Background: Exosomes can transfer information between cells and facilitate tumor development.
Results: PC12 cell-derived exosomes enter into BMSCs through clathrin-mediated endocytosis and macropinocytosis, and decrease the expression of TGFβRII and TPM1 through miR-21.
Conclusion: The results dissect the pathway of exosome internalization and demonstrate their regulation ability.
Significance: These findings enhanced our knowledge of the internalization and function of tumor exosomes.
Keywords: Cell-Cell Interaction, Endocytosis, Exosome, Extracellular Vesicles, microRNA (miRNA)
Abstract
Exosomes are nanoscale membrane vesicles secreted from many types of cells. Carrying functional molecules, exosomes transfer information between cells and mediate many physiological and pathological processes. In this report, utilizing selective inhibitors, molecular tools, and specific endocytosis markers, the cellular uptake of PC12 cell-derived exosomes was imaged by high-throughput microscopy and statistically analyzed. It was found that the uptake was through clathrin-mediated endocytosis and macropinocytosis. Furthermore, PC12 cell-derived exosomes can enter and deliver microRNAs (miRNAs) into bone marrow-derived mesenchymal stromal cells (BMSCs), and decrease the expression level of transforming growth factor β receptor II (TGFβRII) and tropomyosin-1 (TPM1) through miR-21. These results show the pathway of exosome internalization and demonstrate that tumor cell-derived exosomes regulate target gene expression in normal cells.
Introduction
Many types of cells release exosomes (small membrane vesicles (40–100 nm)) into extracellular environments (1). Specific to the parental cell, exosomes carry different types of functional molecules including proteins, soluble factors, mRNAs, and microRNAs (miRNAs)3 (2–4). Attaching on or entering into recipient cells, exosomes can transfer information (5–7). Recent studies have shown that many physiological and pathological processes including antigen presentation and cancer progression are mediated by shuttling exosomes (8–10). Tumor-produced exosomes have drawn more and more attention lately (11–13). Exosomes are involved in autocrine signaling promotion in tumor progression through exchange between tumor cells (14, 15). Neighboring and distant tumor cells can receive information from tumor exosomes (9, 16).
Our previous work used rat pheochromocytoma PC12 cell-derived exosomes as a general model in which to study tumor exosomes (17). The cellular uptake and intracellular trafficking were visualized by real-time fluorescence microscopy and single particle tracking (SPT) (18). However, the endocytic pathway of exosomes has not been further examined. There are multiple pathways that can mediate endocytosis, including phagocytosis, macropinocytosis, clathrin-mediated endocytosis, caveolae-mediated endocytosis, and clathrin- and caveolae-independent endocytosis (19). These pathways lead to different sorting and fate of endocytic cargo. It has been reported that exosomes derived from erythroleukemia cells or oligodendroglia cells were internalized through phagocytosis or macropinocytosis, respectively (20, 21). Recently, Svensson et al. found that glioblastoma cell-derived exosomes were internalized through lipid raft-mediated endocytosis, negatively regulated by caveolin-1 (CAV1) (22). The uptake pathway of exosomes may possibly be cell type specific. Furthermore, oncomiRs, a miRNA that is associated with tumor, may be enclosed in tumor exosomes and delivered to normal cells (15, 16). It is still unknown whether these reduce expression of target gene and facilitate tumor development.
In this study, exosomes were isolated from the culture medium of PC12 cells. By employing selective inhibitors, molecular tools, and endocytosis markers, it was found that the exosome uptake by PC12 cells occurred through clathrin-mediated endocytosis and macropinocytosis. Moreover, using quantitative real-time PCR (RT-PCR) and immunoblot assay, it was demonstrated that PC12 cell-derived exosomes delivered miR-21 into bone marrow-derived mesenchymal stromal cells (BMSCs) and down-regulated the expression levels of their transforming growth factor β receptor II (TGFβRII) and tropomyosin-1 (TPM1). These findings add insights into the endocytic pathway and the biological significance of tumor exosomes.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Rat pheochromocytoma PC12 cells and cardiomyoblast H9C2 cells (Shanghai Cellular Research Institute) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) in a 5% CO2 humidified atmosphere at 37 °C. For exosome purification, cells were cultured for 4 days in 175-cm2 culture flasks with DMEM and exosome-free FBS obtained by ultracentrifugation (200,000 × g for 6 h). For light microscopic analysis, cells were plated on a cover glass. BMSCs from rat bone marrow were extracted as described previously (23). Briefly, the tibias and femurs from 4-week-old Sprague-Dawley rats were dissected. Both ends of the bones were cut down along the epiphysis, then marrow was flushed with 10 ml of α-minimal essential medium (α-MEM) supplemented with 10% FBS contained in one-off syringe with a steel needle. To obtain BMSCs, bone marrow cells were transferred into a culture flask and incubated at 37 °C with 5% CO2. The medium was replaced every 3 days, and most nonadherent cells were removed. Medium and reagents for cell culture were from HyClone Laboratories. 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD) and calcein AM were obtained from Biotium. Carboxyfluorescein diacetatesuccinimidyl ester (CFSE), chlorpromazine (CPZ), genistein, nystatin, methyl-β-cyclodextrin (MβCD), LY294002, FITC-labeled cholera toxin B subunit (FITC-CtxB), FITC-dextran (70 kDa), polystyrene carboxylate-modified fluorescent latex beads (1 μm), and Hoechst 33342 were from Sigma-Aldrich. The μ2 subunit of clathrin adaptor complex AP2, dynamin 2 (DYN2), phosphatase and tensin homolog deleted on chromosome ten (PTEN), TPM1, and GAPDH antibodies, and 5-(n-ethyl-n-isopropyl)-amiloride (EIPA) were purchased from Santa Cruz Biotechnology. Clathrin heavy chain (CHC), CAV1, and TGFβRII antibodies were from Cell Signal Technology.
Exosome Isolation and Labeling
The culture medium from PC12 cells (1 × 108) was collected and isolated as previously described (24). Briefly, the harvested medium was centrifuged at 300 × g for 10 min, 1200 × g for 20 min, and 10,000 × g for 30 min to remove cells and debris. The supernatant was ultracentrifuged at 200,000 × g for 2 h using a Type 70 Ti rotor in an L-80 XP ultracentrifuge (Beckman Coulter). Then the exosome pellet was resuspended in phosphate-buffered saline (PBS). For labeling, the exosome solution was incubated with 5 μg/ml DiD for 30 min. The unincorporated dyes were removed using 300-kDa ultrafiltration tubes (Pall Corp.) and washed in PBS with ultracentrifugation. The concentrated solutions were diluted in PBS. The amount of exosome protein was measured using the Micro BCA Protein Assay Kit (CoWin Biotechnology).
Fluorescence Microscopy
A spinning disk confocal system (Revolution XD, Andor Technology, Northern Ireland) was built on the left port of a Ti-E inverted microscope (Nikon, Japan) with a PFS (perfect focus system) to keep focus plane and a motorized stage (Ludl Electronic Products) to do montage. The confocal images were collected by an electron-multiplying charge-coupled device (EMCCD) iXon DV885 (Andor) with 1004 × 1002 pixels. 405 nm, 491 nm, and 640 nm solid state lasers modulated by AOTF (Acousto-optic tunable filter) were used as the illumination sources for Hoechst 33342, FITC, and DiD, respectively. Fluorescence emission was collected by a 40 × oil-immersion objective (S Fluor, NA. = 1.30, Nikon), passed through EM 452/45, EM 520/15, or EM 685/40 emission filters (Semrock). All images were acquired and processed by iQ v. 2.0 software (Andor).
Uptake Inhibition Studies
To study the pathway of exosome uptake, cells were preincubated with some pharmacological/chemical inhibitors before exosome addition. 1–50 μm CPZ, 10–200 μm genistein, 10–100 μm EIPA, or 10–100 μm LY294002 was applied to pretreat cells at 37 °C for 30 min and were present throughout the experiments, respectively. 5–50 μm nystatin, 10 mm MβCD, or 0.1 mg/ml CtxB was pre-incubated with cells for 60 min and washed excessively prior to exosome addition. For a given experiment, the final concentration of dimethyl sulfoxide (DMSO) was not above 0.1%. As control, cells were cultured in the presence or absence of 0.1% DMSO. To study the influence of K+ depletion to exosome uptake, cells were incubated in a buffer containing 20 mm HEPES, 140 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, 5.5 mm d-glucose, and 0.5% BSA for 1 h at 37 °C. As control, cells were incubated in the buffer supplemented with 10 mm KCl. Calcein AM was applied to test cell viability after 4 h treatment of drugs.
40 μg/ml DiD-labeled exosomes were added to cells, and subsequently kept at 37 °C for 3 h. After washing with PBS, fixed by 2% paraformaldehyde and incubated with 5 μg/ml Hoechst 33342 for 20 min, cells were imaged by confocal microsopy. To quantify the cellular uptake of exosomes, the experiments were repeated three times, and for each individual experiment more than 800 cells were imaged at random locations. All settings of imaging and processing were kept constant, and the relative fluorescence intensities were calculated. The cell numbers were determined by counting the nuclei. The significance of the effects of various treatments versus control was evaluated by Student's t test (p < 0.05). To exclude the disturbance of DiD diffusion, the blocking degree of uptake using CFSE-labeled exosomes by CPZ was evaluated as control. Moreover, to determine whether PC12 cells can phagocytose exosomes, 107/ml latex beads with 40 μg/ml exosomes were incubated with cells at 37 °C for 3 h, and confocal imaging was performed.
RNA Knockdown, Loss-of-Function Mutation, and Rescue Design
Small interfering RNA (siRNA) duplexes against CHC and negative control (NC) were synthesized (GenePharma) according to Refs. 25, 26. The oligos were transfected using jetPRIME (Polyplus, France). Short hairpin RNA (shRNA) targeting sequences for μ2, CAV1, and DYN2 were GATCAAGCGCATGGCAGGCAT, CCAGAAGGGACACACAGTT, and GCTGGTGAAGATGGAGTTTGA separately. The NC sequence was UUCUCCGAACGUGUCACGUUA. All the shRNAs were synthesized and cloned into pLVX-shRNA2 vector (GFP-tagged, Clontech). The μ2 wild type (μ2-WT) and its loss-of-function mutation (μ2-D176A) sequences were obtained from the plasmid μ2-HA-WT (plasmid 32752, Addgene) and μ2-HA-D176A (plasmid 32754, Addgene) contributed by Professor Sorkin (27). The CAV1 wild type (CAV1-WT) and its loss-of-function mutation (CAV1-P132L) sequences were synthesized by NeuronBiotech. The rescue sequences for μ2 (μ2-res) and CAV1 (CAV1-res) with silent mutations resistant to RNA knockdown were customized based on their wild type version (NeuronBiotech). The above sequences were cloned into pLOV-CMV-mCherry vector. All the plasmids were transfected by Lipofectamine 2000 (Invitrogen). Knockdown and rescue efficiency was tested by PCR and Western blots 48 h after transfection. 48 h after transfection, cells were incubated with exosomes for 3 h, and the blocking degree of uptake was evaluated as described above.
Colocalization Studies
To colocalize exosomes with dextran, 40 μg/ml DiD-labeled exosomes with 3 mg/ml FITC-dextran were added to cells. Washing, fixing, nucleus staining, and imaging were carried out after 10 min, 30 min, or 1 h incubation at 37 °C. For colocalization of exosomes with CtxB, incubation of cells with FITC-CtxB (0.1 mg/ml) was performed for 1 h at 4 °C to allow the toxin to bind to lipid raft without substantial internalization. After excessive washing and adding exosomes, cells were shifted to the stagetop incubator on the microscope. Live-cell imaging was performed after 10 min, 30 min, and 1 h.
RT-PCR
Total RNA of cells was extracted by Trizol reagent (Invitrogen). Exosome samples were cracked by repeated freezing and thawing. The first strand synthesis of cDNA was performed using equal amounts of RNA samples, according to M-MLV reserve transcriptase instructions (Promega). β-Actin or U6 was employed as the housekeeping gene for mRNAs or miRNAs analysis. PCR reactions were performed using SYBR Premix ExTaq (TaKaRa, Dalian, China) in a total volume of 20 μl (1.5 μl cDNA samples). RT-PCR was carried out using the 7500 system (Application Biosystems). Relative expression was calculated by the comparative 2−ΔΔCt method. Each sample was assessed at least in triplicate.
Electron Microscopy
A droplet of diluted exosomes was applied onto a carbon-coated copper grid and incubated until dry. Specimens were observed with a JEM-2100 transmission electron microscope (JEOL).
RESULTS
Roles of Clathrin-mediated Endocytosis and Macropinocytosis in Exosome Uptake
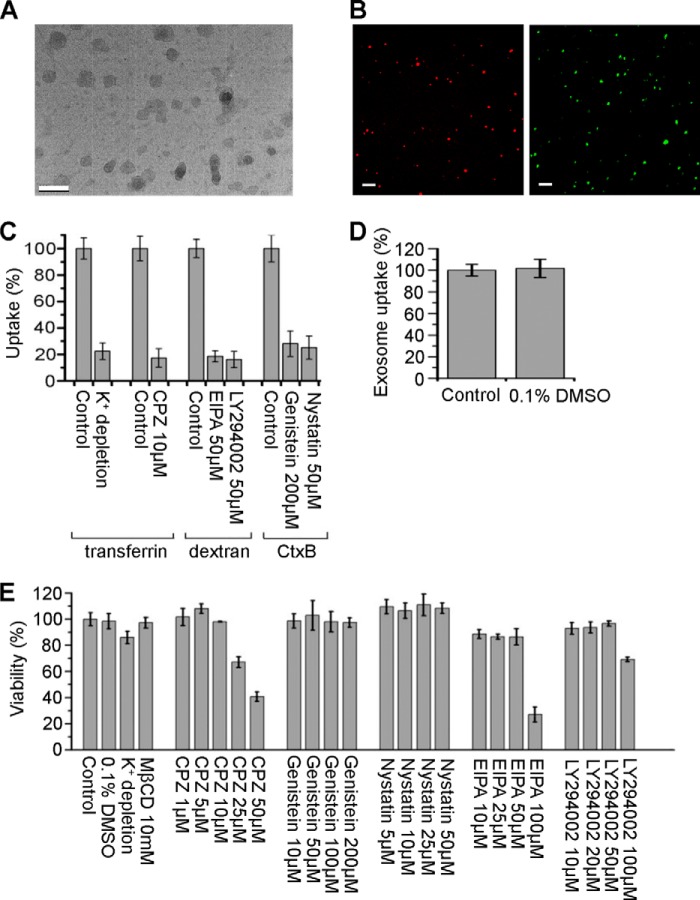
In this work, the culture medium of PC12 cells was sequentially centrifuged according to a generally accepted exosome isolation protocol. The resulting pellet was confirmed consisting of vesicles of 40–100 nm in size by electron microscopy (Fig. 1A). The isolated exosomes were labeled by lipophilic dye DiD or amino-reactive fluorophor CFSE, and detected by fluorescence microscopy (Fig. 1B). Before applying inhibiting treatments to study the uptake pathway, several control experiments were carried out. First, controls of endocytosis inhibition were performed to test the activity of the treatments (Fig. 1C). Second, the effect of solvent was excluded. Results demonstrate that DMSO in concentrations as low as 0.1% did not affect exosome uptake (Fig. 1D). Third, to optimize drug concentrations and to avoid disturbing cells, cell viability was tested under different treatments. The results are shown in Fig. 1E, and the treatments causing a decrease in cell viability of more than 15% were not used.
FIGURE 1.
Exosome images and control for pharmacological experiments. A, image of exosomes under electron microscope (scale bar, 200 nm). (B) DiD (red) or CFSE (green)-labeled exosomes detected by fluorescence microscopy (scale bars, 2 μm). C, PC12 cells were pretreated with various inhibitors or not (as control), and then incubated with FITC-transferrin, FITC-dextran, or FITC-CtxB. Uptake was quantified by determining the fluorescence intensity. D, PC12 cells were incubated with exosomes at 37°C for 3 h in the presence or absence (as control) of 0.1% DMSO. Exosome uptake was quantified by determining the fluorescence intensity. E, viability of PC12 cells under the presence or absence (as control) of various treatments at 37°C for 4 h determined by fluorescence intensity of calcein AM. The values are normalized to the control. Mean ± SD of three independent experiments is shown.
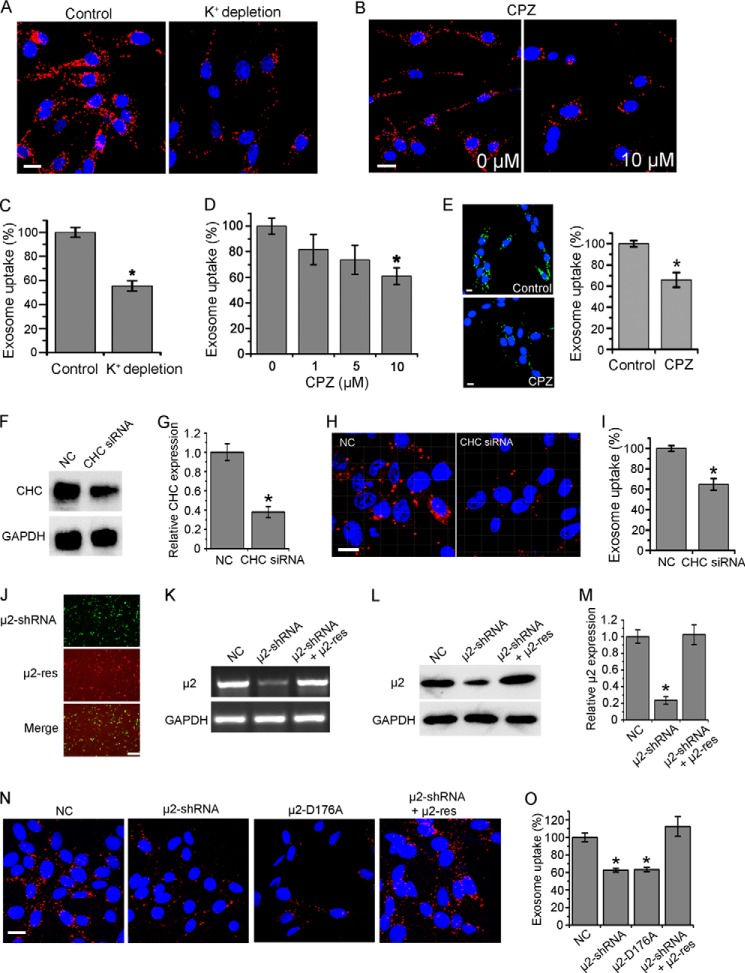
Clathrin-mediated endocytosis (also known as clathrin-dependent endocytosis) is inherently active in all mammalian cells (19). When some inhibiting treatments were applied, exosome uptake of several hours was quantified following previous reports. K+ depletion presents a useful procedure to block clathrin-coated pits (28). Fig. 2, A and C shows that treatment of K+ depletion buffer induced a partial inhibition of exosome uptake, with the percentage of internalized exosomes dropping to 55.5%. Besides, CPZ, a cationic amphipathic drug likely inducing a loss of clathrin, inhibits clathrin-mediated endocytosis (28). As shown in Fig. 2, B and D, CPZ caused a dose-dependent inhibition of exosome uptake with 41% block at 10 μm. To exclude the disturbance of DiD diffusion, CFSE-labeled exosomes were also used to evaluate the uptake inhibition under CPZ treatment. As shown in Fig. 2E, CFSE-labeled exosomes produced similar results as DiD-labeled exosomes. Furthermore, CHC, the basic subunit of clathrin, was knocked down by siRNA successfully (Fig. 2, F and G). Exosome uptake was blocked by about 35.3% in the CHC-negative cells (Fig. 2, H and I). In addition, μ2, the subunit of clathrin adaptor complex AP2, inhibited the assembly of clathrin-coated pits (Fig. 2, J–M). Transfection of μ2-shRNA or loss-of-function mutation μ2-D176A caused a 37.5% or 36.8% decrease of exosome uptake. To further validate these results, a rescue experiment was performed by co-transfection of μ2-shRNA and a shRNA-resistant silent mutation-containing variant of μ2. This shRNA inhibitory effect on exosome uptake could be rescued by μ2-res (Fig. 2, N and O). All the results above indicated that exosome uptake involves clathrin-mediated endocytosis.
FIGURE 2.
Role of clathrin-mediated endocytosis in exosome uptake. A, confocal images of PC12 cells incubated with DiD-labeled exosomes at 37 °C for 3 h in K+ depletion buffer with (as control) or without KCl. B, confocal images of PC12 cells pretreated with CPZ or not for 30 min, and then incubated with exosomes at 37 °C for 3 h. C and D, exosome uptake under various treatments and control was quantified by determining the fluorescence intensity. E, confocal images of PC12 cells pretreated with 10 μm CPZ or not for 30 min, and then incubated with CFSE-labeled exosomes at 37 °C for 3 h. Exosome uptake was quantified by determining the fluorescence intensity. All scale bars above, 15 μm. F, Western blots of CHC after 48 h of treatment of corresponding siRNA. G, CHC expression level was quantified by determining the gray value. H, confocal images of cells pretreated with siRNA against CHC or NC for 48 h, and then incubated with exosomes at 37 °C for 3 h. Scale bar, 20 μm. I, exosome uptake was quantified by determining the fluorescence intensity. J, fluorescence images of PC12 cells transfected with μ2-shRNA (GFP tagged), μ2-res (mCherry tagged), and merged together. Scale bar, 50 μm. PCR (K) and Western blot (L) results of μ2 48 h after transfection of NC, μ2-shRNA, or μ2-shRNA and μ2-res together. M, μ2 expression level was quantified by determining the gray value. N, confocal images of cells 48 h after transfection of NC, μ2-shRNA, μ2-D176A, or μ2-shRNA and μ2-res together, and then incubated with exosomes at 37 °C for 3 h. Scale bar, 20 μm. O, exosome uptake was quantified by determining the fluorescence intensity. In all the confocal images, red refers to DiD-labeled exosomes, and blue indicates nuclei. All the values are normalized to the control. For all the quantification plots, mean ± S.D. of three independent experiments is shown. Values significantly different (p < 0.05) from control are marked with asterisks.
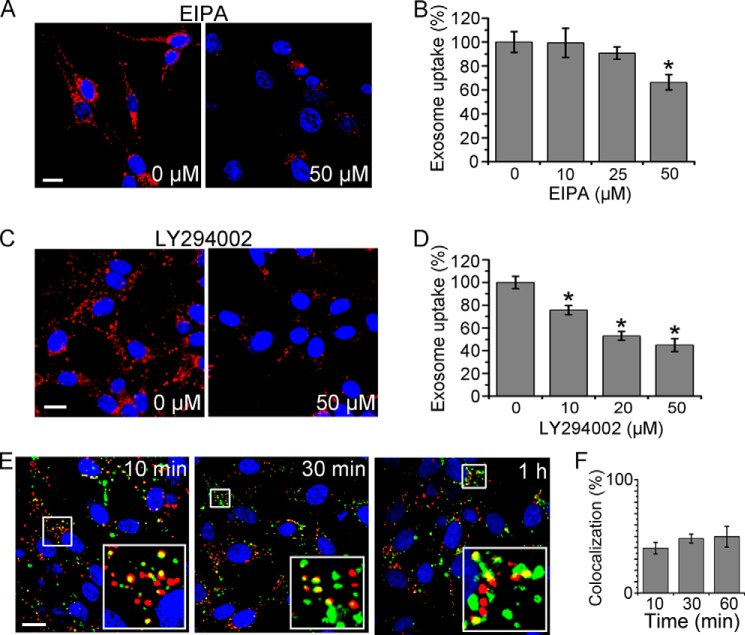
Another main endocytosis pathway, macropinocytosis, was considered. EIPA, a Na+-H+ exchanger inhibitor, and LY294002, a phosphoinositide 3-kinase (PI3K) inhibitor, were used to block macropinocytosis (28). The effects of both inhibitors on exosome uptake by PC12 cells were quantified by fluorescence microscopy, and the uptake efficiency was reduced remarkably and depended on the dose of EIPA and LY294002 (Fig. 3, A–D). Colocalization study of exosomes with dextran at the early time of cellular internalization was performed to provide further evidence. Approximately half of the colocalization emerged 10, 30, or 60 min after internalization initiation (Fig. 3, E and F). All results above indicate that macropinocytosis played a role in exosome uptake.
FIGURE 3.
Role of macropinocytosis in exosome uptake. A and C, confocal images of PC12 cells pretreated with various concentrations of EIPA (A) or LY294002 (C) for 30 min, and then incubated with exosomes at 37 °C for 3 h. B and D, exosome uptake under various treatments and control was quantified by determining the fluorescence intensity. The values are normalized to the control. E, confocal images of PC12 cells incubated with DiD-labeled exosomes (red) and FITC-dextran (green) for 10 min, 30 min, or 1 h. Yellow indicates the colocalization of exosomes and dextran. F, quantification of the colocalization of exosomes and dextran at various time points. In all the images, red refers to DiD-labeled exosomes, and blue indicates nuclei. For all the quantification plots, mean ± SD of three independent experiments is shown. Values significantly different (p < 0.05) from control are marked with asterisks. All the scale bars are 15 μm.
Roles of Caveolae-mediated Endocytosis and Phagocytosis in Exosome Uptake
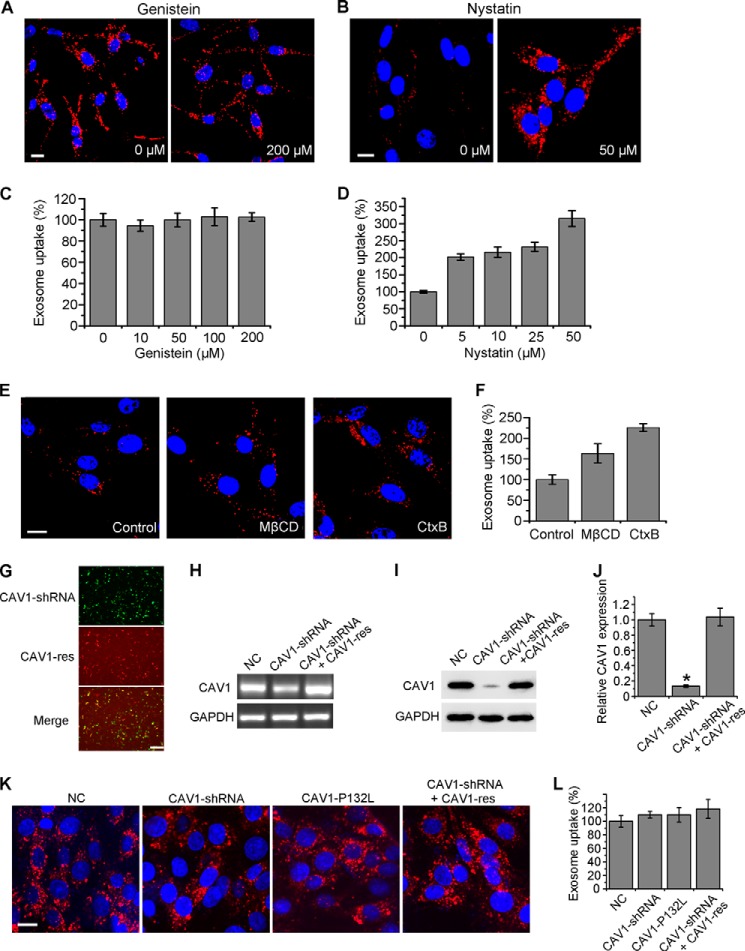
Besides clathrin-mediated endocytosis and macropinocytosis, caveolae-mediated endocytosis is one of the main routes for cellular internalization (19). Disrupting lipid rafts and inhibiting tyrosine protein kinase are two of the most widely used techniques for blocking this endocytic pathway (28). Exosome uptake was analyzed in the presence of known inhibitors, genistein and nystatin. For avoiding cholesterol sequestration and property changes on exosome membrane, nystatin was washed excessively before exosome addition. Fig. 4, A and B show fluorescence images after exosome uptake for 3 h by cells with or without drug treatments. Statistical results are shown in Fig. 4, C and D. The exosome internalization was generally not affected by genistein, even at a concentration as high as 200 μm. Surprisingly, lipid raft disruption by nystatin treatment enhanced exosome uptake. The exposure time for Fig. 4B was shortened to prevent overexposure. Similar results also emerged under lipid raft disruption by another cholesterol depletion-drug MβCD or lipid rafts blocking by CtxB (Fig. 4, E and F). It was interpreted that the plasma membrane became more fluid under these treatments leading to the enhancement of uptake through other pathways (29). Therefore, caveolae-mediated endocytosis cannot play roles in exosome uptake. Moreover, clathrin- and caveolae-independent endocytosis was also not involved in exosome uptake, because this class of pathways, including Arf6-dependent, flotillin-dependent, Cdc42-dependent, and RhoA-dependent endocytosis are all inhibited by genistein and cholesterol depletion (30).
FIGURE 4.
Role of caveolae-mediated endocytosis in exosome uptake. A and B, confocal images of PC12 cells pretreated with various concentrations of genistein for 30 min (A), or nystatin for 1 h and washing (B), and then incubated with exosomes at 37 °C for 3 h. The exposure time for B was shortened to prevent overexposure. C and D, exosome uptake under various treatments was quantified by determining the fluorescence intensity. The values are normalized to the control. E, cells were pretreated with 10 mm MβCD or 0.1 mg/mL CtxB for 1 h and washing, or without any pretreatment (as control), and then incubated with exosomes at 37 °C for 3 h. Confocal imaging was performed afterward. All the scale bars above, 15 μm. F, exosome uptake under various treatments and control was quantified by determining the fluorescence intensity. The values are normalized to the control. G, fluorescence images of PC12 cells transfected with CAV1-shRNA (GFP tagged), CAV1-res (mCherry tagged), and merged together. Scale bar, 50 μm. PCR (H) and Western blot (I) results of CAV1 48 h after transfection of NC, CAV1-shRNA or CAV1-shRNA and CAV1-res together. J, CAV1 expression level was quantified by determining the gray value. K, confocal images of cells 48 h after transfection of NC, CAV1-shRNA, CAV1-P132L, or CAV1-shRNA and CAV1-res together, and then incubated with exosomes at 37 °C for 3 h. Scale bar, 20 μm. L, exosome uptake was quantified by determining the fluorescence intensity. In all the confocal images, red refers to DiD-labeled exosomes, and blue indicates nuclei. For all the quantification plots, mean ± S.D. of three independent experiments is shown. Values significantly different (p < 0.05) from control are marked with asterisks.
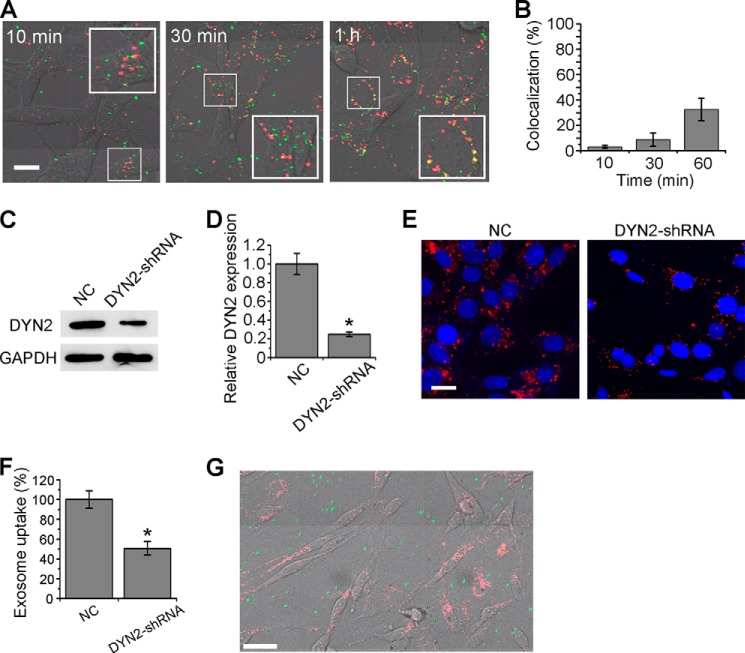
Next, CAV1, the key protein in caveolae-mediated endocytosis, was knocked down by CAV1-shRNA and rescued by co-transfection of CAV1-res (Fig. 4, G–J). The loss-of-function mutation CAV1-P132L was also expressed and tested. Exosome uptake was almost not affected in the CAV1-negative or CAV1-rescued cells (Fig. 4, K and L). Furthermore, the colocalization study of exosomes with CtxB at an early time of cellular internalization was performed to provide further evidence. As shown in Fig. 5, A and B, the colocalization percentages of exosomes and FITC-CtxB were very low after 10 min (3.0%) or 30 min (8.7%) uptake. And the percentage increased to 32.6% after 60 min of incubation, possibly due to the similar sorting target of CtxB and exosomes after cellular uptake. All the results above indicated that the internalization pathway of exosomes was quite different from caveolae-mediated endocytosis.
FIGURE 5.
Colocalization test with CtxB, role of DYN2 and phagocytosis in exosome uptake. A, merging of bright-field images and confocal images of PC12 cells incubated with exosomes and FITC-CtxB (green) for 10 min, 30 min, or 1 h. Yellow indicates the colocalization of exosomes and CtxB (scale bars, 15 μm). B, quantification of the colocalization of exosomes and CtxB at various time points. C, Western blot results of DYN2 48 h after transfection of NC or DYN2-shRNA. D, DYN2 expression level was quantified by determining the gray value. E, confocal images of cells transfected with NC or DYN2-shRNA for 48 h, and then incubated with exosomes at 37 °C for 3 h. Scale bar, 20 μm. F, exosome uptake was quantified by determining the fluorescence intensity. G, merging of bright-field images and confocal images of PC12 cells incubated with exosomes and FITC-labeled latex beads (green) at 37 °C for 3 h. Scale bar, 50 μm. In all the images, red refers to DiD-labeled exosomes, and blue indicates nuclei. For all the quantification plots, mean ± S.D. of three independent experiments is shown.
DYN2, another key protein necessary for both clathrin- and caveolin-mediated endocytosis, but not involved in macropinocytosis, was also tested. Applying DYN2-shRNA, the uptake efficiency was reduced to 50.7% (Fig. 5, C–F), indicating that dynamin-dependent endocytosis was partially involved. In general, phagocytosis is performed by specialized professional cells such as macrophages, neutrophils, or monocytes (19). Experiments were still carried out to exclude phagocytosis during exosome uptake by PC12 cells. Latex beads, a well-known marker of phagocytosis, were added to culture medium, allowing cellular uptake (28). Shown as Fig. 5G, no bead was observed to be internalized into cells at more than 3 h. This result indicated that PC12 cells cannot phagocytose efficiently.
Delivery of miR-21 into BMSCs through PC12 Cell-derived Exosomes
To study the entry of exosomes to normal cells, BMSCs were selected as the recipients of exosomes derived from PC12 cells. After 3 h of incubation with BMSCs, exosomes were detected in the recipient cells by fluorescence microscopy, showing that PC12 cell-derived exosomes enter into BMSCs (Fig. 6A). Our previous work showed that miR-21 has a relatively high copy number in PC12 cells detected by high-throughput sequencing (data not shown). miR-21 is a unique miRNA that is overexpressed in most tumor types (31). Thus, we selected miR-21 to study miRNA transfer through PC12 cell-derived exosomes. Using RT-PCR, it was confirmed that miR-21 was as abundantly expressed in exosomes as it was in cytoplasm; greater than miR-22, miR-26a, miR-214, miR-221, and miR-222. The expression level of miR-21 in BMSCs increased dramatically as early as 3 h after incubation with exosomes (Fig. 6B). Unlike tumor cells, the endogenous miR-21 level in BMSCs was quite low, leading to a rapid, more than 4-fold enhancement of miR-21 level after exosome uptake for 3–12 h. The level recovered at 24 h was possibly caused by miR-21 degradation. Shown in Fig. 6C, the down-regulation of pre-miR-21 levels occurred in BMSCs after a 6–24 h incubation with exosomes. To demonstrate that the increased miR-21 was from exosomes but not from activated endogenous miR-21 during exosome incubation, cardiomyoblast H9C2 cell-derived exosomes were selected as negative controls to be incubated with BMSCs (because of lower miR-21 enclosed). After 3–24 h of incubation, the miR-21 level in BMSCs did not change significantly (Fig. 6D). Together, the miR-21 level in BMSCs was elevated by PC12 cell-derived exosomes, while the pre-miR-21 level was down-regulated. These results showed that PC12 cell-derived exosomes delivered miR-21 into other cell types and suppressed the expression of endogenous pre-miR-21.
FIGURE 6.
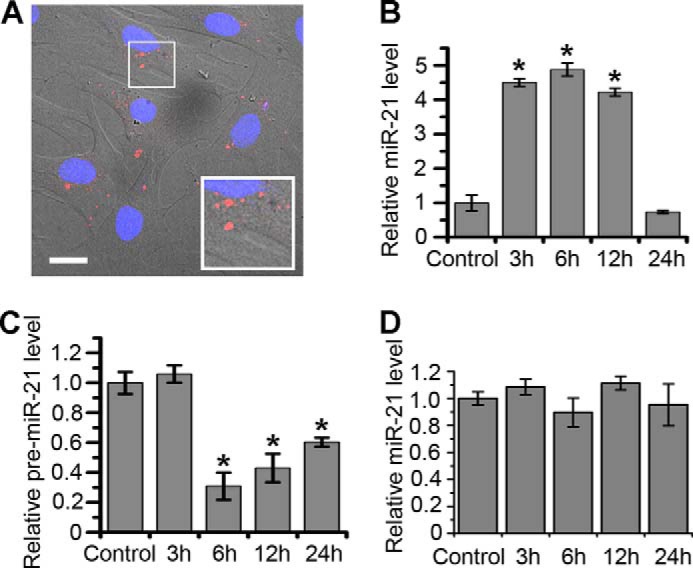
Delivery of miR-21 into BMSCs through exosomes. A, merging of bright-field images and confocal images of BMSCs incubated with DiD-labeled exosomes for 3 h. Red refers to exosomes, and blue indicates nuclei (scale bar, 20 μm). B and C, miR-21 and pre-miR-21 levels in BMSCs incubated with PC12 cell-derived exosomes for 0 (as control), 3, 6, 12, and 24 h according to RT-PCR results. D, miR-21 levels in BMSCs incubated with H9C2 cell-derived exosomes for 0 (as control), 3, 6, 12, and 24 h according to RT-PCR results. For all the quantification plots, values are normalized to the control, and mean ± S.D. of three independent experiments is shown. Values significantly different (p < 0.05) from control are marked with asterisks.
Target Genes of miR-21 in BMSCs Regulated by PC12 Cell-derived Exosomes
miR-21 is one of the most important oncomiRs. To determine biological functions of miR-21 delivered from PC12 cells to BMSCs through exosomes, several target genes were analyzed. TGFβRII, a receptor on BMSCs that respond to an important regulator TGFβ and target gene of miR-21, was selected to be analyzed. Interestingly, the expression level of TGFβRII mRNA decreased substantially after 12 h and 24 h of exosome treatment (Fig. 7A). TGFβRII levels also decreased after exosome treatment, which was revealed by Western blots. At the same time, the antisense oligonucleotides of miR-21 (anti-miR-21) resisted the exosome effect to reduce TGFβRII expression (Fig. 7, B and C). Next, TPM1 as a tumor suppressor and miR-21 target was analyzed. After treatment with exosomes for 24 h, the expression level of TPM1 in BMSCs was reduced significantly. Such effect was resisted by anti-miR-21 (Fig. 7, D and E). Furthermore, PTEN, another important tumor suppressor and gene target of miR-21 was tested. However, the expression level of PTEN mRNA in BMSCs did not change after treatment with exosomes for 24 h (Fig. 7F). The suppression efficiencies for target genes of miR-21 may be cell type specific. These results indicated that PC12 cell-derived exosomes could decrease TGFβRII and TPM1 in BMSCs through miR-21. The above results show an example of information transfer from tumor cells to normal cells.
FIGURE 7.
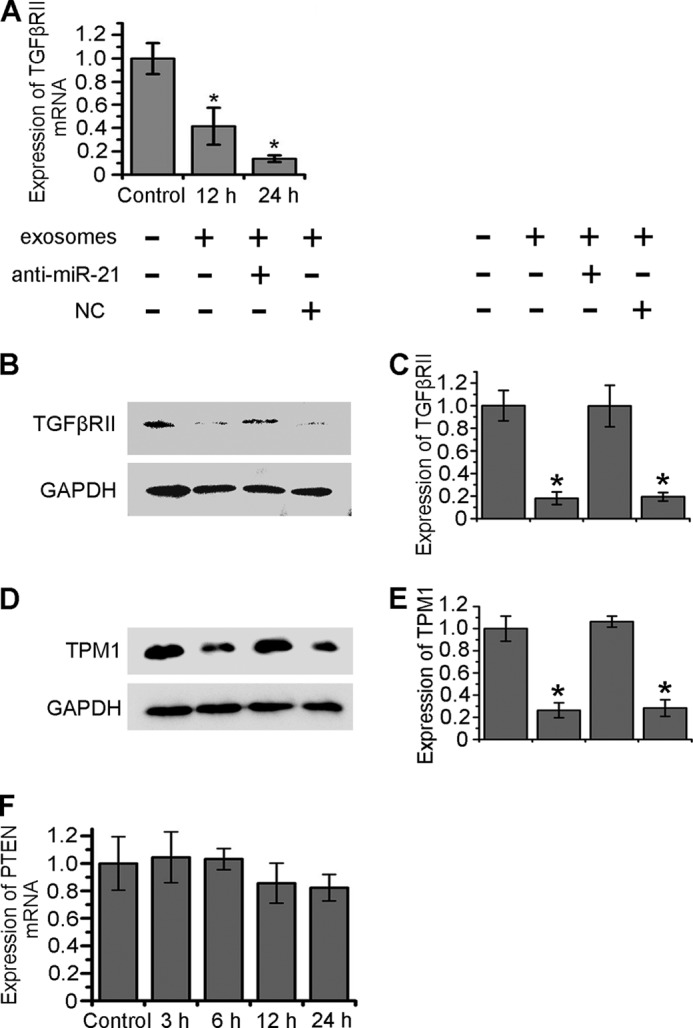
Reduction of TGFβRII and TPM1 expression level in BMSCs through exosomes. A, TGFβRII mRNA levels in BMSCs incubated with exosomes for 0 (as control), 3, 6, 12, and 24 h according to RT-PCR results. B and D, 24 h after transfection of NC or anti-miR-21, Western Blot was performed to analyze the expression of TGFβRII and TPM1 in BMSCs incubated with PC12 cell-derived exosomes at 37 °C for 24 h. C and E, TGFβRII and TPM1 expression levels were quantified by determining the gray value. F, PTEN mRNA levels in BMSCs incubated with exosomes for 0 (as control), 12, and 24 h according to RT-PCR results. For all the quantification plots, values are normalized to the control, and mean ± S.D. of three independent experiments is shown. Values significantly different (p < 0.05) from control are marked with asterisks.
DISCUSSION
The interaction between tumor exosomes and cells plays important roles in tumor progression and immunity (10). Our previous works have shown that PC12 cells internalized exosomes through endocytosis (17, 18). However, the endocytic pathway of exosomes was still unclear. Recently, phosphatidylserine on the exosome surface is reported to activate macropinocytosis in microglia (21). Svensson et al. found that glioblastoma cell-derived exosomes trigger lipid raft-mediated endocytosis through the activation of ERK1/2 signaling pathways that include an important role of heat shock protein 27 (HSP27) (22). Heparan sulfate proteoglycans (HSPGs) were reported to function as receptors of glioblastoma cell-derived exosomes (32). Hence, it is hypothesized that the uptake pathway of exosomes are dependent on the activation of the receptors on the recipient cells by exosomes. In this work, based on experimental data using selective inhibitors, molecular tools, and special endocytosis markers, it was concluded here that clathrin-mediated endocytosis and macropinocytosis were involved in the uptake of PC12 cell-derived exosomes. Both pathways can be mediated by receptor activation (33, 34). Clathrin-mediated endocytosis involves engulfment of receptors associated with their ligands, while macropinocytosis occurs due to membrane ruffles triggered by receptor tyrosine kinases. Specific to the parental cell, many kinds of proteins and lipids on the exosome surface can attach to different receptors, inducing complex signaling. The uptake pathway dependent on the signaling is possibly cell type specific. At least two kinds of signaling activation may be involved in the uptake of PC12 cell-derived exosomes. That was the reason that exosome uptake was at the same time dependent on two different pathways. The signaling pathway within it will be revealed in future studies. Furthermore, the intracellular trafficking occurring after the two endocytic pathways tends to sort toward lysosomes. Exosome proteins may be stored and utilized there. Whether exosome RNAs can escape from degradation and take effect in cells is an interesting question.
Tumor cells secrete exosome-packaging miRNAs, circulating in body fluids (serum, plasma, saliva, urine, and milk), and enter into neighboring and distant cells (15, 35). miRNAs, protected in exosomes against degradation, is an important regulator of gene expression at the post-transcriptional level (36, 37). In this work, exosomes acted as a natural liposome transferring miRNAs from tumor cells to normal cells. First, we confirmed that PC12 cell-derived exosomes enclosed abundant miR-21, an oncomiR overexpressed in most tumor types and associated with breast cancer, prostate cancer, and many other cancers (38, 39). Second, it was found that exosomes entered and transferred miR-21 into BMSCs. Third, TGFβRII and TPM1 expression were down-regulated by exosomes and through miR-21. TGFβRII is often mutated in cancers, and the TGFβ signaling pathway plays complex roles during cancer progression (40, 41). TPM1, which belongs to the class II tumor suppressor genes, modulates cell transformation and tumor cell growth. So the tumor exosomes may regulate cancer progression by reducing TGFβRII and TPM1 in normal cells. Here, our data presents an example of information transfer from tumor cells to normal cells.
In summary, the endocytosis pathway involved in PC12 cell-derived exosome uptake was shown to be clathrin-mediated endocytosis and macropinocytosis. It was demonstrated that exosomes entered and transferred miR-21 into BMSCs, and decreased their TGFβRII and TPM1 expression. These results deepen the understanding of the internalization pathway of tumor exosomes, and verify an effect of tumor exosomes to normal cells.
This work was supported by the National Basic Research Program of China (973 Program: 2013CB932902), NSFC (No. 61071047, 31371003), the Natural Science Foundation of Jiangsu Province (No. BK2012122, BK20130886), and the Specialized Research Fund for the Doctoral Program of Higher Education of China (No. 20130092110030).
- miRNA
- microRNA
- CAV
- caveolin
- BMSC
- bone marrow-derived mesenchymal stromal cell
- TPM
- tropomyosin
- TGF
- transforming growth factor
- DiD
- 3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt
- CFSE
- carboxyfluorescein diacetatesuccinimidyl ester
- CPZ
- chlorpromazine
- CHC
- clathrin heavy chain
- EIPA
- 5-(n-ethyl-n-isopropyl)-amiloride.
REFERENCES
- 1. Stoorvogel W., Kleijmeer M. J., Geuze H. J., Raposo G. (2002) The biogenesis and functions of exosomes. Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 2. Mathivanan S., Ji H., Simpson R. J. (2010) Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 [DOI] [PubMed] [Google Scholar]
- 3. de Gassart A., Géminard C., Hoekstra D., Vidal M. (2004) Exosome secretion: the art of reutilizing nonrecycled proteins? Traffic 5, 896–903 [DOI] [PubMed] [Google Scholar]
- 4. Zhang F., Sun S., Feng D., Zhao W. L., Sui S. F. (2009) A novel strategy for the invasive toxin: hijacking exosome-mediated intercellular trafficking. Traffic 10, 411–424 [DOI] [PubMed] [Google Scholar]
- 5. Chen X., Gao C., Li H., Huang L., Sun Q., Dong Y., Tian C., Gao S., Dong H., Guan D., Hu X., Zhao S., Li L., Zhu L., Yan Q., Zhang J., Zen K., Zhang C. Y. (2010) Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 20, 1128–1137 [DOI] [PubMed] [Google Scholar]
- 6. Miyanishi M., Tada K., Koike M., Uchiyama Y., Kitamura T., Nagata S. (2007) Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439 [DOI] [PubMed] [Google Scholar]
- 7. György B., Szabó T. G., Pásztói M., Pál Z., Misják P., Aradi B., László V., Pállinger E., Pap E., Kittel A., Nagy G., Falus A., Buzás E. I. (2011) Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 68, 2667–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai Z., Zhang W., Yang F., Yu L., Yu Z., Pan J., Wang L., Cao X., Wang J. (2012) Immunosuppressive exosomes from TGF-β1 gene-modified dendritic cells attenuate Th17-mediated inflammatory autoimmune disease by inducing regulatory T cells. Cell Res 22, 607–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iero M., Valenti R., Huber V., Filipazzi P., Parmiani G., Fais S., Rivoltini L. (2008) Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 15, 80–88 [DOI] [PubMed] [Google Scholar]
- 10. Schorey J. S., Bhatnagar S. (2008) Exosome function: from tumor immunology to pathogen biology. Traffic 9, 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shabo I., Svanvik J. (2011) Expression of macrophage antigens by tumor cells. Adv Exp. Med. Biol. 714, 141–150 [DOI] [PubMed] [Google Scholar]
- 12. Vlassov A. V., Magdaleno S., Setterquist R., Conrad R. (2012) Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 1820, 940–948 [DOI] [PubMed] [Google Scholar]
- 13. Staals R. H., Pruijn G. J. (2011) The human exosome and disease. Adv Exp Med Biol 702, 132–142 [DOI] [PubMed] [Google Scholar]
- 14. Karagiannis G. S., Pavlou M. P., Diamandis E. P. (2010) Cancer secretomics reveal pathophysiological pathways in cancer molecular oncology. Mol. Oncol. 4, 496–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen X., Liang H., Zhang J., Zen K., Zhang C. Y. (2012) Horizontal transfer of microRNAs: molecular mechanisms and clinical applications. Protein Cell 3, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X., Liang H., Zhang J., Zen K., Zhang C. Y. (2012) Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 22, 125–132 [DOI] [PubMed] [Google Scholar]
- 17. Tian T., Wang Y., Wang H., Zhu Z., Xiao Z. (2010) Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J. Cell. Biochem. 111, 488–496 [DOI] [PubMed] [Google Scholar]
- 18. Tian T., Zhu Y. L., Hu F. H., Wang Y. Y., Huang N. P., Xiao Z. D. (2013) Dynamics of exosome internalization and trafficking. J. Cell. Physiol. 228, 1487–1495 [DOI] [PubMed] [Google Scholar]
- 19. Conner S. D., Schmid S. L. (2003) Regulated portals of entry into the cell. Nature 422, 37–44 [DOI] [PubMed] [Google Scholar]
- 20. Feng D., Zhao W. L., Ye Y. Y., Bai X. C., Liu R. Q., Chang L. F., Zhou Q., Sui S. F. (2010) Cellular internalization of exosomes occurs through phagocytosis. Traffic 11, 675–687 [DOI] [PubMed] [Google Scholar]
- 21. Fitzner D., Schnaars M., van Rossum D., Krishnamoorthy G., Dibaj P., Bakhti M., Regen T., Hanisch U. K., Simons M. (2011) Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci. 124, 447–458 [DOI] [PubMed] [Google Scholar]
- 22. Svensson K. J., Christianson H. C., Wittrup A., Bourseau-Guilmain E., Lindqvist E., Svensson L. M., Mörgelin M., Belting M. (2013) Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 288, 17713–17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lü L. X., Zhang X. F., Wang Y. Y., Ortiz L., Mao X., Jiang Z. L., Xiao Z. D., Huang N. P. (2013) Effects of Hydroxyapatite-Containing Composite Nanofibers on Osteogenesis of Mesenchymal Stem Cells In vitro and Bone Regeneration In vivo. ACS Appl Mater Interfaces 5, 319–330 [DOI] [PubMed] [Google Scholar]
- 24. Thery C., Amigorena S., Raposo G., Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 25. Yuyama K., Yanagisawa K. (2009) Late endocytic dysfunction as a putative cause of amyloid fibril formation in Alzheimer's disease. J Neurochem 109, 1250–1260 [DOI] [PubMed] [Google Scholar]
- 26. Zhang W., Smith A., Liu J. P., Cheung N. S., Zhou S., Liu K., Li Q. T., Duan W. (2009) GSK3β modulates PACAP-induced neuritogenesis in PC12 cells by acting downstream of Rap1 in a caveolae-dependent manner. Cell Signal. 21, 237–245 [DOI] [PubMed] [Google Scholar]
- 27. Nesterov A., Carter R. E., Sorkina T., Gill G. N., Sorkin A. (1999) Inhibition of the receptor-binding function of clathrin adaptor protein AP-2 by dominant-negative mutant mu2 subunit and its effects on endocytosis. EMBO J. 18, 2489–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khalil I. A., Kogure K., Akita H., Harashima H. (2006) Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 58, 32–45 [DOI] [PubMed] [Google Scholar]
- 29. Gaus K., Le Lay S., Balasubramanian N., Schwartz M. A. (2006) Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 174, 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayor S., Pagano R. E. (2007) Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 8, 603–612 [DOI] [PubMed] [Google Scholar]
- 31. Medina P. P., Nolde M., Slack F. J. (2010) OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467, 86–90 [DOI] [PubMed] [Google Scholar]
- 32. Christianson H. C., Svensson K. J., van Kuppevelt T. H., Li J. P., Belting M. (2013) Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. U.S.A. 110, 17380–17385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rappoport J. Z., Simon S. M., Benmerah A. (2004) Understanding living clathrin-coated pits. Traffic 5, 327–337 [DOI] [PubMed] [Google Scholar]
- 34. Kerr M. C., Teasdale R. D. (2009) Defining macropinocytosis. Traffic 10, 364–371 [DOI] [PubMed] [Google Scholar]
- 35. Chiba M., Kimura M., Asari S. (2012) Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep 28, 1551–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L., Zhu D., Huang L., Zhang J., Bian Z., Chen X., Liu Y., Zhang C. Y., Zen K. (2012) Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One 7, e46957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X., Sun Q., Wang K., Ba Y., Wang Q., Wang D., Yang J., Liu P., Xu T., Yan Q., Zhang J., Zen K., Zhang C. Y. (2010) Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 39, 133–144 [DOI] [PubMed] [Google Scholar]
- 38. O'Day E., Lal A. (2010) MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 12, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ribas J., Lupold S. E. (2010) The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle 9, 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pardali K., Moustakas A. (2007) Actions of TGF-β as tumor suppressor and pro-metastatic factor in human cancer. Biochim. Biophys. Acta 1775, 21–62 [DOI] [PubMed] [Google Scholar]
- 41. Bierie B., Moses H. L. (2006) TGF-beta and cancer. Cytokine Growth Factor Rev. 17, 29–40 [DOI] [PubMed] [Google Scholar]