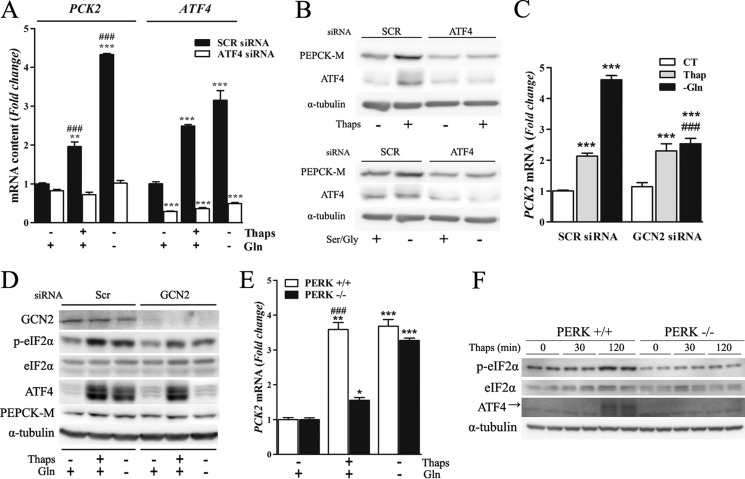
Conclusively, knocking down ATF4 in MCF7 cells abrogated ATF4 induction under ER- tress and brought its expression below basal levels in unstimulated conditions (Fig. 4, A and B). Consequently, ATF4 depletion completely abolished PCK2 induction by thapsigargin and glutamine deprivation and reduced its expression in unstimulated conditions (Fig. 4, A and B).
FIGURE 4.
Induction of PCK2 under amino acid limitation and ER stress depends on GCN2/eIF2α/ATF4 and PERK/eIF2α/ATF4 axis, respectively. A, real-time PCR analysis of PCK2 and ATF4 mRNA levels in MCF7 cells transfected with ATF4 or control (SCR) siRNA and incubated for 8h in control medium or in media either lacking glutamine or media supplemented with thapsigargin (Thaps; 500 nm). B, protein levels of PEPCK-M, ATF4, and α-tubulin were measured by immunoblotting in ATF4 siRNA knockdown cells exposed to 500 nm thapsigargin or to media lacking serine and glycine for 24 h. C, real-time PCR analysis of PCK2 mRNA levels in MCF7 cells transfected with GCN2 or control siRNA and incubated for 8 h with either control medium or in media lacking either glutamine or supplemented with thapsigargin (500 nm). CT, control. D, Western blot analysis of the experiment detailed in C demonstrates GCN2 knockdown with blunted ATF4 induction in response to amino acid depletion but not to ER stress. E, WT and Perk knock-out (Perk−/−) mouse embryonic fibroblasts were exposed to glutamine depletion or 500 nm thapsigargin for 8 h, and expression levels of PEPCK-M were quantified by real-time RT-PCR. F, Western blot analysis showing Perk−/− mouse embryonic fibroblasts blunted eif2α phosphorylation and ATF4 induction in response to thapsigargin. In all cases DMSO was added as vehicle control. The mRNA data are shown as -fold change relative to the control condition and represent the means ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 relative to basal conditions; ###, p < 0.001 relative to same treatment in the other siRNA treatment or genotype. One-way ANOVA, with a Newman-Keuls post-hoc test.
We next studied the participation of GCN2 and PERK kinases involved in the AAR and UPR responses, respectively, in the events leading to PCK2 up-regulation. GCN2 silencing in MCF7 blunted PCK2 mRNA up-regulation in response to glutamine deprivation, but not upon thapsigargin treatment (Fig. 4C). This is consistent with reduced eIF2α phosphorylation and lack of ATF4 induction only under glutamine deprivation conditions (Fig. 4D). Conversely, in PERK knock-out murine embryonic fibroblasts (Perk−/− mouse embryonic fibroblasts) treated with thapsigargin, PCK2 mRNA levels remained unchanged (Fig. 4E), consistent with reduced eIF2α phosphorylation and blunted ATF4 induction (Fig. 4F). On the other hand, glutamine deprivation induced a similar increase of PEPCK-M in both Perk+/+ and Perk−/− cells (Fig. 4E). Taken together, our results demonstrate that amino acid starvation and ER stress triggered a cascade of protein kinases including GCN2, PERK, and eIF2α that mediates ATF4 activation and subsequent PCK2 up-regulation.
PEPCK-M Is Necessary to Tip the Balance toward Survival upon AAR and UPR Activation
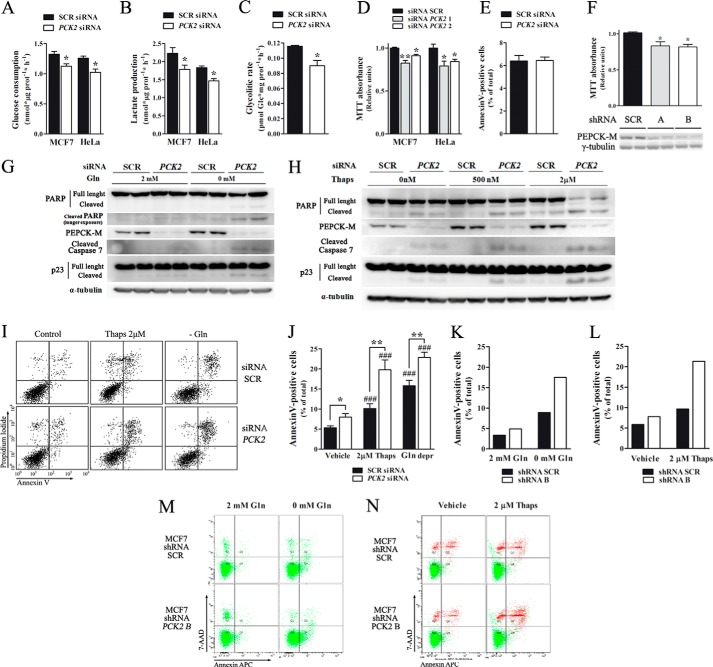
To investigate the impact of PEPCK-M knockdown on cell proliferation in basal, unstressed conditions and under ER stress, we used two strategies: (i) transient siRNA silencing or (ii) stable shRNA upon lentiviral infection. Consistently, either strategy reduced PEPCK-M levels significantly in exponentially growing MCF7 and HeLa cells that correlated with a reduction in glucose consumption (Fig. 5A), lactate production (Fig. 5B) and glycolytic flux (Fig. 5C). Either PCK2 gene silencing strategy afforded significant negative effects on cell proliferation (Fig. 5, D and F) without apoptosis (Fig. 5E), demonstrating that PEPCK-M has a significant role in tumor cells under normal growth conditions.
FIGURE 5.
PCK2 knockdown impairs survival under amino acid deprivation and ER stress conditions. PCK2 knockdown decreases glucose consumption (A) and lactate production (B) in MCF7 and HeLa cells. Glucose/lactate levels were measured in complete media collected after 8 h of cell culture 48 h after PCK2 knockdown. C, glycolytic flux was assayed by measuring the radioactivity of the 3H2O released at the enolase step of glycolysis using [5-3H]glucose after 1 h of incubation. D, MTT proliferation assay of MCF7 cells treated with 2 different siRNA against PCK2 for 2 days in complete media. E, FACS analysis of annexin V in the cells treated as panels A–D demonstrates the absence of apoptosis under normal grown conditions. F, MTT proliferation assay of MCF7 cells expressing two different shRNA against PCK2. PEPCK-M protein levels were determined by Western blot. G and H, 48 h after PCK2 knockdown with siRNA, MCF7 were incubated for 24 h in serum-free media either lacking glutamine or supplemented with thapsigargin (Thaps; 500 and 2 μm), and protein expression was assayed by Western blot. Apoptosis is demonstrated by PARP (poly(ADP-ribose) polymerase), caspase-7, and co-chaperone p23 cleavage. Note PEPCK-M efficient knockdown. I, FACS analysis of annexin V/propidium iodide labeling in Scr (top) and PCK2 siRNA (bottom) of the experiment detailed in G and H (thapsigargin: 2 μm). J, quantitation of annexin V labeling depicted in I (three experiments with four replicates per condition). Data are shown as -fold change relative to the control condition and represent the means ± S.E. of ≥3 independent experiments; * p < 0.05; **, p < 0.01; ***, p < 0.001 relative to SCR; ###, p < 0.001 relative to the basal condition. One-way ANOVA, with a Newman-Keuls post-hoc test, or two-tailed Student's t test (C and E). Annexin V/7-aminoactinomycin D analysis of MCF7 cells expressing shRNA against PCK2 either untreated, glutamine-deprived (K and M) or treated with thapsigargin (2 μm) (L and N). The average result and flow cytometry plots are representative of two experiments.
We next investigated if PCK2 knockdown could decrease the survival capacity of cells subjected to stress stimuli. For this purpose, MCF7 cells treated with siRNA against PCK2 were incubated 24 h with media lacking glutamine or in the presence of thapsigargin (2 μm). Fig. 5, G and H, show that PCK2 silencing produced a significant increase in caspase 7 activation and subsequent cleavage of the caspase-dependent PARP and p23 as compared with control siRNA treatment. Flow cytometry quantitation of apoptotic cells (annexin V-FITC/propidium iodide staining) (Fig. 5I) showed that late apoptotic cells (annexin V-FITC positive/propidium iodide positive, upper right quadrant) in PCK2-silenced cells was statistically increased (Fig. 5J). Stably infected lentiviral hairpin expression PCK2 knockdown identically sensitized MCF7 cells to glutamine deprivation and thapsigargin induced death (Fig. 5, K–N).
DISCUSSION
Based on data available in a large number of public databases and on our own recollection, PEPCK-M expression was in marked contrast to the absence of its cytosolic counterpart, PEPCK-C. This suggests that the mitochondrial activity could provide a selective advantage to growth. Indeed, whereas PEPCK-C is mainly expressed in the liver, kidney, and adipose tissue, PEPCK-M is present in a variety of non-gluconeogenic tissues, including pancreas, brain, leukocytes, heart, or neurons.
We take the presence of PEPCK-M in most neoplastic cells and tumors as an indication of a functional role of the enzyme in the adaptations to cancer cell metabolism. Here we are testing this hypothesis by deciphering both PCK2 transcriptional regulation in response to various stimuli including nutrient deprivation and endoplasmic reticulum stress inducers and its functional role in the regulation of cancer cell homeostasis. Our results identify PEPCK-M as a component of the “amino acid response” and the “unfolded protein response” and suggest that PEPCK-M is important to maintain cell progression and survival, especially under stress conditions.
The study of cancer metabolism, a fundamental part of oncogenesis, should offer potential therapeutic targets (9, 10). Oncogenic transformation processes result in a strong demand for energy to support metabolism and tumor proliferation. Besides the Warburg effect, by which the tumor cells rely on aerobic glycolysis (11), a key element of metabolic adaptation in the tumor is deregulated amino acid requirements. Thus, glutamine is a vital element of tumor metabolism and plays an essential role in protein synthesis and the supply of ATP and metabolic intermediates for the synthesis of nucleotides and antioxidant capacity (12). Cell dependence on glutamine, branded as glutamine addiction by various authors (13–15), is reflected in restricted growth and cell death under conditions of limited or no glutamine. Solid tumors grow at a rate that exceeds the inflow of substrates through the vasculature and are frequently exposed to a microenvironment in which the supply of nutrients and oxygen can be reduced. Increased lactic acidification, reduced oxygen tension or decreased amino acid and glucose availability characteristics of this tumor microenvironment activate pro-survival signaling pathways, including the UPR (16–19), which exhibits a robust resemblance with the amino acid deprivation response (AAR). The inhibition of the UPR leads to inhibition of tumor progression (19–21). The ISR interconnects AAR and UPR as they share downstream effector mechanisms through the phosphorylation of eIF2α and the activation of ATF4 translation (22). These mechanisms allow stressed cells to survive by reprogramming gene expression and metabolic functions, enhancing amino acid uptake, and recycling macromolecules (23, 24).
Here, we demonstrate that ATF4-dependent PCK2 transcription in response to UPR and AAR induction depends on the eIF2α phosphorylation, mediated by GCN2, activated by uncharged tRNA during amino acid limitation, and PERK, activated by problems with protein processing in the endoplasmic reticulum, respectively (22, 25). Both kinases, however, could act as redundant kinases and contribute to PCK2 regulation in different circumstances according to previous data showing that eIF2α is indeed phosphorylated in cells lacking PERK or GCN2 upon UPR and AAR activation, respectively, although with delayed kinetics in comparison with wild type cells (26, 27). Besides PERK, UPR comprises other two integrated signaling cascades that depend on the ER transmembrane proteins IRE1 and ATF6 (28, 29).
Thapsigargin and tunicamycin produce concomitant activation of these three arms. Downstream effectors of IRE1 and ATF6 (full-length) are XBP1 and cleaved ATF6, respectively. XBP1 and ATF6 bind cis-acting elements in ER-stress-responsive genes, driving gene expression changes. XBP1 binds to the unfolded protein response element (UPRE) sequence, TGACTG(G/A), whereas ATF6 binds to the CCACG portion of the ER stress response element (ERSE) sequence, CCAAT-N9-CCACG. For example, PEPCK-C expression in the liver is down-regulated by ATF6 under ER stress (30, 31). ATF6 down-regulates PCK1 expression by competing for CREB binding at the C/EBP binding site with no similarity to PCK2 AARE. Taking into account that (a) PCK2 proximal promoter does not contain putative response elements for these transcription factors and (b) ATF4 silencing produces an almost complete ablation of PEPCK-M induction upon thapsigargin treatment, IRE1 and ATF6 arms do not seem to play a major role per se in PCK2 regulation under these conditions. Also, in addition to PERK and GCN2, other two eIF2α kinases have been identified in mammals: PKR (double-strand RNA activated protein kinase, activated by viral infection) (32) and HRI (heme-regulated inhibitor kinase) expressed predominantly in erythroid cells and regulated by heme (33). Because PKR and HRI role remain poorly defined, we cannot rule out that they may have a secondary role in PCK2 regulation in these or other models, acting as redundant eIF2α kinases.
We show that ATF4-dependent PCK2 gene activation is necessary for the progress of the ISR response, probably by enhancing the flexibility of mitochondrial metabolism, providing an alternative cataplerotic pathway. In the liver and pancreatic β-cell PEPCK-M has been shown to regulate TCA flux and enhance cataplerosis. Notably, liver-specific PEPCK-M overexpression in mice lacking hepatic PEPCK-C improved defects in glucose production and TCA cycle flux (4), showing a preference for lactate as a substrate. Also, PEPCK-M overexpression in the presence of PEPCK-C enhanced glucose production, suggesting a role for PEPCK-M in glucose homeostasis both through its intrinsic gluconeogenic potential and by complementing PEPCK-C, especially in species in which it is highly expressed, such as in humans. Reinforcing this concept, PEPCK-M silencing in rat liver severely reduced glucose production from lactate and amino acids (34). Another example of a role of PEPCK-M in cataplerosis is its involvement in the recycling of citric acid cycle anions in β-cells (5), linking PEPCK-M activity to the recycling of the GTP generated in the citric acid cycle (by succinyl-CoA synthase) to provide continuous TCA function and enhancing insulin secretion. Noteworthy, lactate can be also metabolized in the tumor in these conditions (35) in a reaction that feeds the TCA cycle. Because PEPCK-M cataplerotic activity is favored by lactate metabolism in hepatocytes, one can envision PEPCK-M to feed forward phosphoenolpyruvate into the triose-phosphate pool, substrates for serine and glycerol biosynthesis (36). This pathway could have important consequences for the tumor flexibility toward nutrient availability.
Two putative ATF4 binding ATF-CRE (AARE) sequences are present in the PCK2 proximal promoter region. Although, AARE-1 and AARE-2 are C/EBP-ATF composite sites, described as sequences that bind heterodimers of ATF and C/EBP bZIP transcription factors (37, 38), AARE-1 sequence appears to be the major contributor to the induction of the PCK2 gene during the AAR and UPR responses. Consistently, no evidence of ATF4 binding to the AARE-2 site was obtained by ChIP, although we cannot discard a role of this element in the activation of PCK2 transcription given that the promoter constructs retaining the AARE-2 did not exhibit complete ablation of UPR-mediated activation. Furthermore, reporter assays showed that a shorter construct with no AARE sites starting at nucleotide 288 5′ of exon 1 also demonstrated significant induction. This sequence contains three GC boxes potential SP1 and EGR-1 sites that may play an auxiliary role in PCK2 gene regulation. These could help maintain the level of basal transcription and allow maximal activation of the PCK2 gene following amino acid or glucose limitation as previously described for the ASNS gene for sites that show high similarity to the PCK2 promoter structure (24, 39). However, silencing of ATF4 in unstressed conditions brought down PCK2 mRNA content as well, suggesting a direct role of ATF4 in regulating the basal levels of PCK2 in these cells. The directive role of ATF4 on PCK2 mRNA levels prompted us to examine the expression correlation between PCK2 and ATF4 in tumor cell lines. Both genes are expressed in all cell lines on the NCI-60 data set, a cancer microarray project of 109 human cell lines from a variety of origins. A non-parametric Spearman correlation statistics study between ATF4 and PCK2 normalized RNA counts, allowed us to identify a statistically significant relationship (p < 0.0001; r = 0.55). In addition, all cell lines presented in Table 1 co-expressed ATF4 and PCK2 with significant correlation statistics (p < 0.0001; r = 0.72), pointing to a functional relationship between both genes in tumors
The most remarkable phenotype observed in unstimulated PCK2-silenced cells is a decreased glycolytic flux, as measured by incorporation of [3H]glucose in exponentially growing cells. This is consistent with lower glucose consumption and lactate production. Thus, PEPCK-M can represent a regulatory junction connecting the glycolytic and TCA cycle. However, because down-regulation by shRNA has an impact on cell proliferation (Fig. 5), it is not clear if the reduction of glycolytic flux is secondary to reduced growth or a primary event, more so because reduced PEPCK-M content by siRNA or stable shRNA knockdown in conditions that elicit AAR or UPR has an impact on cell survival. The reason for increased mortality is still elusive, although one might argue that the coordinated nutrient-sensing response requires a dynamic cataplerotic flux from glutamine carbons into phosphoenolpyruvate to enhance the synthesis of amino acids, such as serine/glycine, together with other metabolites required to cope with metabolic stress. In this view, serine synthesis also increases TCA cycle flux through the α-ketoglutarate generated in the glutamate-dependent transamination mediated by phosphoserine aminotransferase (40, 41). Thus, increasing cataplerosis and GTP recycling, both intrinsic PEPCK-M roles, could contribute to maximal rate of serine production.
Interestingly, only PEPCK isozymes can bypass the irreversible reaction catalyzed by pyruvate kinase and convert TCA cycle intermediates to glycolytic intermediates. Thus, most cancer cells must rely on PEPCK-M activity for re-channeling mitochondrial metabolites upstream of PEP into the glycolytic pathway (Fig. 6). It is not entirely clear why the mitochondrial isoform is preferred to its cytosolic counterpart, although PEPCK-M direct connection with the TCA and its reliance on phosphoenolpyruvate export might proof key to its preferential utilization. It is important to note that the mechanism for phosphoenolpyruvate transport across the inner mitochondrial membrane has not been identified yet. Recent studies also claim an important role for phosphoenolpyruvate as a phosphate donor for pyruvate kinase, PKM2, -dependent phosphorylation of target genes in the cytosol and the nucleus of neoplastic cells, (42–44). PKM2 has a paradoxical low kinase activity, effectively limiting the lower part of glycolysis, linking glucose and amino acids metabolism Interestingly, PKM2 promotes tumorigenesis (45), whereas its activators or PKM2 overexpression are suppressive (46). Furthermore, pharmacological activators of PKM2, which reduce carbon flow into the serine biosynthetic pathway, produce a profound dependence on the nonessential amino acid serine for continued cell proliferation. This causes a strong up-regulation of multiple enzymes involved in amino acid metabolic pathways, including PEPCK-M (47). In this sense, it is interesting to view this pathway in coordination with other enzymes, such as Tigar or PFKFB4, that control the flow to the lower glycolytic pathway to account for increased diversion of sugar carbons into the pentose-phosphate pathway (Fig. 6).
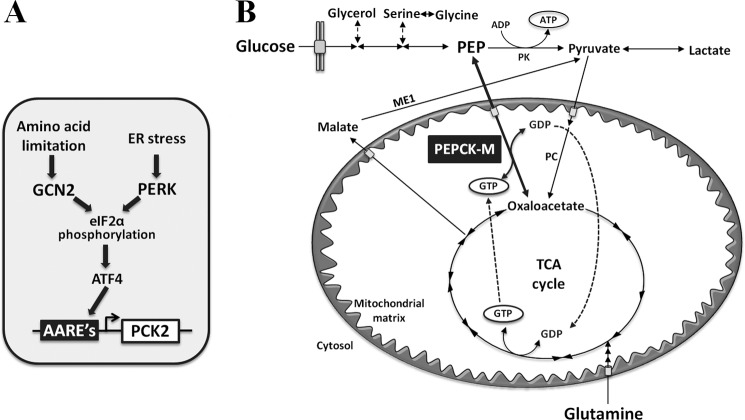
FIGURE 6.
A, schematic diagram of our working hypothesis. Both GCN2 and PERK activation by stress lead to the phosphorylation of eif2α, which in turn increases the translation of ATF4. ATF4 binds to the AARE sequences in the PCK2 gene promoter, enhancing its transcriptional activity. B, schematic diagram of PEPCK-M involvement in cataplerosis and TCA cycle metabolism. PK, pyruvate kinase; PC, pyruvate carboxylase; ME1, malic enzyme 1.
Glucose reduction activates ER stress and up-regulates ATF4, and as demonstrated in the present work, it correspondingly regulates PCK2 mRNA. Our data provide a mechanistic explanation for results by Leithner et al. (48) showing increased PCK2 expression and activity under low glucose in lung cancer cell lines. Phosphoenolpyruvate enrichment from [13C3]lactate was dependent on PEPCK-M presence under these conditions. Also, Park et al. (49) has recently proposed PEPCK-M as a potential predictor of chemoradiation response, as its expression correlates with sensitivity to 5-fluouracyl. Consistent with our data, both studies noted decreased proliferation after PCK2 knockdown.
Overall, we demonstrate that PEPCK-M is a pro-survival, metabolic stress target gene with potential significance to our understanding of cancer cell metabolism. Its precise contribution to stress adaptations needs further examination by metabolic flux analysis. This pathway provides a novel window for therapeutic intervention in cancer.
Acknowledgment
We thank the Research Support Services from the Biology Unit of Bellvitge (University of Barcelona) for technical assistance.
This work was supported by a grant from the Ministerio de Economia and y Competitividad and FEDER (BFU2012-37177 (to J. C. P.)).
- PEPCK
- phosphoenolpyruvate carboxykinase
- PEP
- phosphoenolpyruvate
- ISR
- integrated stress response
- AAR
- amino acid response
- AARE
- amino acid response element
- UPR
- unfolded protein response
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- GCN2
- general control nonderepressible 2
- PERK
- PKR-like endoplasmic reticulum-resident kinase
- ER
- endoplasmic reticulum
- ANOVA
- analysis of variance
- ATF
- activating transcription factor
- CREB
- cAMP-response element (CRE)-binding protein.
REFERENCES
- 1. Chang H. C., Lane M. D. (1966) The enzymatic carboxylation of phosphoenolpyruvate. II. Purification and properties of liver mitochondrial phosphoenolpyruvate carboxykinase. J. Biol. Chem. 241, 2413–2420 [PubMed] [Google Scholar]
- 2. Nordlie R. C., Lardy H. A. (1963) Mammalian liver phosphoneolpyruvate carboxykinase activities. J. Biol. Chem. 238, 2259–2263 [PubMed] [Google Scholar]
- 3. Hanson R. W., Garber A. J. (1972) Phosphoenolpyruvate carboxykinase: I. Its role in gluconeogenesis. Am. J. Clin. Nutr. 25, 1010–1021 [DOI] [PubMed] [Google Scholar]
- 4. Méndez-Lucas A., Duarte J. A., Sunny N. E., Satapati S., He T., Fu X., Bermúdez J., Burgess S. C., Perales J. C. (2013) PEPCK-M expression in mouse liver potentiates, not replaces, PEPCK-C mediated gluconeogenesis. J. Hepatol. 59, 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stark R., Pasquel F., Turcu A., Pongratz R. L., Roden M., Cline G. W., Shulman G. I., Kibbey R. G. (2009) Phosphoenolpyruvate cycling via mitochondrial phosphoenolpyruvate carboxykinase links anaplerosis and mitochondrial GTP with insulin secretion. J. Biol. Chem. 284, 26578–26590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Novellasdemunt L., Obach M., Millán-Ariño L., Manzano A., Ventura F., Rosa J. L., Jordan A., Navarro-Sabate A., Bartrons R. (2012) Progestins activate 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) in breast cancer cells. Biochem. J. 442, 345–356 [DOI] [PubMed] [Google Scholar]
- 7. Novellasdemunt L., Tato I., Navarro-Sabate A., Ruiz-Meana M., Méndez-Lucas A., Perales J. C., Garcia-Dorado D., Ventura F., Bartrons R., Rosa J. L. (2013) Akt-dependent activation of the heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB2) isoenzyme by amino acids. J. Biol. Chem. 288, 10640–10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Sousa-Coelho A. L., Marrero P. F., Haro D. (2012) Activating transcription factor 4-dependent induction of FGF21 during amino acid deprivation. Biochem. J. 443, 165–171 [DOI] [PubMed] [Google Scholar]
- 9. Hsu P. P., Sabatini D. M. (2008) Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 [DOI] [PubMed] [Google Scholar]
- 10. Tennant D. A., Durán R. V., Gottlieb E. (2010) Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10, 267–277 [DOI] [PubMed] [Google Scholar]
- 11. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 12. DeBerardinis R. J., Cheng T. (2010) Q's next: the diverse functions of glutamine in metabolism, cell biology, and cancer. Oncogene 29, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., Thompson C. B. (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wise D. R., Thompson C. B. (2010) Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang C., Sudderth J., Dang T., Bachoo R. M., Bachoo R. G., McDonald J. G., DeBerardinis R. J. (2009) Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 69, 7986–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang G., Yang Z. Q., Zhang K. (2010) Endoplasmic reticulum stress response in cancer: molecular mechanism and therapeutic potential. Am. J. Transl. Res. 2, 65–74 [PMC free article] [PubMed] [Google Scholar]
- 17. Tang X., Lucas J. E., Chen J. L., LaMonte G., Wu J., Wang M. C., Koumenis C., Chi J. T. (2012) Functional interaction between responses to lactic acidosis and hypoxia regulates genomic transcriptional outputs. Cancer Res. 72, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hart L. S., Cunningham J. T., Datta T., Dey S., Tameire F., Lehman S. L., Qiu B., Zhang H., Cerniglia G., Bi M., Li Y., Gao Y., Liu H., Li C., Maity A., Thomas-Tikhonenko A., Perl A. E., Koong A., Fuchs S. Y., Diehl J. A., Mills I. G., Ruggero D., Koumenis C. (2012) ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J. Clin. Invest. 122, 4621–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koumenis C., Naczki C., Koritzinsky M., Rastani S., Diehl A., Sonenberg N., Koromilas A., Wouters B. G. (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 22, 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bobrovnikova-Marjon E., Grigoriadou C., Pytel D., Zhang F., Ye J., Koumenis C., Cavener D., Diehl J. A. (2010) PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene 29, 3881–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bi M., Naczki C., Koritzinsky M., Fels D., Blais J., Hu N., Harding H., Novoa I., Varia M., Raleigh J., Scheuner D., Kaufman R. J., Bell J., Ron D., Wouters B. G., Koumenis C. (2005) ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 24, 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 23. B'chir W., Maurin A. C., Carraro V., Averous J., Jousse C., Muranishi Y., Parry L., Stepien G., Fafournoux P., Bruhat A. (2013) The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 41, 7683–7699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kilberg M. S., Pan Y. X., Chen H., Leung-Pineda V. (2005) Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 25, 59–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sood R., Porter A. C., Olsen D. A., Cavener D. R., Wek R. C. (2000) A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics 154, 787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamanaka R. B., Bennett B. S., Cullinan S. B., Diehl J. A. (2005) PERK and GCN2 contribute to eIF2α phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol. Biol. Cell 16, 5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. (2004) Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 29. Schröder M., Kaufman R. J. (2005) The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 30. Seo H. Y., Kim M. K., Min A. K., Kim H. S., Ryu S. Y., Kim N. K., Lee K. M., Kim H. J., Choi H. S., Lee K. U., Park K. G., Lee I. K. (2010) Endoplasmic reticulum stress-induced activation of activating transcription factor 6 decreases cAMP-stimulated hepatic gluconeogenesis via inhibition of CREB. Endocrinology 151, 561–568 [DOI] [PubMed] [Google Scholar]
- 31. Wang Y., Vera L., Fischer W. H., Montminy M. (2009) The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature 460, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levin D., London I. M. (1978) Regulation of protein synthesis: activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc. Natl. Acad. Sci. U.S.A. 75, 1121–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crosby J. S., Chefalo P. J., Yeh I., Ying S., London I. M., Leboulch P., Chen J. J. (2000) Regulation of hemoglobin synthesis and proliferation of differentiating erythroid cells by heme-regulated eIF-2α kinase. Blood 96, 3241–3248 [PubMed] [Google Scholar]
- 34. Stark R., Guebre-Egziabher F., Zhao X., Feriod C., Dong J., Alves T. C., Ioja S., Pongratz R. L., Bhanot S., Roden M., Cline G. W., Shulman G. I., Kibbey R. G. (2014) A role for mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) in the regulation of hepatic gluconeogenesis. J. Biol. Chem. 289, 7257–7263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Semenza G. L. (2008) Tumor metabolism: cancer cells give and take lactate. J. Clin. Invest. 118, 3835–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kalhan S. C., Hanson R. W. (2012) Resurgence of serine: an often neglected but indispensable amino Acid. J. Biol. Chem. 287, 19786–19791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen H., Pan Y. X., Dudenhausen E. E., Kilberg M. S. (2004) Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J. Biol. Chem. 279, 50829–50839 [DOI] [PubMed] [Google Scholar]
- 38. Barbosa-Tessmann I. P., Chen C., Zhong C., Siu F., Schuster S. M., Nick H. S., Kilberg M. S. (2000) Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 275, 26976–26985 [DOI] [PubMed] [Google Scholar]
- 39. Leung-Pineda V., Kilberg M. S. (2002) Role of Sp1 and Sp3 in the nutrient-regulated expression of the human asparagine synthetase gene. J. Biol. Chem. 277, 16585–16591 [DOI] [PubMed] [Google Scholar]
- 40. Possemato R., Marks K. M., Shaul Y. D., Pacold M. E., Kim D., Birsoy K., Sethumadhavan S., Woo H. K., Jang H. G., Jha A. K., Chen W. W., Barrett F. G., Stransky N., Tsun Z. Y., Cowley G. S., Barretina J., Kalaany N. Y., Hsu P. P., Ottina K., Chan A. M., Yuan B., Garraway L. A., Root D. E., Mino-Kenudson M., Brachtel E. F., Driggers E. M., Sabatini D. M. (2011) Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Amelio I., Markert E. K., Rufini A., Antonov A. V., Sayan B. S., Tucci P., Agostini M., Mineo T. C., Levine A. J., Melino G. (2013) p73 regulates serine biosynthesis in cancer. Oncogene 10.1038/onc.2013.456 [DOI] [PubMed] [Google Scholar]
- 42. Vander Heiden M. G., Locasale J. W., Swanson K. D., Sharfi H., Heffron G. J., Amador-Noguez D., Christofk H. R., Wagner G., Rabinowitz J. D., Asara J. M., Cantley L. C. (2010) Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329, 1492–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao X., Wang H., Yang J. J., Liu X., Liu Z. R. (2012) Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 45, 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang Y., Li X., Yang W., Hawke D. H., Zheng Y., Xia Y., Aldape K., Wei C., Guo F., Chen Y., Lu Z. (2014) PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol. Cell 53, 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Alfred Yung W. K., Lu Z. (2012) PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anastasiou D., Yu Y., Israelsen W. J., Jiang J. K., Boxer M. B., Hong B. S., Tempel W., Dimov S., Shen M., Jha A., Yang H., Mattaini K. R., Metallo C. M., Fiske B. P., Courtney K. D., Malstrom S., Khan T. M., Kung C., Skoumbourdis A. P., Veith H., Southall N., Walsh M. J., Brimacombe K. R., Leister W., Lunt S. Y., Johnson Z. R., Yen K. E., Kunii K., Davidson S. M., Christofk H. R., Austin C. P., Inglese J., Harris M. H., Asara J. M., Stephanopoulos G., Salituro F. G., Jin S., Dang L., Auld D. S., Park H. W., Cantley L. C., Thomas C. J., Vander Heiden M. G. (2012) Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat. Chem. Biol. 8, 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kung C., Hixon J., Choe S., Marks K., Gross S., Murphy E., DeLaBarre B., Cianchetta G., Sethumadhavan S., Wang X., Yan S., Gao Y., Fang C., Wei W., Jiang F., Wang S., Qian K., Saunders J., Driggers E., Woo H. K., Kunii K., Murray S., Yang H., Yen K., Liu W., Cantley L. C., Vander Heiden M. G., Su S. M., Jin S., Salituro F. G., Dang L. (2012) Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem. Biol. 19, 1187–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leithner K., Hrzenjak A., Trotzmuller M., Moustafa T., Kofeler H. C., Wohlkoenig C., Stacher E., Lindenmann J., Harris A. L., Olschewski A., Olschewski H. (2014) PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene 10.1038/onc.2014.47 [DOI] [PubMed] [Google Scholar]
- 49. Park J. W., Kim S. C., Kim W. K., Hong J. P., Kim K. H., Yeo H. Y., Lee J. Y., Kim M. S., Kim J. H., Yang S. Y., Kim D. Y., Oh J. H., Cho J. Y., Yoo B. C. (2014) Expression of phosphoenolpyruvate carboxykinase linked to chemoradiation susceptibility of human colon cancer cells. BMC Cancer 14, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]