Background: Gap1 is a yeast amino acid permease ubiquitylated in response to internal amino acids.
Results: Gap1 is also ubiquitylated under stress conditions, via a different mechanism.
Conclusion: Different signals trigger permease ubiquitylation and degradation via different mechanisms.
Significance: Understanding the signals and mechanisms promoting yeast permease ubiquitylation sheds light on similar pathways controlling membrane transport proteins in more complex organisms.
Keywords: Amino Acid Transport, Arrestin, Endocytosis, TOR Complex (TORC), Stress, Ubiquitin, Yeast
Abstract
Gap1, the yeast general amino acid permease, is a convenient model for studying how the intracellular traffic of membrane transporters is regulated. Present at the plasma membrane under poor nitrogen supply conditions, it undergoes ubiquitylation, endocytosis, and degradation upon activation of the TORC1 kinase complex in response to an increase in internal amino acids. This down-regulation is stimulated by TORC1-dependent phosphoinhibition of the Npr1 kinase, resulting in activation by dephosphorylation of the arrestin-like Bul1 and Bul2 adaptors recruiting the Rsp5 ubiquitin ligase to Gap1. We report here that Gap1 is also down-regulated when cells are treated with the TORC1 inhibitor rapamycin or subjected to various stresses and that a lack of the Tco89 subunit of TORC1 causes constitutive Gap1 down-regulation. Both the Bul1 and Bul2 and the Aly1 and Aly2 arrestin-like adaptors of Rsp5 promote this down-regulation without undergoing dephosphorylation. Furthermore, they act via the C-terminal regions of Gap1 not involved in ubiquitylation in response to internal amino acids, whereas a Gap1 mutant altered in the N-terminal tail and resistant to ubiquitylation by internal amino acids is efficiently down-regulated under stress via the Bul and Aly adaptors. Although the Bul proteins mediate Gap1 ubiquitylation of two possible lysines, Lys-9 and Lys-16, the Aly proteins promote ubiquitylation of the Lys-16 residue only. This stress-induced pathway of Gap1 down-regulation targets other permeases as well, and it likely allows cells facing adverse conditions to retrieve amino acids from permease degradation.
Introduction
About 125 plasma membrane transport proteins have been inventoried in yeast (1). Many of them are subject to tight regulation via mechanisms acting essentially at three levels as follows: gene transcription, intracellular trafficking, and/or intrinsic activity. Ubiquitin (Ub)3 is well established as a central player in transporter regulation at the membrane trafficking level. Covalent attachment of Ub to cell surface transporters typically promotes their sorting to endocytic vesicles, transit through endosomes, and delivery to the lumen of the vacuole (the lysosome of yeast) where they are degraded (2, 3). This ubiquitylation depends on Rsp5, a Ub ligase of the Nedd4 HECT family (4, 5), and human proteins of this family are also known to mediate ubiquitylation and down-regulation of various transport proteins and receptors (6). Further study of specific yeast permeases has shown that they are in fact modified with a short Lys-63-linked Ub chain (7, 8), the type of chain preferentially formed by Rsp5 (9). This Lys-63-Ub chain is particularly important for proper sorting of transporters reaching the limiting membrane of endosomes in vesicles budding into the lumen of this compartment, which is crucial for their final delivery to the vacuolar lumen (10, 11).
The Rsp5 Ub ligase possesses three WW domains enabling it to interact with PPX(Y/F) sequences exposed on its substrate proteins (6). Yet most yeast transporters under Rsp5 control possess no such motif. Actually, Rsp5 binds via its WW domains to PPX(Y/F)-exposing accessory proteins acting as adaptors for transporter ubiquitylation (2, 12). About 10 of these Rsp5 adaptors, named Art (arrestin-related trafficking) proteins, harbor an arrestin motif (13, 14) and resemble a subclass of mammalian arrestin-like proteins, named arrestin-domain-containing proteins (ARRDC) or α-arrestins, also containing PPX(Y/F) motifs (12). The ubiquitylation and down-regulation of yeast plasma membrane transporters can occur in response to diverse signals, ranging from substrate excess, more global changes in nutrient supply conditions, and a variety of stresses (3). Ubiquitylation of a specific permease sometimes involves different adaptors according to the signal inducing its down-regulation (13, 15). Recent work has shown that the ubiquitylation of specific permeases can be triggered by direct regulation of arrestin-like adaptors. For instance, glucose-induced ubiquitylation of the Jen1 lactate permease requires the Rod1/Art4 adaptor, and this protein is activated in response to glucose by dephosphorylation and ubiquitylation (16). Similarly, the Npr1 kinase inhibits, by phosphorylation, the Art1 adaptor, thereby preventing down-regulation of the Can1 arginine permease, and this negative control is relieved after cycloheximide addition (17).
The role of Ub and arrestin-like Rsp5 adaptors in transporter down-regulation is also well illustrated in the case of Gap1, the general amino acid permease (2). This high affinity, broad spectrum amino acid permease (18) is highly stable and active at the plasma membrane of cells grown on a medium with all required nutrients and only a poor nitrogen source. The cells therefore grow, but at a limited rate, and the role of Gap1 is to scavenge any amino acids that might be present for use as a source of nitrogen or for protein synthesis. The high cell surface stability of Gap1 under poor nitrogen-supply conditions is due to the Npr1 kinase (19), which inhibits by phosphorylation the redundant Bul1 and Bul2 arrestin-like adaptors potentially able to recruit the Rsp5 Ub ligase to Gap1 (20, 21). When phosphorylated, the Bul proteins are not active, because they are bound to 14-3-3 proteins. This inhibition is relieved when Npr1 is itself phosphoinhibited by the TORC1 kinase complex in response to an increase in the amino acid internal pool, e.g. after ammonium uptake and conversion to glutamate (Fig. 1) (21). Once Npr1 is inhibited, the Bul adaptors are dephosphorylated in a manner dependent on Sit4, a PP2A-type phosphatase. They dissociate from the 14-3-3 proteins and target Rsp5 to Gap1, which is ubiquitylated (21). This modification consists of a short Lys-63-linked Ub chain linked to one of two possible Ub acceptor lysines, Lys-9 or Lys-16, present in the Gap1 N-terminal tail (8, 10, 20).
FIGURE 1.
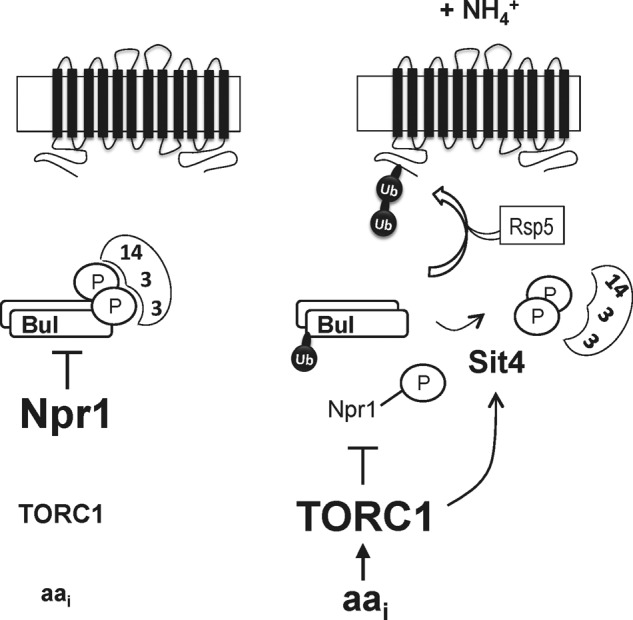
Model for ammonium-induced ubiquitylation of the Gap1 permease. Left, on a poor nitrogen source, the Npr1 kinase is active and phosphorylates the Bul adaptors. These bind to 14-3-3 proteins and are thus inhibited. Hence, Gap1 is not ubiquitylated. Right, upon uptake of ammonium (or amino acids) into the cells, the internal concentration of amino acids increases. This leads to stimulation of the TORC1 kinase complex. Active TORC1 inhibits by phosphorylation the Npr1 kinase and somehow stimulates the Sit4 phosphatase. The Bul adaptors thus undergo dephosphorylation, dissociate from the 14-3-3 proteins, and recruit the Rsp5 Ub ligase to the Gap1 permease, which is ubiquitylated. Npr1 inactivation also induces Rsp5-dependent monoubiquitylation of the Bul proteins, but the role of this modification remains unknown.
In this study, we show that both TORC1 inhibition and various stresses can trigger Gap1 ubiquitylation and down-regulation, via a mechanism differing from that triggered by TORC1 stimulation by amino acids.
EXPERIMENTAL PROCEDURES
Strains and Media
All Saccharomyces cerevisiae strains used in this study (Table 1) derive from the Σ1278b wild type (22). Cells were grown at 29 °C in minimal buffered medium (pH 6.1) (23). The main carbon source was glucose (3%) or galactose (3%). In Gap1, Can1, and Lyp1 endocytosis experiments, the initial nitrogen source of the growth medium was proline (10 mm) or urea (10 mm), and in Fur4 experiments, it was ammonium (20 mm) in the form of (NH4)2SO4. Permease endocytosis was induced by addition of ammonium (20 or 50 mm), rapamycin (200 ng/ml), ethanol (10%), or H2O2 (0.88 mm) for 2 or 3 h or by transferring cells to 37 °C for 1 h. Strains MYC003, MYC004, and MA056 were isolated by insertion of a TCO89- or ALY1-deleting hphMX hygromycin B resistance gene amplified by PCR from plasmid pAG32 used as template. Strains MYC009, MA059, and MA062 were isolated by insertion of a TCO89- or ALY2-deleting kanMX2 geneticin resistance gene amplified by PCR from plasmid pUG06 used as template. Strains JA986 and JA997 were isolated by insertion of a DNA fragment amplified by PCR from plasmids pFA6a-6GLY-FLAG-hphMX and pUG06-HA-kanMX2 used, respectively, as templates. All oligonucleotides used to isolate inserted PCR DNA fragments are available upon request.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| 23344c | ura3 | Lab collection |
| EK008 | gap1Δ ura3 | Lab collection |
| CJ005 | gap1Δ rsp5 (npi1) ura3 | Lab collection |
| 27038a | rsp5 (npi1) ura3 | 4 |
| 41453c | rsp5 (npi1) tco89Δ ura3 | This study |
| FA198 | tco89Δ ura3 | This study |
| JA479 | gap1Δ bul1Δ bul2Δ ura3 | 10 |
| MA056 | gap1Δ aly1Δ ura3 | This study |
| MA059 | gap1Δ aly2Δ ura3 | This study |
| 35101a | gap1Δ aly1Δ aly2Δ ura3 | This study |
| MA062 | gap1Δ bul1Δ bul2Δ aly1Δ aly2Δ ura3 | This study |
| MYC003 | gap1Δ tco89Δ ura3 | This study |
| MYC004 | gap1Δ tco89Δ bul1Δ bul2Δ ura3 | This study |
| MYC009 | gap1Δ tco89Δ aly1Δ aly2Δ ura3 | This study |
| MA025 | gap1Δ BUL1-FLAG bul2Δ ura3 | 21 |
| MA032 | gap1Δ BUL2-HA ura3 | 21 |
| JA986 | ALY1-FLAG ura3 | This study |
| JA997 | ALY2-HA ura3 | This study |
| JA983 | gap1Δ art1Δ ura3 | This study |
| 35246a | gap1Δ tco89Δ art1Δ ura3 | This study |
Plasmids
The plasmids used in this work are listed in Table 2. The additional Gap1 mutant plasmids used here have been described previously (24). Plasmid pMYC016 (Gap1-134–135) was constructed by recombination in yeast between the linearized pMA33 (Gap1-135) plasmid and a DNA fragment, including the VDLD → AAAA substitution mutations (Gap1-134). This DNA fragment was obtained by PCR using pMA33 (Gap1-135) as a template and appropriate oligonucleotides for insertion of the above-mentioned 134 mutations.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Ref. or source |
|---|---|---|
| pFL38 | CEN-ARS (URA3) | 46 |
| pJOD10 | p416 GAL1-GAP1-GFP (URA3) | 19 |
| pCJ038 | p416 GAL1-GAP1K9R,K16R-GFP (URA3) | Lab collection |
| pCJ004 | p416 GAL1-GAP1 (URA3) | Lab collection |
| pCJ034 | p416 GAL1-GAP1K9R,K16R (URA3) | Lab collection |
| pEL003 | YCp GAP1-GFP (URA3) | 45 |
| pEL005 | YCp GAP1K9R,K16R-GFP (URA3) | 45 |
| pEL018 | YCp GAP1K9R-GFP (URA3) | 10 |
| pEL021 | YCp GAP1K16R-GFP (URA3) | 10 |
| pRS426-GST | pRS426-pADH-GST (URA3) | 47 |
| pRS426-GST-BMH2 | pRS426-pADH-GST-BMH2 (URA3) | 47 |
| pMA74 (Gap1-112) | p416 GAL1-GAP1ITIP33AAAA-GFP (URA3) | 24 |
| pMA47 (Gap1-106) | p416 GAL1-GAP1EKNN11AAAA-GFP (URA3) | 24 |
| pMA51 (Gap1-108) | p416 GAL1-GAP1KHNG19AAAA-GFP (URA3) | 24 |
| pMA03 (Gap1-103) | p416 GAL1-GAP1T567A-GFP (URA3) | 24 |
| pMA28 (Gap1-132) | p416 GAL1-GAP1EIAE583AAAA-GFP (URA3) | 24 |
| pMA31 (Gap1-134) | p416 GAL1-GAP1VDLD575AAAA-GFP (URA3) | 24 |
| pMA33 (Gap1-135) | p416 GAL1-GAP1GRRE571AAAA-GFP (URA3) | 24 |
| pMA35 (Gap1-136) | p416 GAL1-GAP1AEKM563AAAA-GFP (URA3) | 24 |
| pNG29 (Gap1-157) | p416 GAL1-GAP1AEQR408AAAA-GFP (URA3) | 24 |
| pNG38 (Gap1-162) | p416 GAL1-GAP1ALAA480AAAA-GFP (URA3) | 24 |
| pNG41 (Gap1-164) | p416 GAL1-GAP1LDEL488AAAA-GFP (URA3) | 24 |
| pNG42 (Gap1-165) | p416 GAL1-GAP1SFK491AAA-GFP (URA3) | 24 |
| pNG53 (Gap1-170) | p416 GAL1-GAP1LVG426AAA-GFP (URA3) | 24 |
| pMYC016 (Gap1-134–135) | p416 GAL1-GAP1GRREVDLD571AAAAAAAA-GFP (URA3) | This study |
| pMA185 | p416 GAL1-CAN1-GFP (URA3) | Lab collection |
| pNAM001 | YCp LYP1-GFP (URA3) | 48 |
| pFL38-gF-GFP | pFL38 GAL1-FUR4-GFP (URA3) | Lab collection |
Yeast Cell Extracts, Immunoblotting, and GST Pulldown
For Western blot analysis, crude cell extracts were prepared and immunoblotted as described previously (4). After transfer to a nitrocellulose membrane (Schleicher & Schüell), mouse anti-HA (12CA5; Roche Diagnostics), anti-FLAG (F1804; Sigma), anti-GFP (Roche Diagnostics), anti-BMH (a kind gift of Sandra Lemmon), or anti-3-phosphoglycerate kinase of yeast (Invitrogen) antibodies were used at 1:10,000, 1:2,000, and 1:5,000 dilution, respectively. Primary antibodies were detected with horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G secondary antibody (GE Healthcare). For the glutathione S-transferase (GST) pulldown experiments, exponentially growing cells were harvested before and 30 min after ammonium (50 mm) addition or 60 min after rapamycin (200 ng/ml) addition. GST pulldown was carried out as described previously (21). Proteins were analyzed by immunoblotting with anti-GST (Invitrogen) or anti-FLAG antibodies.
Alkaline Phosphatase Treatment of Protein Extracts
Crude cell extracts were prepared as described above, and proteins were suspended in SDS buffer (4% SDS, 0.1 m Tris-HCl (pH 6.8), 20% glycerol, 2% 2-mercaptoethanol). The dephosphorylation buffer was 1 m Tris (pH 8.0) and phenylmethylsulfonyl fluoride (PMSF) (1 mm). Then alkaline phosphatase (Roche Diagnostics) was added or not, and the mixture was incubated for 2 h at 37 °C. Proteins were then precipitated with 10% TCA, resuspended in sample buffer, and analyzed by immunoblotting.
Fluorescence Microscopy
Subcellular localization of the Gap1-GFP, Can1-GFP, or Lyp1-GFP protein was analyzed in cells growing exponentially in liquid glucose/proline, galactose/proline, or galactose/urea medium and that of Fur4-GFP was analyzed in cells growing in galactose/ammonium medium. When galactose was used as a carbon source, glucose was added (final concentration, 3%) for 2 h before visualizing cells or for 30 min before addition of rapamycin to arrest Gap1-GFP, Can1-GFP, Lyp1-GFP, or Fur4-GFP neosynthesis. Labeling of the vacuolar membrane with FM4-64 was performed as described previously (25). Cells were deposited on a thin layer of 1% agarose and viewed at room temperature with a fluorescence microscope (Eclipse 80i; Nikon) equipped with a ×60 differential interference contrast NA 1.40 Plan-ApoVc objective (Nikon) and appropriate fluorescence light filter sets. Images were captured with a digital camera (DS-Qi1MC-Nikon) and processed with Photoshop CS (Adobe Systems). For clarity, the images are shown inverted.
Permease Assays
Gap1 activity was determined by measuring the initial uptake rate of 14C-labeled citrulline (75 μm) (22) in exponentially growing cells during the state of balanced growth. Cumulative counts were measured 30, 60, and 90 s after addition of the radioactive substrate and corrected for protein concentration.
RESULTS
Rapamycin Triggers Ubiquitylation and Down-regulation of the Gap1 Permease
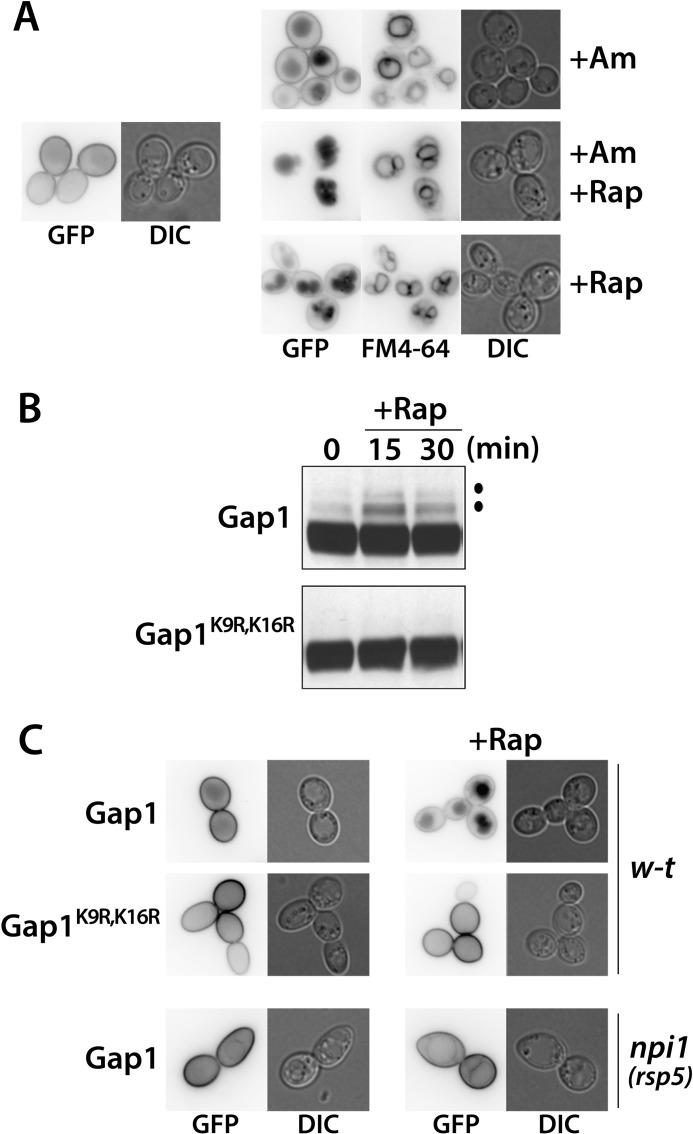
In cells grown on a poor nitrogen source such as proline, the Gap1 permease is present and active at the plasma membrane. Addition of a high concentration of Am results in rapid ubiquitylation and endocytosis of Gap1, followed by its degradation in the vacuole (2, 26). A prediction of the current model for the mechanism of this down-regulation (see Introduction and Fig. 1) is that simultaneous addition of Am and the TORC1 inhibitor Rap should prevent Gap1 ubiquitylation, because the Npr1 kinase is supposed to remain active and thus able to inhibit the Bul proteins by phosphorylation. Yet, when we added Am and Rap to the cells, Gap1-GFP was rapidly targeted to the vacuole, as confirmed by colabeling with the FM4-64 dye (Fig. 2A). Furthermore, the presence of Rap increased the efficiency of Gap1 down-regulation, because the amount of Gap1-GFP subsisting at the cell surface after Am addition dropped to an undetectable level when both Am and Rap were provided to the cells (Fig. 2A). We thus tested the influence of Rap alone. Interestingly, Gap1 was also efficiently down-regulated after Rap addition to cells grown on Pro medium (Fig. 2A). In support of the view that this down-regulation was due to Gap1 ubiquitylation, addition of Rap resulted in the rapid appearance, on immunoblots, of two upper bands above the main Gap1 signal, and these upper bands were not detected with the Gap1K9R,K16R form, in which the two Ub-acceptor lysines are mutated (Fig. 2B). Furthermore, the Gap1K9R,K16R mutant remained stable at the cell surface after Rap addition (Fig. 2C). Rap-induced endocytosis of Gap1 was also impaired in the npi1(rsp5) mutant (Fig. 2C), in which the cellular level of the Rsp5 Ub ligase is strongly reduced (26). This indicates that Rap-induced down-regulation requires Rsp5, as expected. These results thus show that in cells growing on a poor nitrogen source such as proline, TORC1 exerts an activity essential to maintaining Gap1 stable at the plasma membrane by preventing its Rsp5-dependent ubiquitylation. This contrasts with the ability of TORC1 to promote Gap1 ubiquitylation when internal amino acids are abundant.
FIGURE 2.
Rapamycin triggers ubiquitylation and endocytosis of Gap1. A, strain EK008 (gap1Δ ura3) transformed with the pJOD10 (YCpGAL-GAP1-GFP) plasmid was grown on galactose/proline medium. Glucose was added for 2 h to repress Gap1 synthesis, and when indicated, Am (20 mm) and/or Rap (200 ng/ml) was/were added for 2 h. Gap1-GFP localization was examined by fluorescence microscopy. Cells were labeled with FM4-64 to stain the vacuolar membrane. B, strain EK008 (gap1Δ ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP) or pCJ038 (YCpGAL-GAP1K9R,K16R-GFP) was grown on galactose/proline medium. Cells were collected before and 15 and 30 min after Rap (200 ng/ml) addition. Crude cell extracts were immunoblotted with anti-GFP antibodies. C, strains EK008 (gap1Δ ura3) and CJ005 (gap1Δ rsp5(npi1) ura3) were transformed with pJOD10 (YCpGAL-GAP1-GFP) or pCJ038 (YCpGAL-GAP1K9R,K16R-GFP). Growth conditions were as in A. Gap1-GFP localization was examined by fluorescence microscopy. DIC, differential interference contrast.
Rapamycin-induced Down-regulation of Gap1 Is Mediated by the Bul1, Bul2, Aly1, and Aly2 Arrestin-like Adaptors
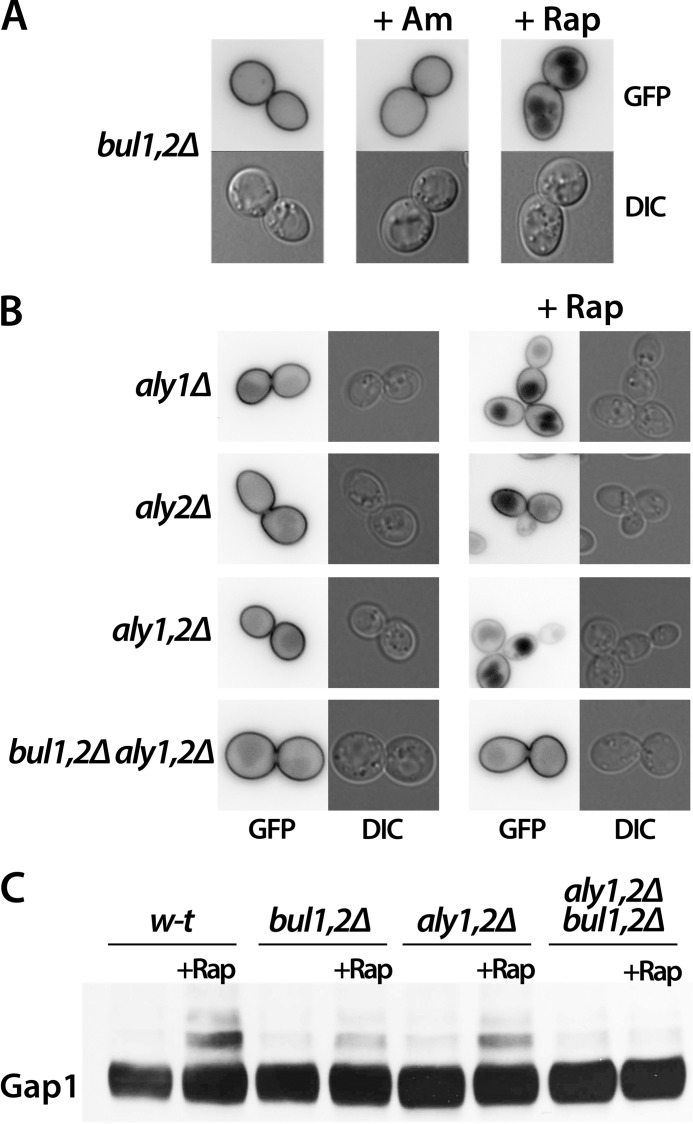
Ubiquitylation of Gap1 induced by Am addition or genetic inactivation of the Npr1 kinase requires the redundant Bul1 and Bul2 arrestin-like adaptors (20, 21). We thus tested whether these Bul proteins are also involved in Rap-induced Gap1 down-regulation. In the bul1Δ bul2Δ mutant, as expected, Gap1 was stable at the plasma membrane after Am addition. After Rap addition, however, the permease of this mutant was internalized and targeted to the vacuole (Fig. 3A). Hence, the Bul proteins are not essential to Rap-induced down-regulation of Gap1, suggesting that other adaptors of Rsp5 are involved. As Rap-induced endocytosis was nevertheless significantly slower in the bul1Δ bul2Δ mutant than in wild-type cells (data not shown), we hypothesized that the Bul proteins might contribute to Rap-induced ubiquitylation and down-regulation of Gap1 along with additional adaptors of Rsp5. About 10 additional arrestin-like adaptors have been inventoried in yeast, and most of them are reported to contribute to Ub-dependent down-regulation of at least some cargoes (12, 27). We first examined bul1Δ bul2Δ strains with additional deletion of the ART1, ART2, or ART8 gene, but in none of them was Gap1 protected against Rap-induced down-regulation (data not shown). Because of a previous report that the Aly1/Art3 and Aly2/Art6 proteins control Gap1 traffic and that the aly1Δ aly2Δ mutant is hypersensitive to Rap-induced growth inhibition (28), we tested the role of these Aly proteins in Rap-induced Gap1 endocytosis. In the aly1Δ and aly2Δ single mutants and the double aly1Δ aly2Δ mutant, Gap1 was normally down-regulated by Rap (Fig. 3B), but in the bul1Δ bul2Δ aly1Δ aly2Δ quadruple mutant, remarkably, Gap1 was resistant to Rap-induced down-regulation (Fig. 3B). Accordingly, Rap-induced ubiquitylation of Gap1 was deficient in this strain, whereas residual ubiquitylation was detectable in the bul1Δ bul2Δ and aly1Δ aly2Δ mutants, in keeping with the efficient endocytosis of Gap1-GFP in these strains (Fig. 3C). In conclusion, the Bul and Aly arrestin-like adaptors contribute concomitantly to Rap-induced ubiquitylation and endocytosis of Gap1.
FIGURE 3.
Bul1, Bul2, Aly1, and Aly2 arrestin-like adaptors contribute to rapamycin-induced ubiquitylation and down-regulation of Gap1. A, strain JA493 (gap1Δ bul1Δ bul2Δ ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP) was grown on galactose/proline medium. Glucose was added for 2 h to repress Gap1 synthesis, and when indicated, Am (20 mm) or Rap (200 ng/ml) was added for 2 h. Gap1-GFP localization was examined by fluorescence microscopy. B, strains MA056 (gap1Δ aly1Δ ura3), MA059 (gap1Δ aly2Δ ura3), 35101a (gap1Δ aly1Δ aly2Δ ura3), and MA062 (gap1Δ bul1Δ bul2Δ aly1Δ aly2Δ ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP) were grown on galactose/proline medium. Glucose was added for 2 h to repress Gap1 synthesis, and when indicated, Rap (200 ng/ml) was added for 2 h. Gap1-GFP localization was examined by fluorescence microscopy. C, strains EK008 (gap1Δ ura3), JA493 (gap1Δ bul1Δ bul2Δ ura3), 35101a (gap1Δ aly1Δ aly2Δ ura3), and MA062 (gap1Δ bul1Δ bul2Δ aly1Δ aly2Δ ura3) transformed with the pJOD10 (YCpGAL-GAP1-GFP) plasmid were grown on galactose/proline medium. Cells were collected before and 10 min after Rap (200 ng/ml) addition. Crude cell extracts were immunoblotted with anti-GFP antibodies. DIC, differential interference contrast.
Bul and Aly Adaptors Remain Phosphorylated and Bul2 Still Binds to 14-3-3 Proteins after Rapamycin Addition
In cells grown on Pro medium (conditions under which Gap1 is not ubiquitylated), the Bul proteins are phosphorylated by Npr1, and this promotes their association with and inhibition by the 14-3-3 proteins. Upon addition of Am, the Bul proteins are dephosphorylated, dissociate from the 14-3-3 proteins, and mediate Rsp5-dependent ubiquitylation of Gap1 (Fig. 1) (21). As the Bul proteins also promote Gap1 ubiquitylation after Rap addition, we tested whether Rap induces their dephosphorylation (Fig. 4). In keeping with previous observations, the Bul1 and Bul2 proteins immunodetected in cells grown on Pro medium each migrated in a gel as two bands, previously shown to correspond to phosphorylated species (21). After addition of Am, each Bul protein double band was converted to a faster migrating band, corresponding to the dephosphorylated form, and an additional, slower migrating band, resulting from partial monoubiquitylation by Rsp5 of the dephosphorylated Bul protein (Fig. 4A) (21). After Rap addition, the typical bands corresponding to the phosphorylated Bul1 and Bul2 proteins were still readily detected, suggesting that Rap-induced Gap1 ubiquitylation is not accompanied by dephosphorylation of the Bul proteins. We therefore considered the possibility that the 14-3-3 proteins might undergo some modification after Rap addition, leading to their dissociation from the Bul proteins. Yet the gel migration profile of the Bmh1 and Bmh2 proteins was unaltered after Rap addition (Fig. 4B), and the same was true when we examined them by two-dimensional gel electrophoresis (data not shown). As the Bul proteins remain phosphorylated and the Bmh proteins do not undergo any detectable modification after Rap addition, we hypothesized that they might remain bound to each other. To test this hypothesis, we pulled down the GST-fused Bmh2 protein and tested its association with Bul2 (Fig. 4C). In keeping with previous observations (21), Bul2-Bmh2 binding was observed in extracts of cells grown on Pro, but not after Am addition. After Rap addition, however, binding was still observed. We noticed that free GST becomes detectable after Rap addition, an effect of unknown origin, but this did not correlate with any reduction of the amount of Bul2 bound to Bmh2. We also immunodetected the Aly1 and Aly2 proteins in cell extracts and found the corresponding signals migrate through a gel more diffusely than the Bul proteins (Fig. 4D). After addition of Rap to the cells, the Aly signals did not undergo any major change in migration. Furthermore, treatment of cell extracts with alkaline phosphatase showed that in the presence or absence of Rap, the Aly proteins are phosphorylated in cells grown on Pro medium (Fig. 4E). From these observations, we conclude that the mechanisms of Am- and Rap-triggered Bul-mediated Gap1 ubiquitylation differ, as the latter involves neither marked dephosphorylation of the Bul proteins nor their complete dissociation from the 14-3-3 proteins. Similarly, the Aly proteins remain phosphorylated after Rap addition, and a previous report states that at least one of them, Aly2, interacts with 14-3-3 proteins (29). It therefore seems that the Bul proteins, and possibly the Aly proteins as well, remain largely bound to 14-3-3 proteins while promoting Rap-induced ubiquitylation of Gap1. We also noticed in the GST pulldown experiment that Rap addition leads to apparition of free GST.
FIGURE 4.
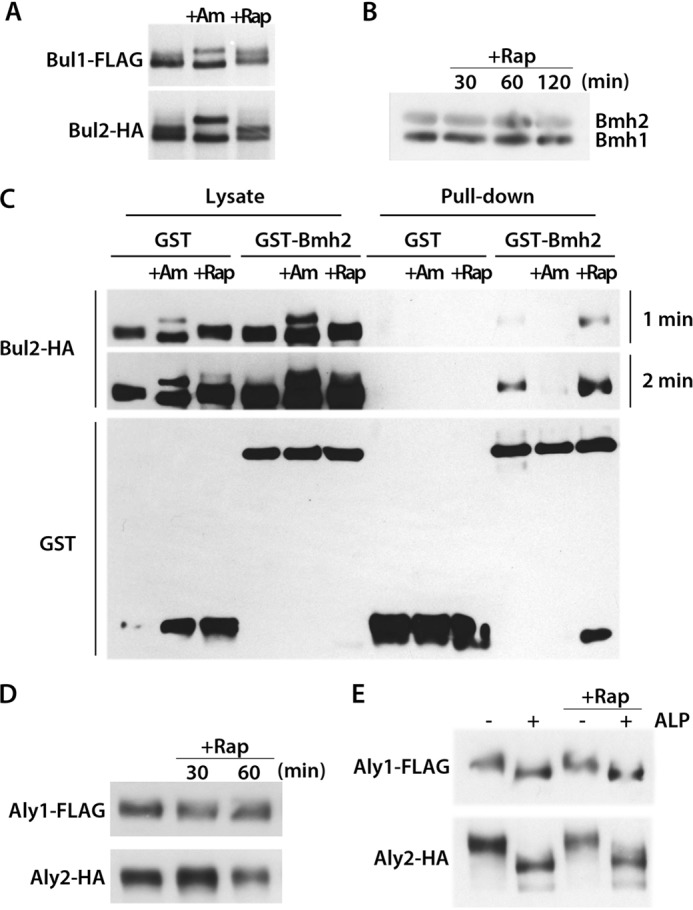
Bul1, Bul2, Aly1, and Aly2 remain phosphorylated and Bul2 still binds to the 14-3-3 protein Bmh2 after rapamycin addition. A, strains MA025 (gap1Δ BUL1-FLAG bul2Δ ura3) and MA032 (gap1Δ BUL2-HA ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP) were grown on galactose/proline medium. Cells were collected before and 30 min after Am (50 mm) or Rap (200 ng/ml) addition. Crude cell extracts were immunoblotted with anti-FLAG or anti-HA antibodies. B, strain 23344c (ura3) transformed with pFL38 (URA3) was grown on glucose/proline medium. Cells were collected before and 30, 60, and 120 min after Rap (200 ng/ml) addition. Crude cell extracts were immunoblotted with anti-Bmh antibodies. C, strain MA032 (gap1Δ BUL2-HA ura3) transformed with pRS426-GST or pRS426-GST-BMH2 was grown on glucose/proline medium. Cells were collected before and 30 or 60 min after Am (50 mm) or Rap (200 ng/ml) addition, respectively. The cells were lysed, and GST was pulled down as described under “Experimental Procedures.” Lysates and pulldown fractions were immunoblotted with anti-GST or anti-HA antibodies. Two exposure times (1 and 2 min) are shown. D, strains JA986 (ALY1-FLAG ura3) and JA997 (ALY2-HA ura3) transformed with pFL38 (URA3) were grown on glucose/proline medium. Cells were collected before and 30 and 60 min after Rap (200 ng/ml) addition. Crude cell extracts were immunoblotted with anti-FLAG or anti-HA antibodies. E, strains and growth conditions were the same as those in D. Crude cell extracts were treated or not with alkaline phosphatase and immunoblotted with anti-FLAG or anti-HA antibodies.
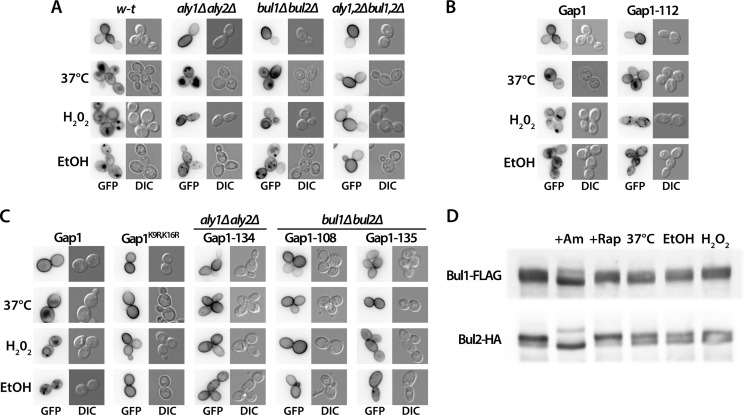
Gap1-112 Mutant, Resistant to Ubiquitylation Mediated by the Dephosphorylated Bul Proteins, Is Normally Down-regulated by Rapamycin
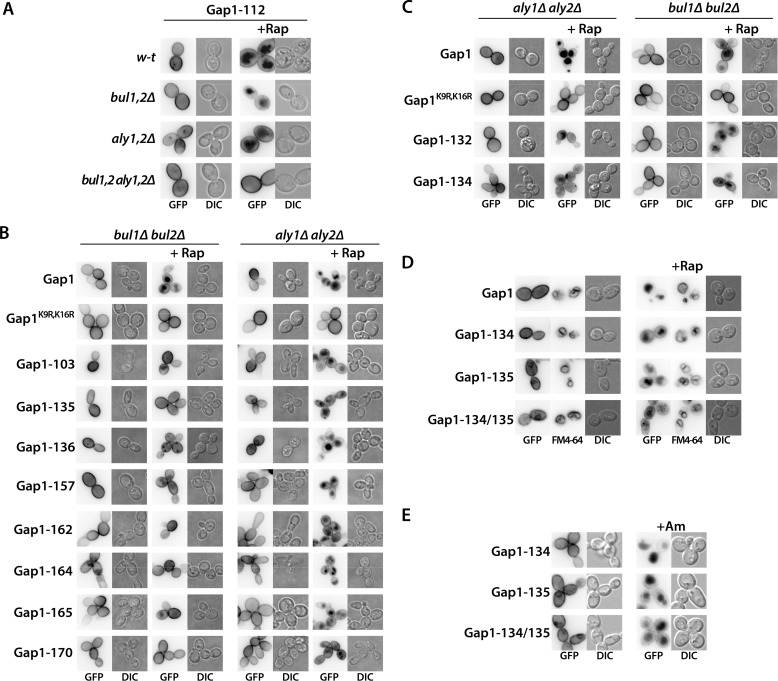
In a recent systematic mutational analysis of the cytosolic regions of Gap1, we identified a region in the N-terminal tail, between residues 20 and 35, essential to Am-induced down-regulation (24). Further analysis of one of the mutants altered in this region, Gap1-112 (32ITIP35 replaced with four alanines) (Fig. 5), showed that it was not ubiquitylated after Am addition. Hence, the Bul proteins having undergone dephosphorylation seem to act through this N-terminal region of Gap1 to promote ubiquitylation of the nearby Lys-9 and Lys-16 residues (24). We thus examined whether this N-terminal region of Gap1 is also important for Rap-induced ubiquitylation via the phosphorylated Bul and Aly adaptors. Interestingly, we found the Gap1-112 mutant to be normally down-regulated by Rap (Fig. 6A), and this down-regulation was efficient in the bul1Δ bul2Δ and aly1Δ aly2Δ mutants but defective in the bul1Δ bul2Δ aly1Δ aly2Δ quadruple mutant. This suggests that both the phosphorylated Bul proteins and phosphorylated Aly proteins promote Gap1 ubiquitylation by acting via one or several Gap1 cytosolic regions different from that used by the dephosphorylated Bul proteins.
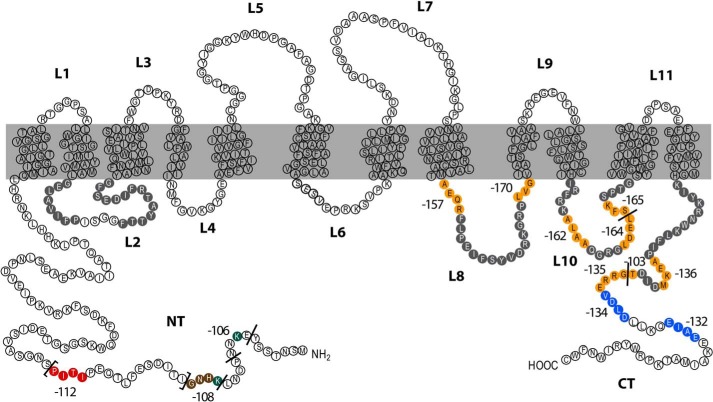
FIGURE 5.
Schematic topology model of the Gap1 protein. Residues shown in gray in the N-terminal tail (NT), C-terminal tail (CT), and intracellular loops (L2, L4, L6, L8, and L10) are those which, when replaced with alanines, cause retention of Gap1 in the ER. Each number close to colored residues corresponds to the Gap1 mutant where these residues have been replaced with alanines. Residues labeled in green in the NT correspond to the ubiquitin-acceptor lysines (Lys-9 and Lys-16). The NT region between brackets and including the four amino acids replaced in the Gap1-112 mutant (red) is the region required for down-regulation mediated by the dephosphorylated Bul proteins in response to internal amino acids. Residues shown in blue identify the Gap1-132 and Gap1-134 mutants resistant to down-regulation mediated by the phosphorylated Bul proteins under stress or in the presence of rapamycin. Residues in brown (Gap1-108) and orange (Gap1-103); Gap1-134; Gap1-136; Gap1-157; Gap1-162; Gap1-164; Gap1-165, and Gap1-170 identify the mutants resistant to down-regulation mediated by the phosphorylated Aly proteins under the same conditions.
FIGURE 6.
Several Gap1 C-terminal mutants resist rapamycin-induced down-regulation via the Bul or Aly adaptors. A, strains EK008 (gap1Δ ura3), JA493 (gap1Δ bul1Δ bul2Δ ura3), 35101a (gap1Δ aly1Δ aly2Δ ura3), and MA062 (gap1Δ bul1Δ bul2Δ aly1Δ aly2Δ ura3) transformed with pMA74 (YCpGAL-GAP1-112-GFP) were grown on galactose/proline medium. Glucose was added for 30 min to repress Gap1 synthesis, and when indicated, Rap (200 ng/ml) was added for 3 h. Gap1-GFP localization was examined by fluorescence microscopy. B, strains JA493 (gap1Δ bul1Δ bul2Δ ura3) and 35101a (gap1Δ aly1Δ aly2Δ ura3) were transformed with pJOD10 (YCpGAL-GAP1-GFP) or pCJ038 (YCpGAL-GAP1K9R,K16R-GFP) or with a plasmid encoding one of the indicated Gap1 mutants. Growth conditions were the same as in A. Gap1-GFP localization was examined by fluorescence microscopy. C, same strains as in B were transformed with pJOD10 (YCpGAL-GAP1-GFP) or pCJ038 (YCpGAL-GAP1K9R,K16R-GFP) or with a plasmid encoding one of the indicated Gap1 mutants. Growth conditions were as in A. Gap1-GFP localization was examined by fluorescence microscopy. D, strain EK008 (gap1Δ ura3) was transformed with pJOD10 (YCpGAL-GAP1-GFP) or a plasmid encoding one of the indicated Gap1 mutants. Growth conditions were as in A. Gap1-GFP localization was examined by fluorescence microscopy. Cells were labeled with FM4-64 to stain the vacuolar membrane. E, same strains as in D (precluding pJOD10) were grown on galactose/proline medium. Glucose was added for 2 h to repress Gap1 synthesis, and when indicated, Am (20 mm) was added for 2 h. Gap1-GFP localization was examined by fluorescence microscopy. DIC, differential interference contrast.
Structural Integrity of Two C-terminal Regions of Gap1 Is Essential to Rapamycin-induced Down-regulation via the Bul and Aly Proteins
To delineate the cytosolic regions of Gap1 that might be involved in ubiquitylation mediated by the phosphorylated Bul and Aly adaptors, we exploited the collection of mutants altered in all predicted cytosolic regions of Gap1, namely the N- and C-terminal tails and five internal loops (Fig. 5) (24). In each of these 64 mutants, 3–4 consecutive amino acids have been replaced with alanines. Among them, 14 were found to be stacked in the endoplasmic reticulum (ER) and were therefore excluded (24). The other 50 mutants were expressed in the bul1Δ bul2Δ and aly1Δ aly2Δ strains and tested for Rap-induced down-regulation. In the bul1Δ bul2Δ strain, eight Gap1 mutants displayed complete or highly significant protection against Rap-induced endocytosis (Fig. 6B). The corresponding mutations were found in the last two internal loops and the first half of the C-terminal tail (Fig. 5). The role of most nearby Gap1 regions could not be examined, because the corresponding mutants failed to exit the ER. The structural integrity of these C-terminal regions of Gap1 is thus important for Aly-mediated targeting of Rsp5 in response to Rap. In the aly1Δ aly2Δ strain, remarkably, these eight mutants were normally down-regulated by Rap (Fig. 6B), indicating that the Bul adaptors can act through a different region of Gap1. We thus analyzed the remaining 42 Gap1 mutants in the aly1Δ aly2Δ strain. None of the examined mutants proved fully resistant to Rap-induced down-regulation, but two of them, Gap1-132 and Gap1-134, displayed partial but significant resistance (Fig. 6C). The residues replaced in these two mutants were also located in the C-terminal tail, but slightly C-terminally with respect to the region required for Aly-mediated down-regulation (Fig. 5). We next combined in the same Gap1 protein the −135 and −134 substitutions, respectively conferring resistance to Aly- and Bul-mediated down-regulation. The resulting Gap1-134–135 mutant expressed in wild-type cells was found partially stacked in the ER, as already seen to some extent for Gap1-135 (Fig. 6D), but a significant proportion of the mutant permease was present at the plasma membrane and active, as judged by its ability to complement the phenotypes of a gap1 strain in growth tests (data not shown). After Rap addition, the Gap1-134 and Gap1-135 single mutants appeared to be down-regulated, as part of the GFP fluorescence initially present at the cell surface was clearly relocated to the vacuole, although this down-regulation was not as efficient as for wild-type Gap1. This diminished efficiency is consistent with the additive effects of the Bul and Aly proteins in Gap1 ubiquitylation and down-regulation (Fig. 2, B and C). However, the Gap1-134–135 variant displayed much more pronounced protection against Rap-induced down-regulation, as shown by much weaker GFP staining of the vacuolar lumen. Importantly, the double mutant was normally down-regulated by Am, suggesting that it is ubiquitylated and that it traffics normally along the endocytic pathway to the vacuole (Fig. 6E). In conclusion, the Gap1 C-terminal region encompassing the last two internal loops and tail is essential for efficient Rap-induced down-regulation mediated by phosphorylated Bul and Aly adaptors, but it is not involved in Am-induced down-regulation mediated by the dephosphorylated Bul proteins.
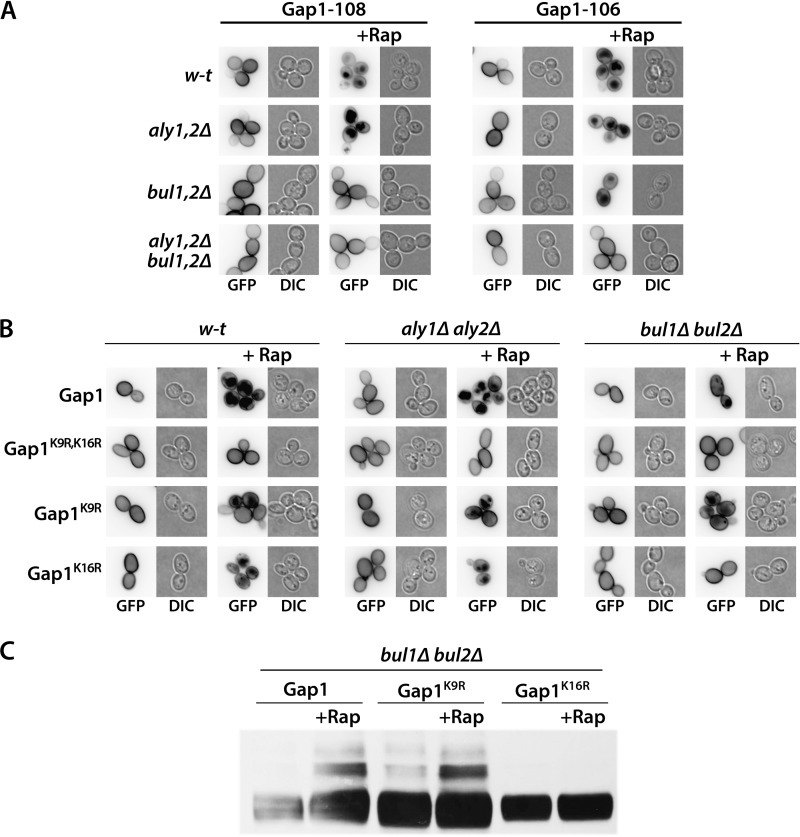
Aly1 and Aly2 Adaptors Promote Gap1 Ubiquitylation on Its Lysine 16 Only
We noticed that another Gap1 mutant, Gap1-108, was strongly protected against Rap-induced down-regulation in the bul1Δ bul2Δ strain, although it was normally down-regulated in the aly1Δ aly2Δ mutant (Fig. 7A). The four amino acids replaced in Gap1-108 (16KHGN19) included Lys-16, which is one of the two target lysines of Gap1 ubiquitylation (20). We thus also examined Gap1-106, as the four residues (8EKNN11) replaced in this mutant included the other target lysine of Gap1 ubiquitylation (Lys-9). This variant, however, displayed normal Rap-induced down-regulation in the wild-type, bul1Δ bul2Δ, and aly1Δ aly2Δ strains (Fig. 7A). These observations raised the interesting possibility that the Aly proteins might mediate Gap1 ubiquitylation on residue Lys-16 only, whereas the Bul adaptors, as reported previously in the context of Am-induced down-regulation, can promote Gap1 ubiquitylation via residue Lys-9 or Lys-16 of the permease. To test this hypothesis, we expressed the Gap1K9R and Gap1K16R variants in the bul1Δ bul2Δ mutant (Fig. 7B). As suspected, Gap1K16R down-regulation was normal in the aly1Δ aly2Δ strain but absent in the bul1Δ bul2Δ mutant. In contrast, Gap1K9R was similarly down-regulated in the bul1Δ bul2Δ and aly1Δ aly2Δ strains. Furthermore, after Rap addition, immunoblot analysis of extracts of bul1Δ bul2Δ cells expressing wild-type Gap1, Gap1K9R, or Gap1K16R revealed upper bands corresponding to ubiquitylated Gap1 forms only for wild-type Gap1 and Gap1K9R (Fig. 7C). The Aly proteins thus promote Rap-induced ubiquitylation of the Gap1 permease on its Lys-16 residue only.
FIGURE 7.
Aly1 and Aly2 adaptors promote Gap1 ubiquitylation on lysine 16 only. A, strains EK008 (gap1Δ ura3), JA493 (gap1Δ bul1Δ bul2Δ ura3), 35101a (gap1Δ aly1Δ aly2Δ ura3), and MA062 (gap1Δ bul1Δ bul2Δ aly1Δ aly2Δ ura3) transformed with pMA51 (YCpGAL-GAP1-108-GFP) or pMA47 (YCpGAL-GAP1-106-GFP) were grown on galactose/proline medium. Glucose was added for 30 min to repress Gap1 synthesis, and when indicated, Rap (200 ng/ml) was added for 3 h. Gap1-GFP localization was examined by fluorescence microscopy. B, strains EK008 (gap1Δ ura3), JA493 (gap1Δ bul1Δ bul2Δ ura3), and 35101a (gap1Δ aly1Δ aly2Δ ura3) transformed with pEL003 (YCp-GAP1-GFP), pEL005 (YCp-GAP1K9R,K16R-GFP), pEL018 (YCp-GAP1K9R-GFP), or pEL021 (YCp-GAP1K16R-GFP) were grown on glucose/proline medium, and when indicated, Rap (200 ng/ml) was added for 3 h. Gap1-GFP localization was examined by fluorescence microscopy. C, strain JA493 (gap1Δ bul1Δ bul2Δ ura3) transformed with pEL003 (YCp-GAP1-GFP), pEL018 (YCp-GAP1K9R-GFP), or pEL021 (YCp-GAP1K16R-GFP) was grown on glucose/proline medium. Cells were collected before and 10 min after Rap (200 ng/ml) addition. Crude cell extracts were immunoblotted with anti-GFP antibodies. DIC, differential interference contrast.
Various Stresses Also Promote Gap1 Down-regulation via the Phosphorylated Bul and Aly Adaptors
Under favorable conditions, TORC1 promotes cell growth by stimulating various functions, including translational initiation and synthesis and assembly of the translational machinery. It simultaneously antagonizes other functions, such as entry into G0 phase, autophagy, and induction of stress programs (30). These TORC1 functions tend to be inhibited under various noxious stress or nutrient starvation conditions (31, 32), and recent studies on fission yeast and mammalian cells indicate that this inhibition involves phosphorylation of a conserved residue of the ATP-binding domain of the TOR kinase (33). As TORC1 activity is clearly essential to maintaining Gap1 stable at the plasma membrane in cells growing on glucose/proline medium, we tested whether certain stresses might also promote Gap1 down-regulation. Cells expressing Gap1-GFP were thus subjected to heat shock (37 °C), oxidative stress (0.88 mm H2O2), or alcoholic stress (10% ethanol). Under all three tested conditions, Gap1 was internalized and targeted to the vacuole (Fig. 8A). Under heat shock and alcoholic stress, the permease was efficiently targeted to the vacuolar lumen, whereas oxidative stress led to its delivery in the vacuolar membrane, suggesting that H2O2 somehow interferes with sorting of Gap1 into the multivesicular body pathway. We next tested the role of the Bul and Aly adaptors. We found, as in Rap-induced down-regulation, that Gap1 endocytosis was impaired only in the bul1Δ bul2Δ aly1Δ aly2Δ mutant (Fig. 8A). Furthermore, stress-induced Gap1 down-regulation also showed the other specific features of Rap-induced down-regulation. In particular, the Gap1-112 variant, which is resistant to Am-induced ubiquitylation but sensitive to Rap-induced down-regulation, was down-regulated under stress (Fig. 8B). Although Gap1K9R,K16R resisted stress-induced down-regulation (Fig. 8C), the Gap1-108 and Gap1–135 mutants, largely insensitive to Rap-triggered Aly-mediated down-regulation, were also strongly protected against down-regulation in the bul1Δ bul2Δ strain subjected to stress (Fig. 8C). Conversely, the Gap1-134 mutant showing resistance to Rap-induced down-regulation mediated by the Bul proteins also displayed significant resistance to stress-induced down-regulation in the aly1Δ aly2Δ strain (Fig. 8C). Finally, cell transfer to various stress conditions, like Rap addition but unlike Am addition, caused no significant change in the gel migration patterns of immunodetected Bul1 and Bul2 (Fig. 8D). In conclusion, Rap addition and transfer to various stress conditions appear to trigger the same Gap1 down-regulation pathway involving the phosphorylated Bul and Aly adaptors.
FIGURE 8.
Stress conditions induce Gap1 down-regulation via the phosphorylated Bul and Aly adaptors. A, strains EK008 (gap1Δ ura3), JA493 (gap1Δ bul1Δ bul2Δ ura3), 35101a (gap1Δ aly1Δ aly2Δ ura3), and MA062 (gap1Δ bul1Δ bul2Δ aly1Δ aly2Δ ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP) were grown on galactose/proline medium. Glucose was added for 2 h to repress Gap1 synthesis, and when indicated, H2O2 (0.88 mm) or ethanol (EtOH) (10%) was added for 2 h or the cells were transferred from 29 to 37 °C for 1 h. Gap1-GFP localization was examined by fluorescence microscopy. B, strain EK008 (gap1Δ ura3) was transformed with pJOD10 (YCpGAL-GAP1-GFP) or pMA74 (YCpGAL-GAP1-112-GFP). Growth and stress conditions were as in A. Gap1-GFP localization was examined by fluorescence microscopy. C, strain EK008 (gap1Δ ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP) or pCJ038 (YCpGAL-GAP1K9R,K16R-GFP), strain 35101a (gap1Δ aly1Δ aly2Δ ura3) transformed with pMA31 (YCpGAL-GAP1-134-GFP), and strain JA493 (gap1Δ bul1Δ bul2Δ ura3) transformed with pMA51 (YCpGAL-GAP1-108-GFP) or pMA33 (YCpGAL-GAP-135-GFP) were grown on galactose/proline medium. Stress conditions were as in A. Gap1-GFP localization was examined by fluorescence microscopy. D, strains MA025 (gap1Δ BUL1-FLAG bul2Δ ura3) and MA032 (gap1Δ BUL2-HA ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP) were grown on galactose/proline medium. Cells were collected before and 30 min after Am (50 mm), Rap (200 ng/ml), H2O2 (0.88 mm), or ethanol (10%) addition or after transfer from 29 to 37 °C. Crude cell extracts were immunoblotted with anti-FLAG or anti-HA antibodies. DIC, differential interference contrast.
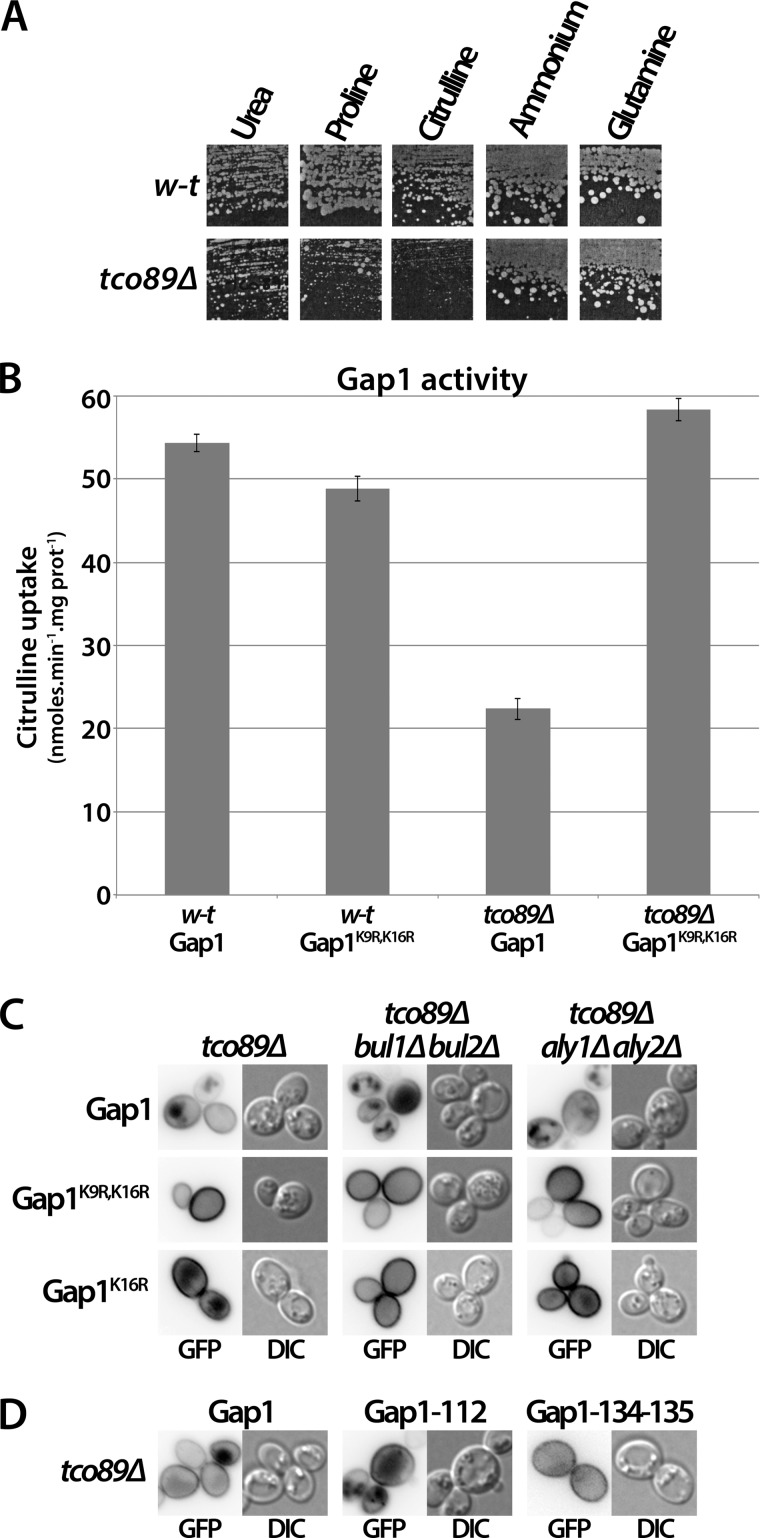
Lack of TORC1 Tco89 Subunit Mimics Stress- or Rapamycin-induced Gap1 Down-regulation
Tco89 is a subunit of the TORC1 complex. Previous studies have shown tco89 mutant cells to be hypersensitive to rapamycin and to resemble in other ways cells with reduced TORC1 activity, suggesting a positive involvement of Tco89 in TORC1 function (34). In the context of our recent work on the role of TORC1 in Gap1 ubiquitylation, we isolated a tco89Δ mutant and found it to display strongly reduced growth on citrulline or proline as the sole nitrogen source but normal growth on glutamine or ammonium medium (Fig. 9A). As citrulline enters cells only via Gap1, poor growth of the tco89Δ mutant on this amino acid could reflect reduced Gap1 activity. This prediction was confirmed by direct assay of [14C]citrulline uptake (Fig. 9B). In contrast, the activity of the nonubiquitylable Gap1K9R,K16R variant was unaltered in the tco89Δ mutant (Fig. 9B), suggesting that a lack of Tco89 causes constitutive Ub-dependent down-regulation of Gap1. In support of this view, Gap1-GFP was found to mislocalize partially to the vacuolar lumen in most tco89Δ cells growing on urea as sole nitrogen source, whereas GFP-fused Gap1K9R,K16R was detected only at the cell surface under these growth conditions (Fig. 9C). We thus hypothesized that a lack of Tco89 results in Gap1 down-regulation via the same pathway as when cells are treated with Rap or subjected to stress. In support of this prediction, Gap1 was still constitutively down-regulated in tco89Δ bul1Δ bul2Δ and tco89Δ aly1Δ aly2Δ cells (Fig. 9C), as expected if Gap1 down-regulation due to a Tco89 deficiency involves both the Bul and Aly adaptors. Our attempts to isolate a tco89Δ aly1Δ aly2Δ bul1Δ bul2Δ quintuple mutant, where Gap1 is predicted to be stable at the plasma membrane, were unsuccessful. We suspect that cumulation of these five mutations might reduce growth too severely, as suggested by the fact that the slow-growth phenotype of the tco89Δ mutant is already strongly exacerbated when the aly1Δ and aly2Δ mutations are also present (data not shown). Nevertheless, an involvement of the Aly adaptors in constitutive down-regulation of Gap1 in the tco89Δ bul1Δ bul2Δ strain is supported by the observation that Gap1K16R, resistant to Rap- and stress-induced down-regulation via the Aly proteins, also resists constitutive down-regulation in the tco89Δ bul1Δ bul2Δ mutant (Fig. 9C). Furthermore, the Gap1-134–135 mutant, resistant to Rap- and stress-induced down-regulation but sensitive to Am-induced down-regulation, shows no down-regulation in the tco89Δ strain (Fig. 9D). Hence, a lack of TORC1 Tco89 subunit mimics the situation of Rap- or stress-induced Gap1 down-regulation.
FIGURE 9.
Lack of Tco89 causes constitutive Gap1 down-regulation via a mechanism involving the Bul and Aly adaptors and the C-terminal tail of Gap1. A, strains 23344c (ura3) and FA198 (tco89Δ ura3) transformed with pFL38 (URA3) were tested for growth on solid medium containing the indicated compound as sole nitrogen source. For each growth condition, both strains were grown on the same plate. B, strains EK008 (gap1Δ ura3) and MYC003 (gap1Δ tco89Δ ura3) transformed with pCJ004 (YCpGAL-GAP1) or pCJ034 (YCpGAL-GAP1K9R,K16R) were grown on galactose/proline medium. Glucose was added for 2 h to repress Gap1 synthesis. The initial uptake rate of 14C-labeled citrulline (75 μm), reflecting Gap1 activity, was then measured. C, strains MYC003 (gap1Δ tco89Δ ura3), MYC004 (gap1Δ tco89Δ bul1Δ bul2Δ ura3), and MYC009 (gap1Δ tco89Δ aly1Δ aly2Δ ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP), pCJ038 (YCpGAL-GAP1K9R,K16R-GFP), or pEL021 (YCpGAP1K16R-GFP) were grown on galactose/urea medium. Glucose was added for 2 h to repress Gap1 synthesis. Gap1-GFP localization was examined by fluorescence microscopy. D, strain MYC003 (gap1Δ tco89Δ ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP), pMA74 (YCpGAL-GAP1-112-GFP), or pMYC016 (YCpGAL-GAP1-134–135-GFP) was grown on raffinose/urea medium. Galactose was added, and the culture was incubated overnight to induce Gap1 synthesis, and then glucose was added for 1.5 h to repress Gap1 synthesis. Gap1-GFP localization was then examined by fluorescence microscopy. DIC, differential interference contrast.
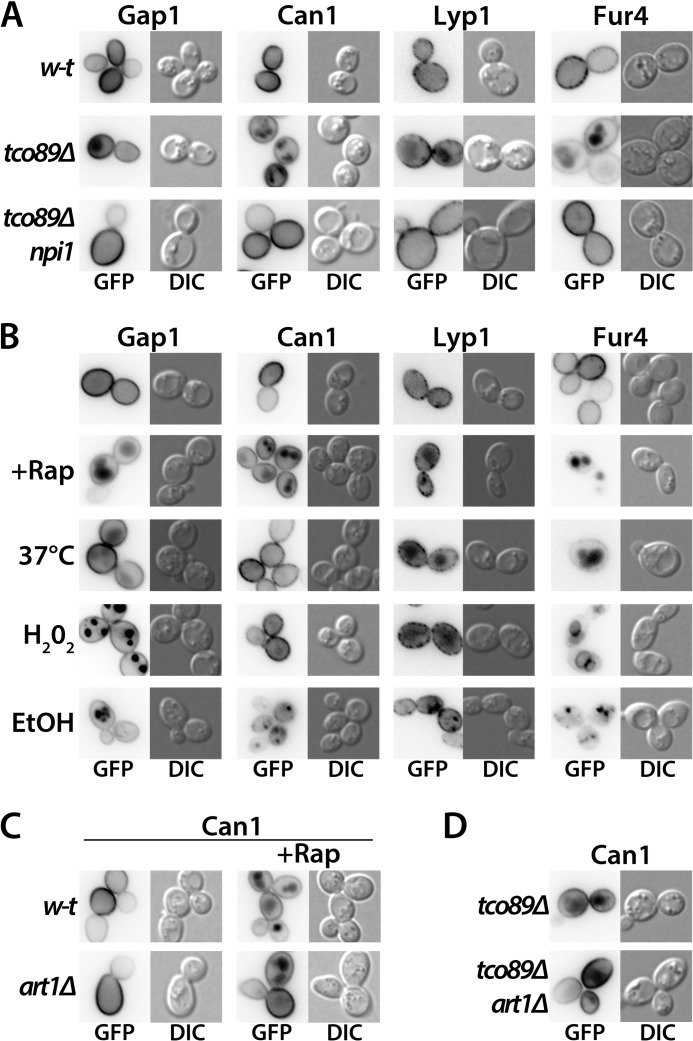
Stress, Rapamycin, or a Lack of Tco89 Promotes Down-regulation of Other Permeases via Arrestin-like Adaptors
Mutations in the TCO89 gene were originally isolated in a screen for mutants with reduced glycerol uptake activity (35). Furthermore, several permeases are reported to undergo down-regulation under various stress or starvation conditions (13–15, 36–39) or upon treatment of cells with Rap (38, 40). Hence, the pathway promoting Gap1 down-regulation upon Rap addition or under stress might target many other plasma membrane transport proteins. In support of this view, we found three other transporters, the arginine permease Can1, the lysine permease Lyp1, and the uracil permease Fur4, present at the cell surface in wild-type cells, to be delivered to the vacuolar lumen in tco89Δ mutant cells but to remain at the cell surface in tco89Δ npi1(rsp5) cells (Fig. 10A). We next tested the influence of Rap, heat shock (37 °C), H2O2, and ethanol on these permeases (Fig. 10B). Fur4 and Lyp1 showed down-regulation under all tested conditions, and Can1 was down-regulated in Rap-, H2O2-, and ethanol-treated cells but not upon shifting to 37 °C. Interestingly, as observed for Gap1, none of the permeases internalized from the cell surface after H2O2 treatment was efficiently delivered to the vacuolar lumen, suggesting that a step of the multivesicular body sorting pathway is not functional under oxidative stress. Furthermore, the permeases internalized after cell treatment with ethanol often accumulated in a perivacuolar compartment likely corresponding to late endosomes, suggesting that multivesicular body sorting is also less efficient under these conditions. We further found Rap-induced Can1 and Fur4 down-regulation to be impaired in the rsp5(npi1) mutant but normal in the bul1Δ bul2Δ aly1Δ aly2Δ strain, suggesting that other arrestin-like adaptors may target Rsp5 to these permeases after Rap addition (data not shown). As in a previous study, Art1 was found to mediate cycloheximide-induced endocytosis of Can1 (13, 17), so we tested the involvement of Art1 in Rap-induced Can1 down-regulation. We found Can1 down-regulation to occur in the art1Δ mutant, albeit less efficiently than in the wild type. Furthermore, an additional art1Δ mutation did not prevent the constitutive delivery of Can1 to the vacuole caused by the tco89Δ mutation (Fig. 10D). These observations suggest that additional arrestin-like adaptors promote Can1 down-regulation in response to Rap or when Tco89 is lacking.
FIGURE 10.
Can1, Lyp1, and Fur4 permeases are down-regulated by Tco89 depletion, rapamycin addition, or stress. A, strains EK008 (gap1Δ ura3), MYC003 (gap1Δ tco89Δ ura3), and 41453c (tco89Δ rsp5(npi1) ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP), pMA185 (YCpGAL1-CAN1-GFP), pNAM001 (YCp-LYP1-GFP), or pFL38-gF-GFP (YCpGAL1-FUR4-GFP) were grown on galactose/urea medium, except for strain 41453c, which was grown on galactose/proline and strains harboring the Fur4 plasmid, which were grown on galactose/ammonium. Glucose was added for 2 h to repress permease synthesis, and cells were visualized by fluorescence microscopy. B, strain EK008 (gap1Δ ura3) transformed with pJOD10 (YCpGAL-GAP1-GFP), pMA185 (YCpGAL1-CAN1-GFP), pNAM001 (YCp-LYP1-GFP), or pFL38-gF-GFP (YCpGAL1-FUR4-GFP) was grown on galactose/proline medium except for strains with the Fur4 plasmid, which were grown on galactose/ammonium. Glucose was added for 2 h to repress permease synthesis, and when indicated, Rap (200 ng/ml), H2O2 (0.88 mm), or ethanol (10%) was added for 2 h or cells were transferred from 29 to 37 °C for 1 h. The cells were then visualized by fluorescence microscopy. C, strains EK008 (gap1Δ ura3) and JA983 (gap1Δ art1Δ ura3) transformed with pMA185 (YCpGAL1-CAN1-GFP) were grown on galactose/ammonium medium. Glucose was added for 30 min to repress Can1 synthesis, and when indicated, Rap (200 ng/ml) was added for 3 h. Can1-GFP localization was then examined by fluorescence microscopy. D, strains MYC003 (gap1Δ tco89Δ ura3) and 35246a (gap1Δ tco89Δ art1Δ ura3) transformed with pMA185 (YCpGAL1-CAN1-GFP) plasmid were grown on galactose/urea medium. Glucose was added for 2 h to repress Can1 synthesis. Can1-GFP localization was then examined by fluorescence microscopy. DIC, differential interference contrast.
DISCUSSION
In this study, we describe a novel pathway promoting ubiquitylation and down-regulation of the yeast Gap1 permease. This pathway is stimulated when the TORC1 kinase complex of cells growing on a poor nitrogen source (e.g. proline) is inhibited by rapamycin and is constitutively active in a mutant lacking the Tco89 subunit of TORC1. It is also stimulated in cells subjected to various stresses (a shift from 29 to 37 °C, addition of H2O2 or ethanol). As previous reports describe TORC1 inhibition in response to stress (31–33), it seems likely that the stress conditions tested here cause Gap1 down-regulation by inhibiting TORC1. Thus, there emerge two different mechanisms for triggering Gap1 ubiquitylation and down-regulation as follows: one involving TORC1 activation by internal amino acids and a different one elicited by stress or direct TORC1 inhibition (Fig. 11).
FIGURE 11.
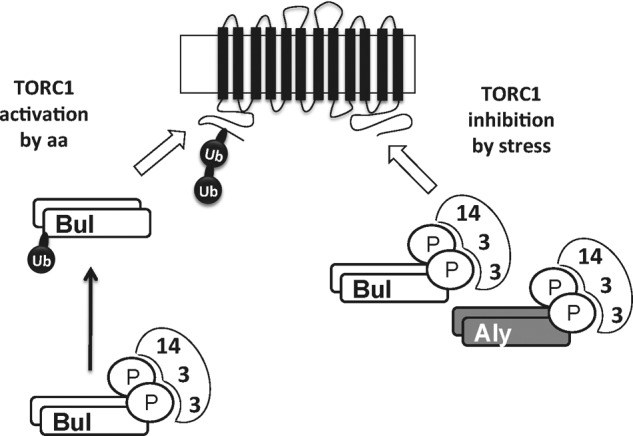
Model for amino acid- and stress-induced ubiquitylation of Gap1 permease. In cells growing on a poor nitrogen source, the TORC1 kinase complex is active for its ability to somehow prevent the phosphorylated Bul and Aly proteins (bound to 14-3-3 proteins) from promoting Gap1 ubiquitylation. When TORC1 is inhibited by rapamycin, stress conditions, or lack of its Tco89 subunit, this negative control is thus relieved, and the Bul and Aly proteins (remaining phosphorylated and bound to 14-3-3 proteins) act through C-terminal regions of Gap1 to induce its Rsp5-dependent ubiquitylation on Lys-9 or Lys-16 residues (Bul proteins) or only the Lys-16 residue (Aly proteins) in the permease's N-terminal tail. When the internal concentration of amino acids increases, active TORC1 further gains the ability to promote the dephosphorylation of the Bul proteins (which also undergo monoubiquitylation). These dephosphorylated Bul proteins dissociate from the 14-3-3 proteins, thereby gaining the ability to induce Gap1 ubiquitylation by acting through an N-terminal region of the permease close to the Lys-9 and Lys-16 residues.
These two mechanisms differ in several ways. First, stress-induced down-regulation involves the Aly1 and Aly2 adaptors of Rsp5 in addition to Bul1 and Bul2, as it is abolished only in the bul1Δ bul2Δ aly1Δ aly2Δ quadruple mutant. In a previous study, the Aly1 and Aly2 proteins were found to control Gap1 trafficking, but in a manner opposite to what we observe under stress conditions (28); the authors found Gap1 to be less abundant at the plasma membrane of aly1Δ aly2Δ cells, suggesting a role of Aly proteins in Gap1 recycling from endosomes to the cell surface. This phenotype was observed in ammonium-grown and not in proline-grown cells, and we have also failed to detect any role of the Aly proteins in Gap1 traffic in cells grown on proline medium. This suggests the interesting possibility that specific arrestin-like adaptors of Rsp5 might control permease traffic in opposite manners according to the growth conditions. This putative dual role of the Aly proteins in Gap1 trafficking is not shared by the Bul adaptors, which promote Gap1 ubiquitylation and endocytosis in response to either internal amino acids or stress.
Second, although the Bul proteins alone can promote efficient Gap1 down-regulation under stress, they remain largely phosphorylated, and Bul2 (at least) remains bound to 14-3-3 proteins. This suggests that the Bul adaptors mediate stress-induced Gap1 ubiquitylation even if they are phosphorylated by the Npr1 kinase and associated with 14-3-3 proteins (Fig. 11). This is likely true of the Aly proteins as well, because they also remain phosphorylated after rapamycin addition. Furthermore, Aly2 was found in a large scale study to bind to 14-3-3 proteins (29), and its phosphorylation is reported to depend on Npr1 (28). The Aly1 protein is also phosphorylated, and its dephosphorylation is mediated by the calcineurin/PP2B phosphatase, but these modifications were reported not to alter Gap1 sorting (49). Dephosphorylation and dissociation from the 14-3-3 proteins is thus a mode of Bul protein activation occurring specifically in response to an increase in the internal amino acid pool, whereas stimulation of Bul action under stress apparently relies on another mechanism. Bul stimulation in response to internal amino acids is also accompanied by monoubiquitylation of a fraction of the dephosphorylated Bul proteins (21), a modification shared by several other arrestin-like adaptors of Rsp5 (12). Although the role of this Bul ubiquitylation remains unclear, it is worth noting that stress does not seem to stimulate this modification (as no upper band becomes detectable when Bul1 or Bul2 is immunodetected after rapamycin addition, for example). Thus, Bul-dependent Gap1 down-regulation might occur independently of Bul ubiquitylation.
A third difference between the two mechanisms concerns the permease regions interacting with the Rsp5 adaptors; the dephosphorylated Bul proteins fail to down-regulate Gap1 mutants such as Gap1-112, harboring substitutions in an N-terminal region close to the Ub acceptor lysines. This mutant, however, is subject to normal stress- and rapamycin-induced Bul- and/or Aly adaptor-mediated down-regulation. In contrast, we have identified several substitutions in the C-terminal loops and tail of Gap1, which confer full protection against stress-induced Aly-dependent Gap1 down-regulation and reduce the efficiency of Bul-mediated down-regulation, and these C-terminal substitutions do not impair ammonium-induced down-regulation. It thus seems that the dephosphorylated Bul proteins on the one hand and the phosphorylated Aly and Bul proteins on the other hand recognize different Gap1 regions exposed to the cytosol. Our data also reveal a difference between Bul- and Aly-promoted Rsp5-mediated Gap1 ubiquitylation. When Rsp5 is targeted to Gap1 by the Bul proteins in response to amino acids or stress signals, Gap1 ubiquitylation can occur on two possible lysines, Lys-9 or Lys-16, but when Rsp5 is targeted to Gap1 by the Aly proteins, only Lys-16 is accessible for ubiquitylation. This suggests that Rsp5 is not similarly oriented with respect to the acceptor lysines when recruited by the Bul or Aly proteins or that the Aly proteins bound to Gap1 somehow occlude the Lys-9 residue, making it less accessible to Rsp5.
Our results further show that at least some of the conditions promoting Aly- and Bul-mediated Gap1 down-regulation, including a lack of the Tco89 subunit of TORC1, also trigger endocytosis of the Can1, Lyp1, and Fur4 permeases. Hence, the Gap1 down-regulation pathway discovered here may well target many other plasma membrane proteins. Consistently, previous reports describe situations in which Rsp5-dependent permease endocytosis occurs under adverse conditions as follows: nutrient starvation, stationary phase, and inhibition of protein synthesis in the case of the Fur4 uracil permease (41) and the iron transporter Ftr1 (37); nitrogen starvation for the Hxt7 hexose transporter (42); TORC1 inhibition, tryptophan starvation (40), or high hydrostatic pressure (43) for the Tat2 tryptophan permease; and heat shock or starvation for other amino acid permeases (Can1, Lyp1, and Mup1) (13, 37, 39). Furthermore, different arrestin-like adaptors of Rsp5, or different specific adaptor combinations, are reported to promote stress-induced down-regulation of different permeases (2, 13, 15, 27).
The precise mechanisms triggering arrestin-dependent down-regulation in response to stress remain to be determined. One proposed model is that adverse conditions cause permease misfolding, thus favoring permease recognition by specific Rsp5 adaptors (39, 44). Although it is likely that specific stresses such as high temperature can cause permease misfolding, it is hard to see how permease misfolding could explain our data showing that several permeases are constitutively down-regulated in tco89Δ mutant cells. It is more likely that Gap1 is properly folded, as the nonubiquitylable Gap1K9R,K16R form is fully active in tco89Δ cells. Furthermore, we have previously reported that Gap1 misfolding (causing an actual loss of activity) due to inhibition of sphingolipid biogenesis does indeed promote its Rsp5-dependent endocytosis, but via a different mechanism, because no less than seven N-terminal lysines in addition to Lys-9 and Lys-16 had to be mutagenized to stabilize the permease at the cell surface (45). For these reasons, we rather favor a model where one or several intracellular pathways stimulating the action of arrestin-like adaptors and Rsp5 is/are activated under adverse conditions. The mechanisms of this stimulation, however, remain unknown. Although our experiments show that the Bul and Aly proteins remain phosphorylated and that Bul2 still binds to 14-3-3 proteins after rapamycin addition, the putative rapamycin-induced stimulation of Bul and Aly Rsp5 adaptor functions might be triggered by other post-translational modifications, including subtle phosphorylation not detectable in our experiments. Alternatively, Gap1 ubiquitylation might be stimulated by certain modifications of other proteins, e.g. the 14-3-3 proteins known to bind to many different Rsp5 adaptors (29), although we detected no change in the gel migration profiles of Bmh proteins after rapamycin addition, for example. Another possibility is that an unknown factor regulating the Bul and Aly proteins and many other arrestins might itself be controlled by stress conditions. For instance, such a protein might inhibit multiple Rsp5 adaptors, and its negative action might be relieved under adverse conditions.
Our results also raise the question of why Gap1 and so many other permeases are down-regulated under adverse conditions. A recent paper reports the interesting observation that several stresses promoting the down-regulation of permeases also destabilize Lst1, a negative regulator of the ESCRT machinery (37). As this system is essential to delivering ubiquitylated membrane proteins to the vacuolar lumen, its stimulation by Lst1 degradation coupled to massive permease endocytosis very likely contributes, together with autophagy, to maintenance of a high cytosolic pool of amino acids, important for the rapid synthesis of proteins involved in adaptation to stress. Hence, the Gap1 down-regulation pathway described in this work is very likely part of a general response of the cell to adverse conditions. Its further molecular dissection, including elucidation of the mechanism stimulating the function of Bul and Aly adaptors, could thus shed new light on the molecular machinery involved in the adaptation of cells to stress.
Acknowledgments
We are grateful to Catherine Jauniaux for her invaluable contribution to isolating strains and plasmids; Stephan Vissers and Fadi Abdel-Sater for their contribution to initial experiments using the tco89Δ mutant, and André Feller for his contribution to two-dimensional gel experiments. We also thank Sandra Lemmon for the anti-Bmh2 antibody, the members of the laboratory for fruitful discussions, and Lydia Spedale for technical assistance.
This work was supported by Fonds National de la Recherche Scientifique Grant 3.4.592.08.F and Actions de Recherche Concertées Grant AUWB 2010-15-2 of the Fédération Wallonie-Bruxelles.
- Ub
- ubiquitin
- ER
- endoplasmic reticulum
- Am
- ammonium
- Rap
- rapamycin.
REFERENCES
- 1. Brohée S., Barriot R., Moreau Y., André B. (2010) YTPdb: a wiki database of yeast membrane transporters. Biochim. Biophys. Acta 1798, 1908–1912 [DOI] [PubMed] [Google Scholar]
- 2. Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 3. Dupré S., Urban-Grimal D., Haguenauer-Tsapis R. (2004) Ubiquitin and endocytic internalization in yeast and animal cells. Biochim. Biophys. Acta 1695, 89–111 [DOI] [PubMed] [Google Scholar]
- 4. Hein C., Springael J. Y., Volland C., Haguenauer-Tsapis R., André B. (1995) NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18, 77–87 [DOI] [PubMed] [Google Scholar]
- 5. Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M. (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 92, 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 7. Galan J. M., Haguenauer-Tsapis R. (1997) Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16, 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Springael J. Y., Galan J. M., Haguenauer-Tsapis R., André B. (1999) NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J. Cell Sci. 112, 1375–1383 [DOI] [PubMed] [Google Scholar]
- 9. Kim H. C., Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lauwers E., Jacob C., André B. (2009) K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Erpapazoglou Z., Froissard M., Nondier I., Lesuisse E., Haguenauer-Tsapis R., Belgareh-Touzé N. (2008) Substrate- and ubiquitin-dependent trafficking of the yeast siderophore transporter Sit1. Traffic 9, 1372–1391 [DOI] [PubMed] [Google Scholar]
- 12. Becuwe M., Herrador A., Haguenauer-Tsapis R., Vincent O., Léon S. (2012) Ubiquitin-mediated regulation of endocytosis by proteins of the arrestin family. Biochem. Res. Int. 2012, 242764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 14. Nikko E., Sullivan J. A., Pelham H. R. (2008) Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9, 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nikko E., Pelham H. R. (2009) Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Becuwe M., Vieira N., Lara D., Gomes-Rezende J., Soares-Cunha C., Casal M., Haguenauer-Tsapis R., Vincent O., Paiva S., Léon S. (2012) A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J. Cell Biol. 196, 247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. MacGurn J. A., Hsu P.-C., Smolka M. B., Emr S. D. (2011) TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell 147, 1104–1117 [DOI] [PubMed] [Google Scholar]
- 18. Grenson M., Hou C., Crabeel M. (1970) Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J. Bacteriol. 103, 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Craene J. O., Soetens O., Andre B. (2001) The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276, 43939–43948 [DOI] [PubMed] [Google Scholar]
- 20. Soetens O., De Craene J. O., Andre B. (2001) Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276, 43949–43957 [DOI] [PubMed] [Google Scholar]
- 21. Merhi A., André B. (2012) Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol. Cell. Biol. 32, 4510–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grenson M., Mousset M., Wiame J. M., Bechet J. (1966) Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim. Biophys. Acta 127, 325–338 [DOI] [PubMed] [Google Scholar]
- 23. Jacobs P., Jauniaux J. C., Grenson M. (1980) A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J. Mol. Biol. 139, 691–704 [DOI] [PubMed] [Google Scholar]
- 24. Merhi A., Gérard N., Lauwers E., Prévost M., André B. (2011) Systematic mutational analysis of the intracellular regions of yeast Gap1 permease. PLoS ONE 6, e18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikko E., Marini A. M., André B. (2003) Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J. Biol. Chem. 278, 50732–50743 [DOI] [PubMed] [Google Scholar]
- 26. Springael J. Y., André B. (1998) Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol. Biol. Cell 9, 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacGurn J. A., Hsu P.-C., Emr S. D. (2012) Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81, 231–259 [DOI] [PubMed] [Google Scholar]
- 28. O'Donnell A. F., Apffel A., Gardner R. G., Cyert M. S. (2010) α-Arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 21, 3552–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kakiuchi K., Yamauchi Y., Taoka M., Iwago M., Fujita T., Ito T., Song S.-Y., Sakai A., Isobe T., Ichimura T. (2007) Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry 46, 7781–7792 [DOI] [PubMed] [Google Scholar]
- 30. Loewith R., Hall M. N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Broach J. R., De Virgilio C., Hall M. N., Loewith R. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674 [DOI] [PubMed] [Google Scholar]
- 32. Binda M., Péli-Gulli M.-P., Bonfils G., Panchaud N., Urban J., Sturgill T. W., Loewith R., De Virgilio C. (2009) The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563–573 [DOI] [PubMed] [Google Scholar]
- 33. Hálová L., Du W., Kirkham S., Smith D. L., Petersen J. (2013) Phosphorylation of the TOR ATP binding domain by AGC kinase constitutes a novel mode of TOR inhibition. J. Cell Biol. 203, 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reinke A., Anderson S., McCaffery J. M., Yates J., 3rd, Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K. P., Powers T. (2004) TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752–14762 [DOI] [PubMed] [Google Scholar]
- 35. Holst B., Lunde C., Lages F., Oliveira R., Lucas C., Kielland-Brandt M. C. (2000) GUP1 and its close homologue GUP2, encoding multimembrane-spanning proteins involved in active glycerol uptake in Saccharomyces cerevisiae. Mol. Microbiol. 37, 108–124 [DOI] [PubMed] [Google Scholar]
- 36. Galan J. M., Moreau V., Andre B., Volland C., Haguenauer-Tsapis R. (1996) Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 271, 10946–10952 [DOI] [PubMed] [Google Scholar]
- 37. Jones C. B., Ott E. M., Keener J. M., Curtiss M., Sandrin V., Babst M. (2012) Regulation of membrane protein degradation by starvation-response pathways. Traffic 13, 468–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Snowdon C., van der Merwe G. (2012) Regulation of Hxt3 and Hxt7 turnover converges on the Vid30 complex and requires inactivation of the Ras/cAMP/PKA pathway in Saccharomyces cerevisiae. PLoS ONE 7, e50458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao Y., Macgurn J. A., Liu M., Emr S. (2013) The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. Elife 2, e00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beck T., Schmidt A., Hall M. N. (1999) Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146, 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Volland C., Urban-Grimal D., Géraud G., Haguenauer-Tsapis R. (1994) Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269, 9833–9841 [PubMed] [Google Scholar]
- 42. Krampe S., Boles E. (2002) Starvation-induced degradation of yeast hexose transporter Hxt7p is dependent on endocytosis, autophagy and the terminal sequences of the permease. FEBS Lett. 513, 193–196 [DOI] [PubMed] [Google Scholar]
- 43. Abe F., Iida H. (2003) Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2. Mol. Cell. Biol. 23, 7566–7584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Babst M. (2014) Quality control: Quality control at the plasma membrane: One mechanism does not fit all. J. Cell Biol. 205, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lauwers E., Grossmann G., André B. (2007) Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell 18, 3068–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. (1991) A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7, 609–615 [DOI] [PubMed] [Google Scholar]
- 47. Mayordomo I., Regelmann J., Horak J., Sanz P. (2003) Saccharomyces cerevisiae 14-3-3 proteins Bmh1 and Bmh2 participate in the process of catabolite inactivation of maltose permease. FEBS Lett. 544, 160–164 [DOI] [PubMed] [Google Scholar]
- 48. Ghaddar K., Krammer E.-M., Mihajlovic N., Brohée S., André B., Prévost M. (2014) Converting the yeast arginine Can1 permease to a lysine permease. J. Biol. Chem. 289, 7232–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Donnell A. F., Huang L., Thorner J., Cyert M. S. (2013) A calcineurin-dependent switch controls the trafficking function of α-arrestin Aly1/Art6. J. Biol. Chem. 288, 24063–24080 [DOI] [PMC free article] [PubMed] [Google Scholar]